The Basics and the Advancements in Diagnosis of Bacterial Lower Respiratory Tract Infections
Abstract
:1. Who is at Risk of Lower Respiratory Tract Infections (LRTIs) and Their Outcomes?
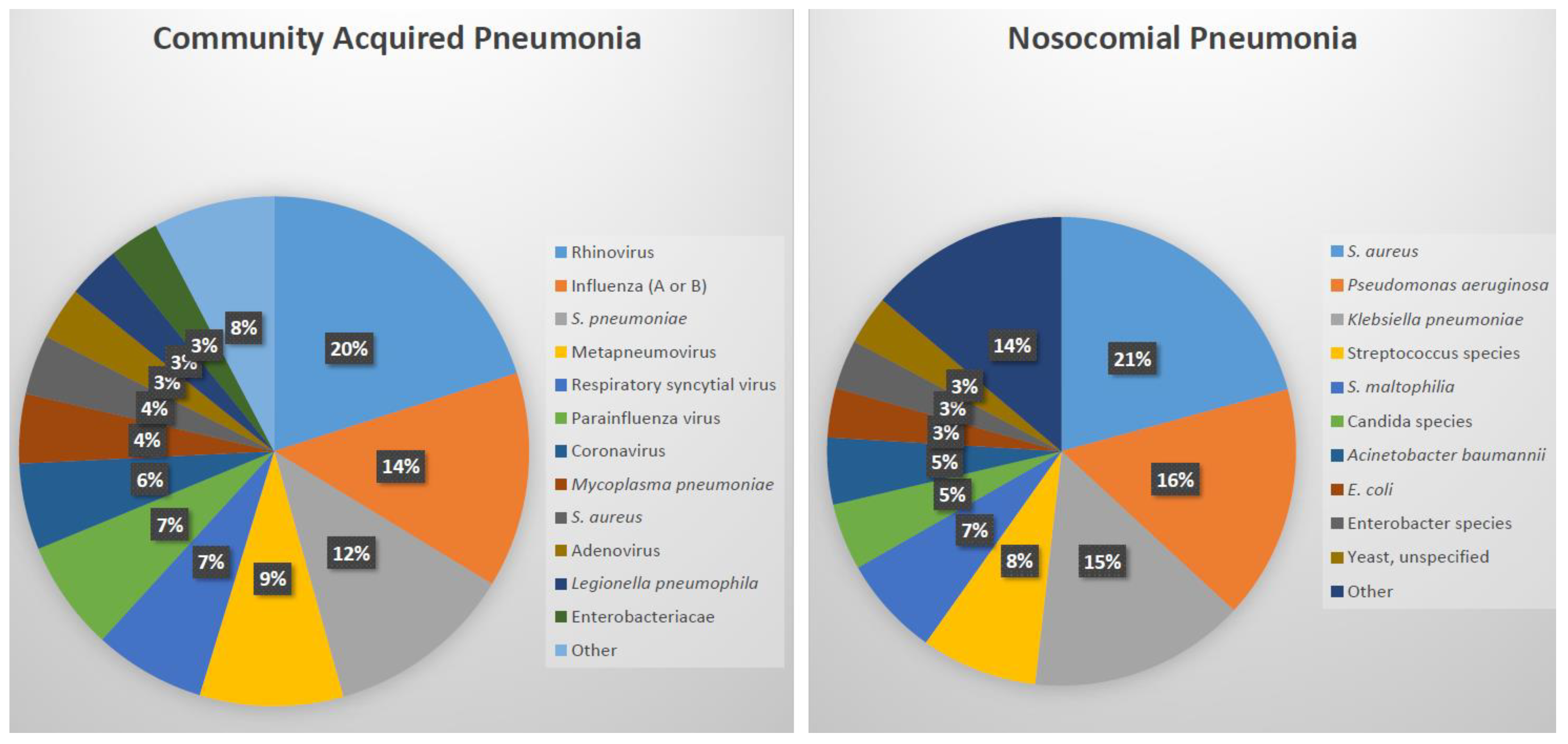
2. What Causes Bacterial Lower Respiratory Tract Infections?
3. How to Differentiate Bacterial from Viral LRTI at the Point of Care?
4. How to Determine the Bacterial Pathogen of LRTIs?
5. Why are Diagnostics Important for LRTIs? (Antibiotic Stewardship)
Funding
Conflicts of Interest
References
- GBD 2016 Lower Respiratory Infections Collaborators. Estimates of the global, regional, and national morbidity, mortality, and aetiologies of lower respiratory infections in 195 countries, 1990–2016: A systematic analysis for the Global Burden of Disease Study 2016. Lancet Inf. Dis. 2018, 18, 1191–1210. [Google Scholar]
- Liapikou, A.; Torres, A. Emerging drugs for nosocomial pneumonia. Expert Opin. Emerg. Drugs 2016, 21, 331–341. [Google Scholar] [CrossRef]
- Musher, D.M.; Abers, M.S.; Bartlett, J.G. Evolving understanding of the causes of pneumonia in adults, with special attention to the role of pneumococcus. Clin. Infect. Dis. 2017, 65, 1736–1744. [Google Scholar] [CrossRef] [PubMed]
- Hendaus, M.A.; Jomha, F.A.; Alhammadi, A.H. Virus-induced secondary bacterial infection: A concise review. Ther. Clin. Risk Manag. 2015, 11, 1265–1271. [Google Scholar] [CrossRef]
- Self, W.H.; Williams, D.J.; Zhu, Y.; Ampofo, K.; Pavia, A.T.; Chappell, J.D.; Hymas, W.C.; Stockmann, C.; Bramley, A.M.; Schneider, E.; et al. Respiratory viral detection in children and adults comparing asymptomatic controls and patients with community-acquired pneumonia. J. Infect. Dis. 2016, 213, 584–591. [Google Scholar] [CrossRef]
- Jain, S.; Self, W.H.; Wunderink, R.G.; Fakhran, S.; Balk, R.; Bramley, A.M.; Reed, C.; Grijalva, C.G.; Anderson, E.J.; Courtney, D.M.; et al. Community-acquired pneumonia requiring hospitalization among U.S. adults. N. Engl. J. Med. 2015, 373, 415–427. [Google Scholar] [CrossRef] [PubMed]
- Habib, M.; Porter, B.D.; Satzke, C. Capsular serotyping of Streptococcus pneumoniae using the Quellung reaction. J. Vis. Exp. 2014, 84, 51208. [Google Scholar] [CrossRef]
- Available online: https://www.europeanlung.org/en/lung-disease-and-information/lung-diseases/acute-lower-respiratory-infections (accessed on 28 February 2019).
- Juvén, T.; Mertsola, J.; Waris, M.; Leinonen, M.; Meurman, O.; Roivainen, M.; Eskola, J.; Saikku, P.; Ruuskanen, O. Etiology of community-acquired pneumonia in 254 hospitalized children. Pediatr. Infect. Dis. J. 2000, 19, 293–298. [Google Scholar] [CrossRef]
- Magill, S.S.; Edwards, J.R.; Bamberg, W.; Beldvas, Z.G.; Dumyati, G.; Kainer, M.A.; Lynfield, R.; Maloney, M.; McAllister-Hollod, L.; Nadle, J.; et al. Emerging Infections Program Healthcare-Associated Infections and Antimicrobial Use Prevalence Survey Team. Multistate point-prevalence survey of health care-associated infections. N. Engl. J. Med. 2014, 370, 1198–1208. [Google Scholar] [CrossRef]
- Sopena, N.; Sabria, M.; Neunos Study Group. Multicenter study of hospital-acquired pneumonia in non-ICU patients. Chest 2005, 127, 213–219. [Google Scholar] [CrossRef] [PubMed]
- Weiner, L.M.; Webb, A.K.; Limbago, B.; Dudeck, M.A.; Patel, J.; Kallen, A.J.; Edwards, J.R.; Sievert, D.M. Antimicrobial-resistant pathogens associated with healthcare-associated infections: Summary of data reported to the National Healthcare Safety Network at the Centers for Disease Control and Prevention, 2011–2014. Infect. Control Hosp. Epidemiol. 2016, 37, 1288–1301. [Google Scholar] [CrossRef] [PubMed]
- Kalil, A.C.; Metersky, M.L.; Klompas, M.; Muscedere, M.; Sweeney, D.A.; Palmer, L.B.; Napolitano, L.M.; O’Grady, N.P.; Bartlett, J.B.; Carratalà, J.; et al. Management of adults with hospital-acquired and ventilator-associated pneumonia: 2016 clinical practice guidelines by the Infectious Diseases Society of America and the American Thoracic Society. Clin. Infect. Dis. 2016, 63, e61–e111. [Google Scholar] [CrossRef]
- Albert, R.H. Diagnosis and treatment of acute bronchitis. Am. Fam. Physician 2010, 82, 1345–1350. [Google Scholar]
- Kinkade, S.; Long, N.A. Acute bronchitis. Am. Fam. Physician 2016, 94, 560–565. [Google Scholar]
- Available online: https://www.cdc.gov/rsv/high-risk/infants-young-children.html (accessed on 25 March 2019).
- Teepe, J.; Broekhuizen, B.D.L.; Loens, K.; Lammens, C.; Ieven, M.; Goossens, H.; Little, P.; Butler, C.C.; Coenen, S.; Godycki-Cwirko, M.; et al. Disease course of lower respiratory tract infection with a bacterial cause. Ann. Fam. Med. 2016, 14, 534–539. [Google Scholar] [CrossRef]
- Cals, J.W.; Hopstaken, R.M.; Butler, C.C.; Hood, K.; Severens, J.L.; Dinant, G.-J. Improving management of patients with acute cough by C-reactive protein point of care testing and communication training (IMPAC3T): Study protocol of a cluster randomised controlled trial. BMC Fam. Pract. 2007, 8, 15. [Google Scholar] [CrossRef]
- Huang, D.T.; Yealy, D.M.; Filbin, M.R.; Brown, A.M.; Chang, C.-C.H.; Doi, Y.; Donnino, M.W.; Fine, J.; Fine, M.J.; Fischer, M.A.; et al. Procalcitonin-guided use of antibiotics for lower respiratory tract infection. N. Engl. J. Med. 2018, 379, 236–249. [Google Scholar] [CrossRef] [PubMed]
- Aabenhus, R.; Jensen, J.U.; Jørgensen, K.J.; Hróbjartsson, A.; Bjerrum, L. Biomarkers as point-of-care tests to guide prescription of antibiotics in patiens with acute respiratory infections in primary care. Cochrane Database Syst. Rev. 2014, 11, CD010130. [Google Scholar]
- Andreeva, E.; Melbye, H. Usefulness of C-reactive protein testing in acute cough/respiratory tract infection: An open cluster-randomized clinical trial with C-reactive protein testing in the intervention group. BMC Fam. Pract. 2014, 15, 80. [Google Scholar] [CrossRef]
- Meisner, M. Update on procalcitonin measurements. Ann. Lab. Med. 2014, 34, 263–273. [Google Scholar] [CrossRef]
- Schuetz, P.; Wirz, Y.; Sager, R.; Christ-Crain, M.; Stolz, D.; Tamm, M.; Bouadma, L.; E Luyt, C.; Wolff, M.; Chastre, J.; et al. Procalcitonin to initiate or discontinue antibiotics in acute respiratory tract infections. Cochrane Database Syst. Rev. 2017, 10, CD007498. [Google Scholar] [CrossRef]
- Briel, M.; Schuetz, P.; Mueller, B.; Young, J.; Schild, U.; Nusbaumer, C.; Périat, P.; Bucher, H.C.; Christ-Crain, M. Procalcitonin-guided antibiotic use vs. a standard approach for acute respiratory tract infections in primary care. Arch. Intern. Med. 2008, 168, 2000–2007. [Google Scholar] [CrossRef] [PubMed]
- Available online: https://www.biomerieux-diagnostics.com/vidasr-brahms-pct (accessed on 15 February 2019).
- Available online: https://clinicaltrials.gov/ct2/show/NCT03341273 (accessed on 15 February 2019).
- Cooke, J.; Butler, C.; Hopstaken, R.; Dryden, M.S.; McNulty, C.; Hurding, S.; Moore, M.; Livermore, D.M. Narrative review of primary care point-of-care testing (POCT) and antibacterial use in respiratory tract infection (RTI). BMJ Open Respir. Res. 2015, 2, e000086. [Google Scholar] [CrossRef]
- Stein, M.; Lipman-Arens, S.; Oved, K.; Cohen, A.; Bamberger, E.; Navon, R.; Boico, O.; Friedman, T.; Etshtein, L.; Paz, M.; et al. A novel host-protein assay outperforms routine parameters for distinguishing between bacterial and viral lower respiratory tract infections. Diagn. Microbiol. Infect. Dis. 2018, 90, 206–213. [Google Scholar] [CrossRef] [PubMed]
- Sambursky, R.; Shapiro, N. Evaluation of a combined MxA and CRP point-of-care immunoassay to identify viral and/or bacterial immune response in patients with acute febrile respiratory infection. Eur. Clin. Respir. J. 2015, 2, 28245. [Google Scholar] [CrossRef] [PubMed]
- Ashkenazi-Hoffnung, L.; Oved, K.; Navon, R.; Friedman, T.; Boico, O.; Paz, M.; Kronenfeld, G.; Etshtein, L.; Cohen, A.; Gottlieb, T.M.; et al. A host-protein signature is superior to other biomarkers for differentiating between bacterial and viral disease in patients with respiratory infection and fever without source: A prospective observational study. Eur. J. Clin. Microbiol. Infect. Dis. 2018, 37, 1361–1371. [Google Scholar] [CrossRef] [PubMed]
- Dasaraju, P.V.; Liu, C. Infections of the Respiratory System. In Medical Microbiology, 4th ed.; Baron, S., Ed.; University of Texas Medical Branch at Galveston: Galveston, TX, USA, 1996; Chapter 93. [Google Scholar]
- Loens, K.; Van Heirstraeten, L.; Malhotra-Kumar, S.; Goossens, H.; Ieven, M. Optimal sampling sites and methods for detection of pathogens possibly causing community-acquired lower respiratory tract infections. J. Clin. Microbiol. 2009, 47, 21–31. [Google Scholar] [CrossRef]
- Cvitkovic Spik, V.; Beovic, B.; Pokorn, M.; Drole Torkar, A.; Vidmar, D. Improvement of pneumococcal pneumonia diagnostics by the use of rt-PCR on plasma and respiratory samples. Scand. J. Infect. Dis. 2013, 45, 731–737. [Google Scholar] [CrossRef]
- Zilberberg, M.D.; Nathanson, B.H.; Sulham, K.; Fan, W.; Shorr, A.F. Multidrug resistance, inappropriate empiric therapy, and hospital mortality in Acinetobacter baumannii pneumonia and sepsis. Crit. Care 2016, 20, 221. [Google Scholar] [CrossRef]
- Available online: https://www.bruker.com/products/mass-spectrometry-and-separations/maldi-biotyper-systems.html (accessed on 21 March 2019).
- Available online: https://www.biomerieux-diagnostics.com/vitekr-ms-0 (accessed on 21 March 2019).
- Available online: https://curetis.com/products/unyvero-a50-system/ (accessed on 21 March 2019).
- Available online: https://www.biomerieux-diagnostics.com/biofire-filmarray-pneumonia-panel (accessed on 20 February 2019).
- Available online: http://www.fast-trackdiagnostics.com/human-line/products/ftd-respiratory-pathogens-33/ (accessed on 21 March 2019).
- Available online: http://www.pathofinder.com/products/multifinder/respifinder-22 (accessed on 21 March 2019).
- Available online: https://www.luminexcorp.com/respiratory-pathogens-flex-test (accessed on 21 March 2019).
- Available online: https://en.vircell.com/diseases/37-mycoplasma-pneumoniae/ (accessed on 21 March 2019).
- Available online: https://www.zeusscientific.com/products/zeus-elisa-test-systems/zeus-elisa-m-pneumoniae-igg-test-system (accessed on 21 March 2019).
- Hee Kim, M.; Young Kang, S.; Lee, W.I. Comparison of Two Enzyme Immunoassays for Detecting Mycoplasma pneumonia. Lab. Med. 2012, 43, 74–77. [Google Scholar] [CrossRef]
- Available online: https://cvi.asm.org/content/8/3/588 (accessed on 21 March 2019).
- Available online: https://www.labsystemsdx.com/products/mifa/chlamydia-pneumoniae-mif-assays (accessed on 21 March 2019).
- Available online: https://www.alere.com/en/home/product-details/binaxnow-legionella.html (accessed on 21 March 2019).
- Available online: http://www.sascientific.com/products/tag/Legionella (accessed on 21 March 2019).
- Available online: https://www.trinitybiotech.com/products/legionella-urinary-antigen-eia/ (accessed on 21 March 2019).
- Available online: http://www.meridianbioscience.com/diagnostic-products/respiratory/tru/tru-legionella.aspx (accessed on 21 March 2019).
- Available online: https://www.alere.com/en/home/product-details/binaxnow-streptococcus-pneumoniae.html (accessed on 21 March 2019).
- Patel, R. MALDI-TOF MS for the diagnosis of infectious disease. Clin. Chem. 2015, 61, 100–111. [Google Scholar] [CrossRef] [PubMed]
- Lévesque, S.; Dufresne, P.J.; Soualhine, H.; Domingo, M.-C.; Bekal, S.; Lefebvre, B.; Tremblay, C. A side by side comparison of Bruker Biotyper and VITEK MS: Utility of MALDI-TOF MS technology for microorganism identification in a public health reference laboratory. PLoS ONE 2015, 10, e0144878. [Google Scholar] [CrossRef]
- Available online: https://www.biomerieux-diagnostics.com/etestr (accessed on 20 February 2019).
- Available online: https://seluxdx.com (accessed on 20 February 2019).
- Charalampous, T.; Richardson, H.; Kay, G.L.; Baldan, R.; Jeanes, C.; Rae, D.; Grundy, S.; Turner, D.J.; Wain, J.; Leggett, R.M.; et al. Rapid diagnosis of lower respiratory infection using nanopore-based clinical metagenomics. bioRxiv 2018, 387548. [Google Scholar] [CrossRef]
- Available online: https://www.accessdata.fda.gov/cdrh_docs/reviews/DEN170047.pdf (accessed on 20 February 2019).
- Dekker, J.P. Metagenomics for Clinical Infectious Disease Diagnostics Steps Closer to Reality. J. Clin. Microbiol. 2018, 56, e00850-18. [Google Scholar] [CrossRef] [PubMed]
- Charalampous, T.; Richardson, H.; Kay, G.; Baldan, R.; Jeanes, C.; Turner, D.; Wain, J.; Livermore, D.; Leggett, R.; O’Grady, J. Diagnosis of lower respiratory tract infections using nanopore sequencing. Eur. Respir. J. 2018, 52, PA5308. [Google Scholar]
- Llor, C.; Bjerrum, L. Antimicrobial resistance: Risk associated with antibiotic overuse and initiatives to reduce the problem. Adv. Drug Saf. 2014, 5, 229–241. [Google Scholar] [CrossRef] [PubMed]
- Gross, A.E.; Van Schooneveld, T.C.; Olsen, K.M.; Rupp, M.E.; Bui, T.H.; Forsung, E.; Kalil, A.C. Epidemiology and predictors of multidrug-resistant community-acquired and health care-associated pneumonia. Antimicrob. Agents Chemother. 2014, 58, 5262–5268. [Google Scholar] [CrossRef]
- Seligman, R.; Ramos-Lima, L.F.; Oliveira Vdo, A.; Sanvicente, C.; Sartori, J.; Pacheco, E.F. Risk factors for infection with multidrug-resistant bacteria in non-ventilated patients with hospital-acquired pneumonia. J. Bras. Pneumol. 2013, 39, 339–348. [Google Scholar] [CrossRef]
- Fleming-Dutra, K.E.; Hersh, A.L.; Shapiro, D.J.; Bartoces, M.; Enns, E.A.; File, T.M.; Finkelstein, J.A.; Gerber, J.S.; Hyun, D.Y.; Linder, J.A.; et al. Prevalence of inappropriate antibiotic prescriptions among US ambulatory care visits, 2010–2011. JAMA 2016, 315, 1864–1873. [Google Scholar] [CrossRef]
| C-Reactive Protein Value (mg/L) | Antibiotic Therapy |
|---|---|
| <20 | Withhold in most patients |
| 21–99 | Further assessment needed to determine |
| ≥100 | Strongly encouraged |
| Procalcitonin Value (ng/mL) | |
| < 0.10 ng/mL | Strongly discouraged |
| 0.10–0.25 ng/mL | Discouraged |
| 0.26–0.50 ng/mL | Encouraged |
| > 0.50 ng/mL | Strongly encouraged |
| Diagnostic | Pathogens | Company | FDA Approved | Sample Type | Time |
|---|---|---|---|---|---|
| Matrix-assisted laser desorption/ionisation time-of-flight mass spectrometry (MALDI-TOF MS) | |||||
| MALDI Biotyper CA; microflex LT/SH MALDI-MS; IVD MALDI Biotyper; MALDI Biotyper smart System; MBT smart CA System [35] | Bacterial, Fungal; 333 species or species groups | Brukner Daltonics | Yes | Induced or expectorated sputum, nasal aspirates or washes, nasopharyngeal (NP) swabs or aspirates, throat washes or swabs, bronchoscopic specimens | Within minutes of analyzing a single colony from isolate |
| Vitek MS [36] | Bacterial, Fungal; 1316 species, includes Brucella, Candida auris, Elizabethkingia anophelis | bio Mérieux | Yes | Induced or expectorated sputum, nasal aspirates or washes, NP swabs or aspirates, throat washes or swabs, bronchoscopic specimens | Within minutes of analyzing a single colony from isolate |
| Nucleic acid amplification tests - Polymerase chain reaction | |||||
| Unyvero A50 System [37] | >30 Gram-positive/Gram-negative bacteria and 10 antibiotic resistance markers or toxins | Curetis AG | Yes | Induced or expectorated sputum, nasal aspirates or washes, NP swabs or aspirates, throat washes or swabs, bronchoscopic specimens | <5 h |
| BIOFIRE FilmArray System Pneumonia Panel plus [38] | 11 Gram-negative, 4 Gram-positive and 3 atypical bacteria, 9 viruses, 7 genetic markers of antibiotic resistance | bio Mérieux | Yes | Induced or expectorated sputum; endotracheal aspirates, bronchoscopic specimens | 1 h |
| Multiplex one-step RT-PCR - FTD Respiratory pathogens 33 [39] | 22 viruses, 11 bacterial pathogens | Fast Track Diagnostics | Yes | Induced or expectorated sputum; endotracheal aspirates, bronchoscopic specimens | <2 h |
| Multiplex ligation-dependent probe amplification - RespiFinder 22 [40] | 18 viruses, 4 bacterial pathogens | PathoFinder | Yes | Induced or expectorated sputum; endotracheal aspirates, bronchoscopic specimens | <2 h |
| VERIGENE Respiratory Pathogens Flex Test [41] | 13 viral and 3 bacterial (Bordetella sp) targets | Nanosphere/Luminex Corporation | Yes | Induced or expectorated sputum; endotracheal aspirates, bronchoscopic specimens | <2 h |
| Serological tests | |||||
| Enzyme immunoassay | Mycoplasma pneumoniae IgG and IgM | Vircell [42]/Zeus [43,44] | Yes | 5 mL serum | <2 h |
| Microimmunofluorescent stain | Chlaymydia pneumoniae | MRL Diagnostics [45]/Labsystems [46] | Yes | 5 mL serum | <2 h |
| Urine antigen tests | |||||
| Enzyme immunoassay - BinaxNOW Legionella Urinary Antigen Card [47] | Legionella pneumophilia (for serogroup 1) | Alere/Abbott | Yes | 10 mL of urine | <1 h |
| Lateral Flow Assay - SAS Legionella [48] | Legionella pneumophilia (for serogroup 1) | SA Scientific | Yes | 10 mL of urine | <1 h |
| Lateral Flow Assay - Bartels Legionella urinary antigen [49] | Legionella pneumophilia (for serogroup 1) | Trinity Biotech | Yes | 10 mL of urine | <1 h |
| Lateral Flow Assay - Meridian Tru Legionella Assay [50] | Legionella pneumophilia (for serogroup 1) | Meridian BioSciences | Yes | 10 mL of urine | <1 h |
| Enzyme immunoassay – BinaxNow Streptococcus pneumoniae Antigen Card [51] | S. pneumoniae | Alere/Abbott | Yes | 10 mL of urine | <1 h |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Noviello, S.; Huang, D.B. The Basics and the Advancements in Diagnosis of Bacterial Lower Respiratory Tract Infections. Diagnostics 2019, 9, 37. https://doi.org/10.3390/diagnostics9020037
Noviello S, Huang DB. The Basics and the Advancements in Diagnosis of Bacterial Lower Respiratory Tract Infections. Diagnostics. 2019; 9(2):37. https://doi.org/10.3390/diagnostics9020037
Chicago/Turabian StyleNoviello, Stephanie, and David B. Huang. 2019. "The Basics and the Advancements in Diagnosis of Bacterial Lower Respiratory Tract Infections" Diagnostics 9, no. 2: 37. https://doi.org/10.3390/diagnostics9020037
APA StyleNoviello, S., & Huang, D. B. (2019). The Basics and the Advancements in Diagnosis of Bacterial Lower Respiratory Tract Infections. Diagnostics, 9(2), 37. https://doi.org/10.3390/diagnostics9020037




