Advanced Deep Learning Approaches in Detection Technologies for Comprehensive Breast Cancer Assessment Based on WSIs: A Systematic Literature Review
Abstract
1. Introduction
- 1.
- What types of datasets are used for comprehensive breast cancer assessment using WSIs?
- 2.
- What are the main challenges associated with comprehensive breast cancer assessment using WSIs?
- 3.
- How do WSIs impact the accuracy and reliability of advanced deep learning approaches for comprehensive breast cancer assessment?
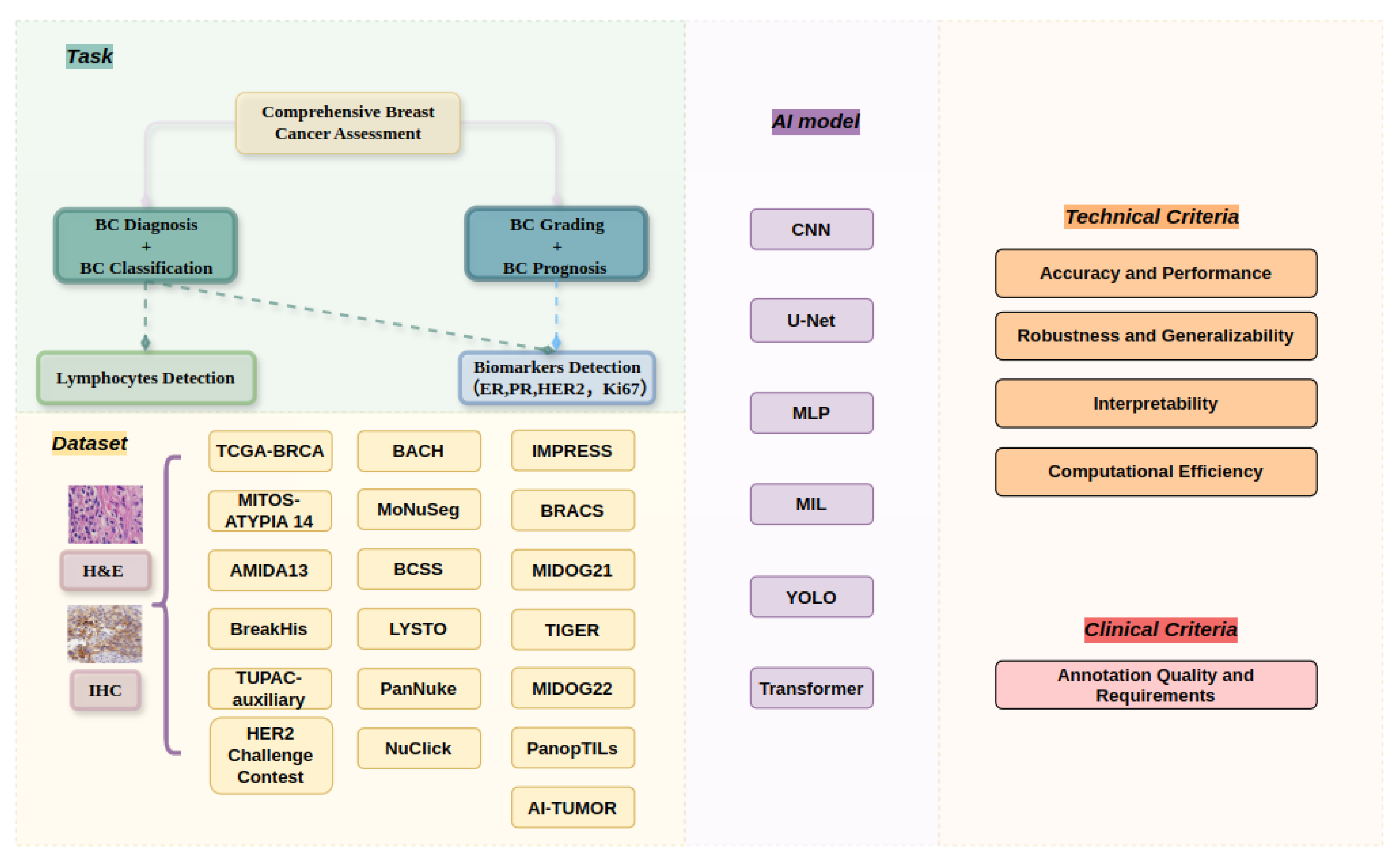
2. Methods
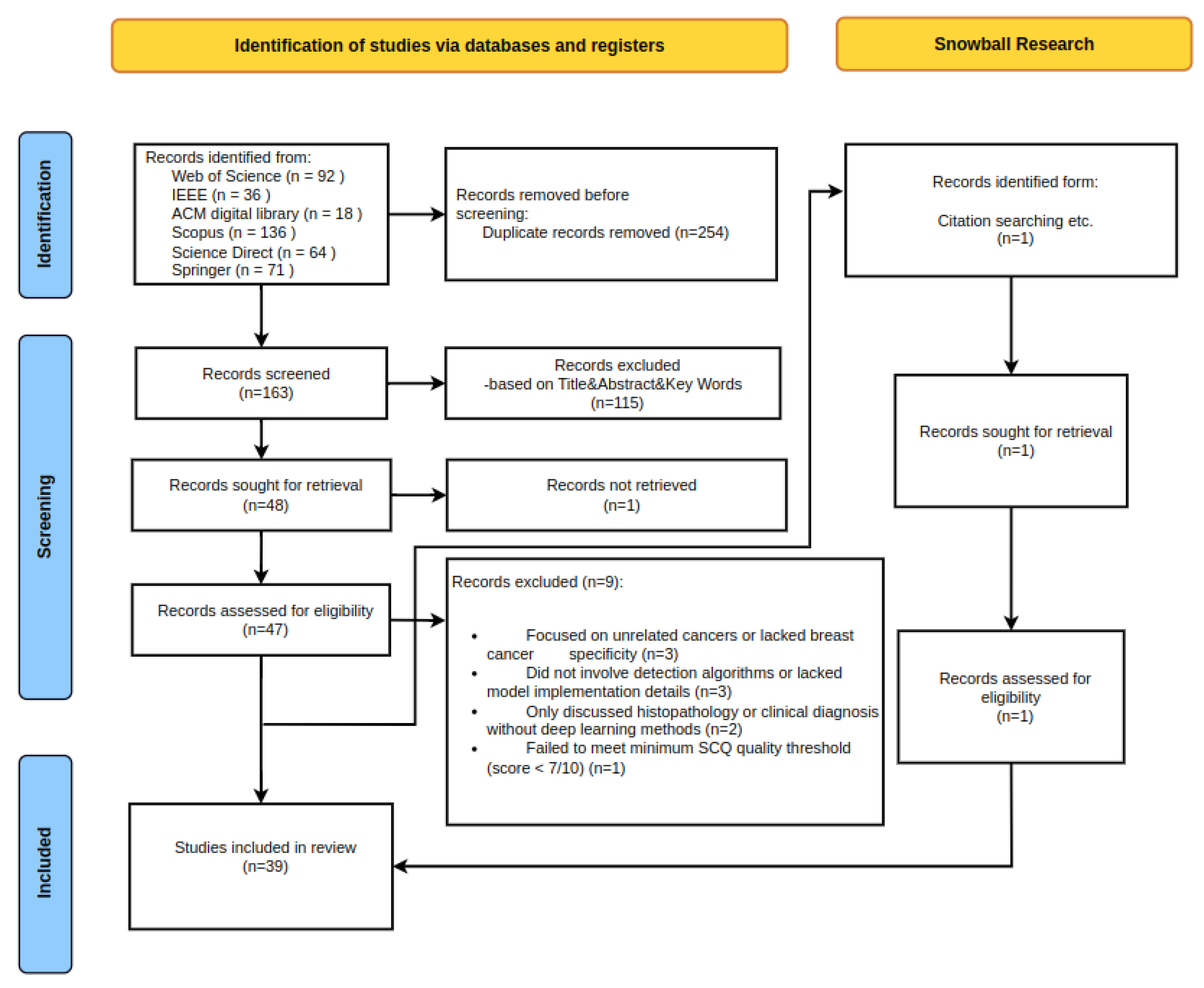
2.1. Data Sources and Search Strategy
- “breast cancer detection” AND “deep learning”
- “breast cancer diagnosis” AND “deep learning”
- “convolutional neural networks” AND “breast cancer”
- “lymphocytes detection” OR “biomarkers detection” AND “breast cancer”
- “H&E stained images” AND “deep learning” AND “breast cancer”
- “immunohistochemistry” AND “deep learning” AND “breast cancer”
- “automated breast cancer diagnosis” OR “AI in breast cancer screening”
2.2. Selection Criteria
2.3. Quality Assessment
2.4. Data Extraction and Synthesis
3. Results and Meta-Analysis
3.1. Overview of Selected Studies
3.2. Research Question 1: What Types of Datasets Are Employed for Comprehensive Breast Cancer Assessment Using WSIs?
- 1.
- Scale and Origin:
- 2.
- Research Focus and Annotation Granularity:
- Datasets cover a wide range of anatomical structures, such as nuclei in MoNuSeg and tumor-infiltrating lymphocytes in PanopTILs, to specific cellular structures.
- Increasing model sophistication is reflected in the variation in annotation detail from whole-slide to pixel-level [71].
- 3.
- Multi-modal Integration:
- 4.
- Ethical Considerations and Diversity:
- More recent datasets, such as AI-TUMOR, highlight the diversity of patient demographics and the use of ethical data collection techniques [74].
3.3. Research Question 2: What Are the Main Challenges Associated with Comprehensive Breast Cancer Assessment Using WSIs?
3.4. Research Question 3: How Do WSIs Affect the Accuracy and Reliability of Advanced Deep Learning Approaches for Comprehensive Breast Cancer Assessment?
4. Criteria for Comprehensive Breast Cancer-Assessment WSI Algorithms
- 1.
- Accuracy and Performance Metrics: According to [91], sensitivity, specificity, accuracy, recall, AUC, and F1 score are essential for accurately identifying cancer cells while reducing false positives.
- 2.
- Robustness and Generalizability: Algorithms must manage common problems like noise and artifacts while operating consistently over a range of datasets, scanners, and staining processes [92].
- 3.
- Interpretability and Explainability: Clinical trust depends on model openness and error analysis capabilities [93].
- 4.
- Computational Efficiency: Algorithms should be appropriate for a range of computational contexts, with respectable processing speeds and efficient resource consumption [94].
4.1. Baseline Models for Detection Technologies Applied in Comprehensive Breast Cancer Assessment Based on WSI Algorithms
4.2. Optimizing and Improving Existing Baselines Based on Evaluation Criteria
4.2.1. Enhancing Model Performance
4.2.2. Improving Robustness and Generalizability
4.2.3. Increasing Interpretability and Explainability
4.2.4. Optimizing Computational Efficiency
4.2.5. Addressing Data Quality and Annotation Challenges
5. Discussion and Potential Solutions for Improving WSIs for Breast Cancer Cell Detection
6. Conclusions, Implication, and Recommendations for Future Research
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| PRISM | Preferred Reporting Items for Systematic Reviews and Meta-Analyses |
| WSI | Whole Slide Image |
| WSIs | Whole Slide Images |
| ER | Estrogen Receptor |
| PR | rogesterone Receptor |
| HER2 | Human Epidermal Growth Factor Receptor 2 |
| Ki-67 | Ki-67 Antigen |
| TIL | Tumor-Infiltrating Lymphocyte |
| H&E | Hematoxylin and Eosin |
| IHC | Immunohistochemistry |
| TNBC | Triple-Negative Breast Cancer |
| LD | Lymphocyte Detection |
| BD | Biomarker Detection |
| SCQ | Systematic Quality Criteria |
| SLR | Systematic Literature Review |
| HPF | High-Power Field |
| CNN | Convolutional Neural Network |
| GAN | Generative Adversarial Network |
| MLP | Multilayer Perceptron |
| MIL | Multiple Instance Learning |
| YOLO | You Only Look Once |
| CAD | Computer-Aided Diagnosis |
| ROI | Region of Interest |
| HIF | Human-Interpretable Features |
References
- Faroughi, F.; Fathnezhad-Kazemi, A.; Sarbakhsh, P. Factors Affecting Quality of Life in Women With Breast Cancer: A Path Analysis. BMC Women’s Health 2023, 23, 578. [Google Scholar]
- Khairi, S.S.M.; Bakar, M.A.A.; Alias, M.A.; Bakar, S.A.; Liong, C.Y.; Rosli, N.; Farid, M. Deep learning on histopathology images for breast cancer classification: A bibliometric analysis. Healthcare 2021, 10, 10. [Google Scholar] [CrossRef] [PubMed]
- Saifullah, S.; Dreżewski, R. Enhancing breast cancer diagnosis: A CNN-based approach for medical image segmentation and classification. In International Conference on Computational Science; Springer: Berlin/Heidelberg, Germany, 2024; pp. 155–162. [Google Scholar]
- Mahmoud, R.; Ordóñez-Morán, P.; Allegrucci, C. Challenges for triple negative breast cancer treatment: Defeating heterogeneity and cancer stemness. Cancers 2022, 14, 4280. [Google Scholar] [CrossRef]
- El Bairi, K.; Haynes, H.R.; Blackley, E.; Fineberg, S.; Shear, J.; Turner, S.; De Freitas, J.R.; Sur, D.; Amendola, L.C.; Gharib, M.; et al. The tale of TILs in breast cancer: A report from the international immuno-oncology biomarker working group. NPJ Breast Cancer 2021, 7, 150. [Google Scholar] [PubMed]
- Pauzi, S.H.M.; Saari, H.N.; Roslan, M.R.; Azman, S.N.S.S.K.; Tauan, I.S.; Rusli, F.A.M.; Aizuddin, A.N. A comparison study of HER2 protein overexpression and its gene status in breast cancer. Malays. J. Pathol. 2019, 41, 133–138. [Google Scholar]
- Finkelman, B.S.; Zhang, H.; Hicks, D.G.; Turner, B.M. The evolution of Ki-67 and breast carcinoma: Past observations, present directions, and future considerations. Cancers 2023, 15, 808. [Google Scholar] [CrossRef]
- Zahari, S.; Syafruddin, S.E.; Mohtar, M.A. Impact of the Cancer Cell Secretome in driving breast Cancer progression. Cancers 2023, 15, 2653. [Google Scholar] [CrossRef]
- Gupta, R.; Le, H.; Van Arnam, J.; Belinsky, D.; Hasan, M.; Samaras, D.; Kurc, T.; Saltz, J.H. Characterizing immune responses in whole slide images of cancer with digital pathology and pathomics. Curr. Pathobiol. Rep. 2020, 8, 133–148. [Google Scholar]
- Roostee, S.; Ehinger, D.; Jönsson, M.; Phung, B.; Jönsson, G.; Sjödahl, G.; Staaf, J.; Aine, M. Tumour immune characterisation of primary triple-negative breast cancer using automated image quantification of immunohistochemistry-stained immune cells. Sci. Rep. 2024, 14, 21417. [Google Scholar] [CrossRef]
- Liu, Y.; Han, D.; Parwani, A.V.; Li, Z. Applications of artificial intelligence in breast pathology. Arch. Pathol. Lab. Med. 2023, 147, 1003–1013. [Google Scholar]
- Shakhawat, H.; Hossain, S.; Kabir, A.; Mahmud, S.H.; Islam, M.M.; Tariq, F. Review of artifact detection methods for automated analysis and diagnosis in digital pathology. In Artificial Intelligence for Disease Diagnosis and Prognosis in Smart Healthcare; CRC Press: Boca Raton, FL, USA, 2023; pp. 177–202. [Google Scholar]
- Li, X.; Li, C.; Rahaman, M.M.; Sun, H.; Li, X.; Wu, J.; Yao, Y.; Grzegorzek, M. A comprehensive review of computer-aided whole-slide image analysis: From datasets to feature extraction, segmentation, classification and detection approaches. Artif. Intell. Rev. 2022, 55, 4809–4878. [Google Scholar] [CrossRef]
- Wu, Y.; Cheng, M.; Huang, S.; Pei, Z.; Zuo, Y.; Liu, J.; Yang, K.; Zhu, Q.; Zhang, J.; Hong, H.; et al. Recent advances of deep learning for computational histopathology: Principles and applications. Cancers 2022, 14, 1199. [Google Scholar] [CrossRef] [PubMed]
- Carriero, A.; Groenhoff, L.; Vologina, E.; Basile, P.; Albera, M. Deep Learning in Breast Cancer Imaging: State of the Art and Recent Advancements in Early 2024. Diagnostics 2024, 14, 848. [Google Scholar] [CrossRef] [PubMed]
- Duggento, A.; Conti, A.; Mauriello, A.; Guerrisi, M.; Toschi, N. Deep computational pathology in breast cancer. Semin. Cancer Biol. 2021, 72, 226–237. [Google Scholar] [CrossRef]
- Luo, L.; Wang, X.; Lin, Y.; Ma, X.; Tan, A.; Chan, R.; Vardhanabhuti, V.; Chu, W.C.W.; Cheng, K.T.; Chen, H. Deep learning in breast cancer imaging: A decade of progress and future directions. IEEE Rev. Biomed. Eng. 2024, 18, 130–151. [Google Scholar] [CrossRef]
- Al-Thelaya, K.; Gilal, N.U.; Alzubaidi, M.; Majeed, F.; Agus, M.; Schneider, J.; Househ, M. Applications of discriminative and deep learning feature extraction methods for whole slide image analysis: A survey. J. Pathol. Inform. 2023, 14, 100335. [Google Scholar] [CrossRef]
- Hulsen, T. Explainable Artificial Intelligence (XAI): Concepts and Challenges in Healthcare. AI 2023, 4, 652–666. [Google Scholar] [CrossRef]
- Mahmood, T.; Li, J.; Pei, Y.; Akhtar, F.; Imran, A.; Rehman, K.U. A brief survey on breast cancer diagnostic with deep learning schemes using multi-image modalities. IEEE Access 2020, 8, 165779–165809. [Google Scholar] [CrossRef]
- Mridha, M.F.; Hamid, M.A.; Monowar, M.M.; Keya, A.J.; Ohi, A.Q.; Islam, M.R.; Kim, J.M. A comprehensive survey on deep-learning-based breast cancer diagnosis. Cancers 2021, 13, 6116. [Google Scholar] [CrossRef]
- Wen, Z.; Wang, S.; Yang, D.M.; Xie, Y.; Chen, M.; Bishop, J.; Xiao, G. Deep learning in digital pathology for personalized treatment plans of cancer patients. Semin. Diagn. Pathol. 2023, 40, 109–119. [Google Scholar] [CrossRef]
- Rabilloud, N.; Allaume, P.; Acosta, O.; De Crevoisier, R.; Bourgade, R.; Loussouarn, D.; Rioux-Leclercq, N.; Khene, Z.E.; Mathieu, R.; Bensalah, K.; et al. Deep learning methodologies applied to digital pathology in prostate cancer: A systematic review. Diagnostics 2023, 13, 2676. [Google Scholar] [CrossRef] [PubMed]
- Page, M.J.; McKenzie, J.E.; Bossuyt, P.M.; Boutron, I.; Hoffmann, T.C.; Mulrow, C.D.; Shamseer, L.; Tetzlaff, J.M.; Akl, E.A.; Brennan, S.E.; et al. The PRISMA 2020 statement: An updated guideline for reporting systematic reviews. BMJ 2021, 372, n71. [Google Scholar] [CrossRef] [PubMed]
- Knobloch, K.; Yoon, U.; Vogt, P.M. Preferred reporting items for systematic reviews and meta-analyses (PRISMA) statement and publication bias. J. Cranio-Maxillofac. Surg. 2011, 39, 91–92. [Google Scholar] [CrossRef] [PubMed]
- Tricco, A.C.; Lillie, E.; Zarin, W.; O’Brien, K.K.; Colquhoun, H.; Levac, D.; Moher, D.; Peters, M.D.; Horsley, T.; Weeks, L.; et al. PRISMA extension for scoping reviews (PRISMA-ScR): Checklist and explanation. Ann. Intern. Med. 2018, 169, 467–473. [Google Scholar] [CrossRef]
- Swiderska-Chadaj, Z.; Gallego, J.; Gonzalez-Lopez, L.; Bueno, G. Detection of Ki67 hot-spots of invasive breast cancer based on convolutional neural networks applied to mutual information of H&E and Ki67 whole slide images. Appl. Sci. 2020, 10, 7761. [Google Scholar] [CrossRef]
- George, K.; Faziludeen, S.; Sankaran, P.; Joseph, P.K. Breast cancer detection from biopsy images using nucleus guided transfer learning and belief based fusion. Comput. Biol. Med. 2020, 124, 103954. [Google Scholar] [CrossRef]
- Krithiga, R.; Geetha, P. Deep learning based breast cancer detection and classification using fuzzy merging techniques. Mach. Vis. Appl. 2020, 31, 63. [Google Scholar] [CrossRef]
- Evangeline, I.K.; Precious, J.G.; Pazhanivel, N.; Kirubha, S.P.A. Automatic Detection and Counting of Lymphocytes from Immunohistochemistry Cancer Images Using Deep Learning. J. Med Biol. Eng. 2020, 40, 735–747. [Google Scholar] [CrossRef]
- Geread, R.S.; Sivanandarajah, A.; Brouwer, E.R.; Wood, G.A.; Androutsos, D.; Faragalla, H.; Khademi, A. piNET: An automated proliferation index calculator framework for Ki67 breast cancer images. Cancers 2021, 13, 11. [Google Scholar] [CrossRef]
- Krijgsman, D.; van Leeuwen, M.B.; van der Ven, J.; Almeida, V.; Vlutters, R.; Halter, D.; Kuppen, P.J.K.; van de Velde, C.J.H.; Wimberger-Friedl, R. Quantitative whole slide assessment of tumor-infiltrating CD8-positive lymphocytes in ER-positive breast cancer in relation to clinical outcome. IEEE J. Biomed. Health Inform. 2021, 25, 381–392. [Google Scholar] [CrossRef]
- Gamble, P.; Jaroensri, R.; Wang, H.; Tan, F.; Moran, M.; Brown, T.; Flament-Auvigne, I.; Rakha, E.A.; Toss, M.; Dabbs, D.J.; et al. Determining breast cancer biomarker status and associated morphological features using deep learning. Commun. Med. 2021, 1, 14. [Google Scholar] [CrossRef] [PubMed]
- Lu, X.; You, Z.; Sun, M.; Wu, J.; Zhang, Z. Breast cancer mitotic cell detection using cascade convolutional neural network with U-Net. Math. Biosci. Eng. 2021, 18, 673–695. [Google Scholar] [CrossRef] [PubMed]
- Budginaite, E.; Morkunas, M.; Laurinavicius, A.; Treigys, P. Deep Learning Model for Cell Nuclei Segmentation and Lymphocyte Identification in Whole Slide Histology Images. Informatica 2021, 32, 23–40. [Google Scholar] [CrossRef]
- Narayanan, P.L.; Raza, S.E.A.; Hall, A.H.; Marks, J.R.; King, L.; West, R.B.; Hernandez, L.; Guppy, N.; Dowsett, M.; Gusterson, B.; et al. Unmasking the immune microecology of ductal carcinoma in situ with deep learning. NPJ Breast Cancer 2021, 7, 19. [Google Scholar] [CrossRef]
- Diao, J.A.; Wang, J.K.; Chui, W.F.; Mountain, V.; Gullapally, S.C.; Srinivasan, R.; Mitchell, R.N.; Glass, B.; Hoffman, S.; Rao, S.K.; et al. Human-interpretable image features derived from densely mapped cancer pathology slides predict diverse molecular phenotypes. Nat. Commun. 2021, 12, 1613. [Google Scholar] [CrossRef]
- Negahbani, F.; Sabzi, R.; Jahromi, B.P.; Firouzabadi, D.; Movahedi, F.; Shirazi, M.K.; Majidi, S.; Dehghanian, A. PathoNet introduced as a deep neural network backend for evaluation of Ki-67 and tumor-infiltrating lymphocytes in breast cancer. Sci. Rep. 2021, 11, 8489. [Google Scholar] [CrossRef]
- Diaz Guerrero, R.E.; Oliveira, J.L. Improvements in lymphocytes detection using deep learning with a preprocessing stage. In Proceedings of the 2021 IEEE 34th International Symposium on Computer-Based Medical Systems (CBMS), Aveiro, Portugal, 7–9 June 2021; pp. 178–182. [Google Scholar] [CrossRef]
- Schirris, Y.; Engelaer, M.; Panteli, A.; Horlings, H.M.; Gavves, E.; Teuwen, J. WeakSTIL: Weak whole-slide image level stromal tumor infiltrating lymphocyte scores are all you need. Proc. SPIE Med. Imaging: Digit. Comput. Pathol. 2022, 12039, 120390B. [Google Scholar] [CrossRef]
- Ektefaie, Y.; Yuan, W.; Dillon, D.A.; Lin, N.U.; Golden, J.A.; Kohane, I.S.; Yu, K.H. Integrative multiomics-histopathology analysis for breast cancer classification. NPJ Breast Cancer 2021, 7, 147. [Google Scholar] [CrossRef]
- Zafar, M.M.; Rauf, Z.; Sohail, A.; Khan, A.R.; Obaidullah, M.; Khan, S.H.; Lee, Y.S.; Khan, A. Detection of tumour infiltrating lymphocytes in CD3 and CD8 stained histopathological images using a two-phase deep CNN. Photodiagnosis Photodyn. Ther. 2022, 37, 102676. [Google Scholar] [CrossRef]
- Zhang, X.; Zhu, X.; Tang, K.; Zhao, Y.; Lu, Z.; Feng, Q. DDTNet: A dense dual-task network for tumor-infiltrating lymphocyte detection and segmentation in histopathological images of breast cancer. Med. Image Anal. 2022, 78, 102415. [Google Scholar] [CrossRef]
- Chen, Y.; Li, H.; Janowczyk, A.; Toro, P.; Corredor, G.; Whitney, J.; Lu, C.; Koyuncu, C.F.; Mokhtari, M.; Buzzy, C.; et al. Computational pathology improves risk stratification of a multi-gene assay for early stage ER+ breast cancer. NPJ Breast Cancer 2023, 9, 40. [Google Scholar] [CrossRef] [PubMed]
- Ali, M.L.; Rauf, Z.; Khan, A.; Sohail, A.; Ullah, R.; Gwak, J. CB-HVT Net: A Channel-Boosted Hybrid Vision Transformer Network for Lymphocyte Detection in Histopathological Images. IEEE Access 2023, 11, 115740–115750. [Google Scholar] [CrossRef]
- Huang, J.; Li, H.; Wan, X.; Li, G. Affine-Consistent Transformer for Multi-Class Cell Nuclei Detection. In Proceedings of the 2023 International Conference on Computer Vision (ICCV 2023), Paris, France, 2–3 October 2023; pp. 21327–21336. [Google Scholar] [CrossRef]
- Jiao, Y.; van der Laak, J.; Albarqouni, S.; Li, Z.; Tan, T.; Bhalerao, A.; Cheng, S.; Ma, J.; Pocock, J.; Pluim, J.P.W.; et al. LYSTO: The Lymphocyte Assessment Hackathon and Benchmark Dataset. IEEE J. Biomed. Health Inform. 2024, 28, 1161–1172. [Google Scholar] [CrossRef] [PubMed]
- Shah, H.A.; Kang, J.M. An Optimized Multi-Organ Cancer Cells Segmentation for Histopathological Images Based on CBAM-Residual U-Net. IEEE Access 2023, 11, 111608–111621. [Google Scholar] [CrossRef]
- Yosofvand, M.; Khan, S.Y.; Dhakal, R.; Nejat, A.; Moustaid-Moussa, N.; Rahman, R.L.; Moussa, H. Automated Detection and Scoring of Tumor-Infiltrating Lymphocytes in Breast Cancer Histopathology Slides. Cancers 2023, 15, 3635. [Google Scholar] [CrossRef]
- Huang, P.W.; Ouyang, H.; Hsu, B.Y.; Chang, Y.R.; Lin, Y.C.; Chen, Y.A.; Hsieh, Y.H.; Fu, C.C.; Li, C.F.; Lin, C.H.; et al. Deep-learning based breast cancer detection for cross-staining histopathology images. Heliyon 2023, 9, e13171. [Google Scholar] [CrossRef]
- Rauf, Z.; Sohail, A.; Khan, S.H.; Khan, A.; Gwak, J.; Maqbool, M. Attention-guided multi-scale deep object detection framework for lymphocyte analysis in IHC histological images. Microscopy 2023, 72, 27–42. [Google Scholar] [CrossRef]
- Wang, X.; Zhang, J.; Yang, S.; Xiang, J.; Luo, F.; Wang, M.; Zhang, J.; Yang, W.; Huang, J.; Han, X. A generalizable and robust deep learning algorithm for mitosis detection in multicenter breast histopathological images. Med. Image Anal. 2023, 84, 102703. [Google Scholar] [CrossRef]
- Ryu, J.; Puche, A.V.; Shin, J.; Park, S.; Brattoli, B.; Lee, J.; Jung, W.; Cho, S.I.; Paeng, K.; Ock, C.Y.; et al. OCELOT: Overlapped Cell on Tissue Dataset for Histopathology. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Paris, France, 2–3 October 2023; pp. 23902–23912. [Google Scholar] [CrossRef]
- Rauf, Z.; Khan, A.R.; Sohail, A.; Alquhayz, H.; Gwak, J.; Khan, A. Lymphocyte detection for cancer analysis using a novel fusion block based channel boosted CNN. Sci. Rep. 2023, 13, 14047. [Google Scholar] [CrossRef]
- Makhlouf, S.; Wahab, N.; Toss, M.; Ibrahim, A.; Lashen, A.G.; Atallah, N.M.; Ghannam, S.; Jahanifar, M.; Lu, W.; Graham, S.; et al. Evaluation of Tumour Infiltrating Lymphocytes in Luminal Breast Cancer Using Artificial Intelligence. Br. J. Cancer 2023, 129, 1747–1758. [Google Scholar] [CrossRef]
- Aswolinskiy, W.; Munari, E.; Horlings, H.M.; Mulder, L.; Bogina, G.; Sanders, J.; Liu, Y.H.; van den Belt-Dusebout, A.W.; Tessier, L.; Balkenhol, M.; et al. PROACTING: Predicting Pathological Complete Response to Neoadjuvant Chemotherapy in Breast Cancer from Routine Diagnostic Histopathology Biopsies with Deep Learning. Breast Cancer Res. 2023, 25, 142. [Google Scholar] [CrossRef] [PubMed]
- Genc-Nayebi, N.; Abran, A. A systematic literature review: Opinion mining studies from mobile app store user reviews. J. Syst. Softw. 2017, 125, 207–219. [Google Scholar] [CrossRef]
- Kabir, S.; Vranic, S.; Al Saady, R.M.; Khan, M.S.; Sarmun, R.; Alqahtani, A.; Abbas, T.O.; Chowdhury, M.E.H. The utility of a deep learning-based approach in Her-2/neu assessment in breast cancer. Expert Syst. Appl. 2024, 238, 122051. [Google Scholar] [CrossRef]
- Li, Z.; Li, W.; Mai, H.; Zhang, T.; Xiong, Z. Enhancing Cell Detection in Histopathology Images: A ViT-Based U-Net Approach. In Graphs in Biomedical Image Analysis, and Overlapped Cell on Tissue Dataset for Histopathology, 5th MICCAI Workshop, Proceedings of the 5th International Workshop on Graphs in Biomedical Image Analysis (GRAIL) / Workshop on Overlapped Cell on Tissue—Cell Detection from Cell-Tissue Interaction Challenge (OCELOT), MICCAI 2023, Vancouver, BC, Canada, 23 September–4 October 2023; Ahmadi, S.A., Pereira, S., Eds.; Lecture Notes in Computer Science; Springer: Berlin/Heidelberg, Germany, 2024; Volume 14373, pp. 150–160. [Google Scholar] [CrossRef]
- Millward, J.; He, Z.; Nibali, A. Dense Prediction of Cell Centroids Using Tissue Context and Cell Refinement. In Graphs in Biomedical Image Analysis, and Overlapped Cell on Tissue Dataset for Histopathology, 5th MICCAI Workshop, Proceedings of the 5th International Workshop on Graphs in Biomedical Image Analysis (GRAIL) and the Workshop on Overlapped Cell on Tissue (OCELOT), MICCAI 2023, Vancouver, BC, Canada, 23 September–4 October 2023; Ahmadi, S.A., Pereira, S., Eds.; Lecture Notes in Computer Science; Springer: Berlin/Heidelberg, Germany, 2024; Volume 14373, pp. 138–149. [Google Scholar] [CrossRef]
- Karol, M.; Tabakov, M.; Markowska-Kaczmar, U.; Fulawka, L. Deep Learning for Cancer Cell Detection: Do We Need Dedicated Models? Artif. Intell. Rev. 2024, 57, 53. [Google Scholar] [CrossRef]
- Lakshmanan, B.; Anand, S.; Raja, P.S.V.; Selvakumar, B. Improved DeepMitosisNet Framework for Detection of Mitosis in Histopathology Images. Multimed. Tools Appl. 2023, 83, 43303–43324. [Google Scholar] [CrossRef]
- Hoerst, F.; Rempe, M.; Heine, L.; Seibold, C.; Keyl, J.; Baldini, G.; Ugurel, S.; Siveke, J.; Gruenwald, B.; Egger, J.; et al. CellViT: Vision Transformers for Precise Cell Segmentation and Classification. Med. Image Anal. 2024, 94, 103143. [Google Scholar] [CrossRef]
- Marzouki, A.; Guo, Z.; Zeng, Q.; Kurtz, C.; Loménie, N. Optimizing Lymphocyte Detection in Breast Cancer Whole Slide Imaging through Data-Centric Strategies. arXiv 2024, arXiv:2405.13710. [Google Scholar]
- Liu, S.; Amgad, M.; More, D.; Rathore, M.A.; Salgado, R.; Cooper, L.A.D. A panoptic segmentation dataset and deep-learning approach for explainable scoring of tumor-infiltrating lymphocytes. NPJ Breast Cancer 2024, 10, 52. [Google Scholar]
- Keele, S. Guidelines for Performing Systematic Literature Reviews in Software Engineering; Technical Report, Version 2.3, EBSE-2007-01; Software Engineering Group, School of Computer Science and Mathematics; Keele University: Keele, UK; Department of Computer Science, University of Durham: Durham, UK, 2007. [Google Scholar]
- Tafavvoghi, M.; Bongo, L.A.; Shvetsov, N.; Busund, L.T.R.; Møllersen, K. Publicly available datasets of breast histopathology H&E whole-slide images: A scoping review. J. Pathol. Inform. 2024, 15, 100363. [Google Scholar]
- Abdel-Nabi, H.; Ali, M.; Awajan, A.; Daoud, M.; Alazrai, R.; Suganthan, P.N.; Ali, T. A Comprehensive Review of the Deep Learning-Based Tumor Analysis Approaches in Histopathological Images: Segmentation, Classification and Multi-Learning Tasks. Clust. Comput. 2023, 26, 3145–3185. [Google Scholar] [CrossRef]
- Basu, A.; Senapati, P.; Deb, M.; Rai, R.; Dhal, K.G. A survey on recent trends in deep learning for nucleus segmentation from histopathology images. Evol. Syst. 2024, 15, 203–248. [Google Scholar]
- Wang, L.; Pan, L.; Wang, H.; Liu, M.; Feng, Z.; Rong, P.; Chen, Z.; Peng, S. DHUnet: Dual-branch hierarchical global–local fusion network for whole slide image segmentation. Biomed. Signal Process. Control 2023, 85, 104976. [Google Scholar]
- Liu, S.; Amgad, M.; Rathore, M.A.; Salgado, R.; Cooper, L.A.D. A panoptic segmentation approach for tumor-infiltrating lymphocyte assessment: Development of the MuTILs model and PanopTILs dataset. MedRxiv 2022. [Google Scholar] [CrossRef]
- Mondello, A.; Dal Bo, M.; Toffoli, G.; Polano, M. Machine learning in onco-pharmacogenomics: A path to precision medicine with many challenges. Front. Pharmacol. 2024, 14, 1260276. [Google Scholar]
- Rydzewski, N.R.; Shi, Y.; Li, C.; Chrostek, M.R.; Bakhtiar, H.; Helzer, K.T.; Bootsma, M.L.; Berg, T.J.; Harari, P.M.; Floberg, J.M.; et al. A platform-independent AI tumor lineage and site (ATLAS) classifier. Commun. Biol. 2024, 7, 314. [Google Scholar]
- Liu, Y.; He, X.; Yang, Y. Tumor immune microenvironment-based clusters in predicting prognosis and guiding immunotherapy in breast cancer. J. Biosci. 2024, 49, 19. [Google Scholar]
- Ahmed, F.; Abdel-Salam, R.; Hamnett, L.; Adewunmi, M.; Ayano, T. Improved Breast Cancer Diagnosis through Transfer Learning on Hematoxylin and Eosin Stained Histology Images. arXiv 2023, arXiv:2309.08745. [Google Scholar]
- Nasir, E.S.; Parvaiz, A.; Fraz, M.M. Nuclei and glands instance segmentation in histology images: A narrative review. Artif. Intell. Rev. 2023, 56, 7909–7964. [Google Scholar]
- National Cancer Institute. TCGA-BRCA Project. Available online: https://portal.gdc.cancer.gov/projects/TCGA-BRCA (accessed on 18 July 2024).
- Veta, M.; Van Diest, P.J.; Willems, S.M.; Wang, H.; Madabhushi, A.; Cruz-Roa, A.; Gonzalez, F.; Larsen, A.B.L.; Vestergaard, J.S.; Dahl, A.B.; et al. Assessment of algorithms for mitosis detection in breast cancer histopathology images. Med. Image Anal. 2015, 20, 237–248. [Google Scholar]
- Spanhol, F.A.; Oliveira, L.S.; Petitjean, C.; Heutte, L. A dataset for breast cancer histopathological image classification. IEEE Trans. Biomed. Eng. 2015, 63, 1455–1462. [Google Scholar] [CrossRef]
- Qaiser, T.; Mukherjee, A.; Reddy Pb, C.; Munugoti, S.D.; Tallam, V.; Pitkäaho, T.; Lehtimäki, T.; Naughton, T.; Berseth, M.; Pedraza, A.; et al. Her 2 challenge contest: A detailed assessment of automated HER 2 scoring algorithms in whole slide images of breast cancer tissues. Histopathology 2018, 72, 227–238. [Google Scholar] [CrossRef] [PubMed]
- Aresta, G.; Araújo, T.; Kwok, S.; Chennamsetty, S.S.; Safwan, M.; Alex, V.; Marami, B.; Prastawa, M.; Chan, M.; Donovan, M.; et al. Bach: Grand challenge on breast cancer histology images. Med. Image Anal. 2019, 56, 122–139. [Google Scholar] [CrossRef] [PubMed]
- Kumar, N.; Verma, R.; Anand, D.; Zhou, Y.; Onder, O.F.; Tsougenis, E.; Chen, H.; Heng, P.A.; Li, J.; Hu, Z.; et al. A Multi-Organ Nucleus Segmentation Challenge. IEEE Trans. Med. Imaging 2020, 39, 1380–1391. [Google Scholar] [CrossRef] [PubMed]
- Ortega-Ruiz, M.A.; Roman-Rangel, E.; Reyes-Aldasoro, C.C. Multiclass Semantic Segmentation of Immunostained Breast Cancer Tissue with a Deep-Learning Approach. medRxiv 2022. [Google Scholar] [CrossRef]
- Gamper, J.; Koohbanani, N.A.; Benes, K.; Graham, S.; Jahanifar, M.; Khurram, S.A.; Azam, A.; Hewitt, K.; Rajpoot, N. Pannuke dataset extension, insights and baselines. arXiv 2020, arXiv:2003.10778. [Google Scholar]
- Koohbanani, N.A.; Jahanifar, M.; Tajadin, N.Z.; Rajpoot, N. NuClick: A deep learning framework for interactive segmentation of microscopic images. Med. Image Anal. 2020, 65, 101771. [Google Scholar] [CrossRef]
- Jeretic, P.; Warstadt, A.; Bhooshan, S.; Williams, A. Are natural language inference models IMPPRESsive? Learning IMPlicature and PRESupposition. arXiv 2020, arXiv:2004.03066. [Google Scholar]
- Brancati, N.; Anniciello, A.M.; Pati, P.; Riccio, D.; Scognamiglio, G.; Jaume, G.; De Pietro, G.; Di Bonito, M.; Foncubierta, A.; Botti, G.; et al. BRACS: A Dataset for BReAst Carcinoma Subtyping in H&E Histology Images. arXiv 2021, arXiv:2111.04740. [Google Scholar]
- Grand Challenge. Tiger Challenge. 2022. Available online: https://tiger.grand-challenge.org/ (accessed on 18 July 2024).
- Aubreville, M.; Stathonikos, N.; Donovan, T.A.; Klopfleisch, R.; Ammeling, J.; Ganz, J.; Wilm, F.; Veta, M.; Jabari, S.; Eckstein, M.; et al. Domain generalization across tumor types, laboratories, and species—Insights from the 2022 edition of the Mitosis Domain Generalization Challenge. Med. Image Anal. 2024, 94, 103155. [Google Scholar] [CrossRef]
- Madoori, P.K.; Sannidhi, S.; Anandam, G. Artificial Intelligence and Computational Pathology: A comprehensive review of advancements and applications. Perspectives 2023, 11, 30. [Google Scholar] [CrossRef]
- Kumar, N.; Gupta, R.; Gupta, S. Whole slide imaging (WSI) in pathology: Current perspectives and future directions. J. Digit. Imaging 2020, 33, 1034–1040. [Google Scholar] [CrossRef] [PubMed]
- Wagner, S.J.; Matek, C.; Boushehri, S.S.; Boxberg, M.; Lamm, L.; Sadafi, A.; Winter, D.J.E.; Marr, C.; Peng, T. Built to last? Reproducibility and reusability of deep learning algorithms in computational pathology. Mod. Pathol. 2024, 37, 100350. [Google Scholar] [CrossRef] [PubMed]
- Nasarian, E.; Alizadehsani, R.; Acharya, U.R.; Tsui, K.L. Designing interpretable ML system to enhance trust in healthcare: A systematic review to proposed responsible clinician-AI-collaboration framework. Inf. Fusion 2024, 108, 102412. [Google Scholar] [CrossRef]
- Hossain, M.S.; Shahriar, G.M.; Syeed, M.M.M.; Uddin, M.F.; Hasan, M.; Hossain, M.S.; Bari, R. Tissue artifact segmentation and severity assessment for automatic analysis using WSI. IEEE Access 2023, 11, 21977–21991. [Google Scholar] [CrossRef]


| Inclusion Criteria | Exclusion Criteria |
|---|---|
| Articles related to advanced deep learning-based techniques for comprehensive breast cancer assessment, focusing on diagnosis, classification, grading, and prognosis | Duplicates |
| Emphasis on peer-reviewed journal articles and conference papers | Articles not addressing breast cancer detection, classification, grading, or prognosis strategies |
| Studies employing WSI techniques and related fields, particularly those focusing on breast cancer diagnosis and prognosis | Studies not related to WSI techniques and their application in breast cancer assessment |
| Articles that meet rigorous quality-assessment standards and have a clear focus on the two major tasks of breast cancer assessment | Articles not meeting quality-assessment standards |
| Articles addressing breast cancer-detection techniques through WSIs, focusing on key points like biomarkers and lymphocytes | Studies not utilizing WSIs or not focusing on the specified key points for breast cancer assessment |
| Source | Year | Task | Model | Result | Dataset and Scale |
|---|---|---|---|---|---|
| 1 [27] | 2020 | BD | AlexNet | F1 score = 0.73 | Private Dataset 50 Ki67 WSIs 50 H&E WSIs |
| 2 [28] | 2020 | LD | NucTraL + BCF | Accuracy = 0.9691 | BreakHis Benign: 637 WSIs Malignant: 1390 WSIs |
| 3 [29] | 2020 | LD | Deep-CNN + FSRM | Accuracy = 98.8%, F1 score = 0.967 | BreakHis Benign: 2480 WSIs Malignant: 5429 WSIs |
| 4 [30] | 2020 | LD | Faster R-CNN | F1 score: Scattered lymphocytes: 0.9615 Agglomerated lymphocytes: 0.8645 Artifact area: 0.8197 | LYSTO 1228 WSIs |
| 5 [31] | 2021 | BD | U-Net-piNET | F1 score: Ki67-: 0.868; Ki67+: 0.804 | Private Dataset 1142 WSIs |
| 6 [32] | 2021 | BD | U-Net | Pearson correlation coefficient of r = 0.95 | Private Dataset 200 WSIs |
| 7 [33] | 2021 | BD | DLS | AUC = 0.60 | TCGA-BRCA 3274 WSIs |
| 8 [34] | 2021 | LD | UBCNN | - | MoNuSeg 120 WSIs |
| 9 [35] | 2021 | LD | Micro-Net + MLP | F1 score = 0.87 | TCGA-BRCA 4281 nuclei images |
| 10 [36] | 2021 | LD | UNMaSk | F1 score: H&E: 0.856 IHC: 0.9037 | Private Dataset 178 WSIs |
| 11 [37] | 2021 | LD | CNN | AUROC = 0.864 | TCGA-BRCA 7075WSIs |
| 12 [38] | 2021 | LD & BD | PathoNet | F1 score = 0.7928 | SHIDC-BC-Ki-67 2357 WSIs |
| 13 [39] | 2021 | LD | SegNet + U-Net | F1 score = 0.91 | BCa 100 ROIs |
| 14 [40] | 2021 | LD | WeakSTIL | - | TCGA-BRCA 286 WSIs |
| 15 [41] | 2021 | LD | CNN | - | TCGA-BRCA 2358 WSIs |
| 16 [42] | 2022 | LD | TDC-LC | F1 score = 0.892 | LYSTO 19663 patches |
| 17 [43] | 2022 | LD | DDTNet | F1 score = 0.907 | TCGA-BRCA 865 WSIs |
| 18 [44] | 2023 | BD | GAN+CNN+U-Net | - | Private Dataset 321 samples |
| 19 [45] | 2023 | LD | CB-HVT Net | F1 score = 0.88 | LYSTO & NuClick 21312 images |
| 20 [46] | 2023 | LD | AC-Former | F1 score = 0.796 | TCGA-BRCA 452 images |
| 21 [47] | 2023 | LD | - | - | LYSTO 83 WSIs |
| 22 [48] | 2023 | LD | CBAM- Residual U-Net | F1 score = 0.9 | TNBC 81 WSIs |
| 23 [49] | 2023 | LD | U-Net +Mask R-CNN | F1 score = 0.941 | Private Dataset 63 WSIs |
| 24 [50] | 2023 | LD | MobileNetV2 +U-Net | F1 score = 0.927 | Private Dataset 30 WSIs |
| 25 [51] | 2023 | LD | DC-Lym-AF | F1 score = 0.84 | LYSTO & NuClick 871 images |
| 26 [52] | 2023 | LD | FMDet | F1 score = 0.773 | MIDOG21 MITOS-ATYPIA 14, AMIDA13, TUPAC-auxiliary 403 WSIs |
| 27 [53] | 2023 | LD | DeepLabV3 | F1 score = 71.23 | OCELOT, TIGER 17041patches |
| 28 [54] | 2023 | LD | BCF-Lym-Detector | F1 score: LYSTO: 0.93 NuClick: 0.84 | LYSTO & NuClick 16326 images |
| 29 [55] | 2023 | LD | CNN | F1 score: Immune cell: 0.82 Tumour cell: 0.92 Stromal cell: 0.81 | Private Dataset 2549 images |
| 30 [56] | 2023 | LD | PROACTING | AUC = 0.88 | IMPRESS 1053 images |
| 31 [57] | 2024 | BD | Based on CNN | AUC = 0.72 | TCGA-BRCA 14435 images |
| 32 [58] | 2024 | BD | ViT | F1 score = 0.9269 | Her 2 challenge contest 172 WSIs |
| 33 [59] | 2024 | LD | Cell-Tissue- ViT | F1 score = 0.7243 | OCELOT 400 Patches |
| 34 [60] | 2024 | LD | Tissue Context + Cell Refinement | F1-score = 0.7473 | OCELOT 400 Patches |
| 35 [61] | 2024 | LD | PathoNet | F1 score: SHIDC-B-Ki-67: 0.842 LSOC-Ki-67: 0.909 | SHIDC-BC-Ki-67, LSOC-Ki-67 2447 WSIs |
| 36 [62] | 2024 | LD | DeepMitosisNet | F1 score = 0.93 | MITOS-ATYPIA 14 2127 images |
| 37 [63] | 2024 | LD | CellViT | F1 score = 0.81 | PanNuke 7959 images |
| 38 [64] | 2024 | LD | YOLOv5 | F1 score = 0.7438 | TIGER 1879 ROIs |
| 39 [65] | 2024 | LD | MuTILs | AUROC = 93.0 | PanopTILs 1317 ROIs |
| Checklist for Quality Assessment | SCQ No |
|---|---|
| Is the report coherent and easy to read? | SCQ1 |
| Is the research’s purpose well-defined? | SCQ2 |
| Is the procedure for gathering data clearly laid out? | SCQ3 |
| Have the settings of diversity been thoroughly examined? | SCQ4 |
| Can the study’s conclusions be trusted? | SCQ5 |
| Is there a connection between the information, analysis, and conclusion? | SCQ6 |
| Is the process of experimentation and approach transparent? | SCQ7 |
| Are the research methods sufficiently documented? | SCQ8 |
| If they are credible, are they important? | SCQ9 |
| Is it possible to duplicate the research findings? | SCQ10 |
| Name | Year | Key Features | Contribution | Link |
|---|---|---|---|---|
| TCGA-BRCA [77] | - | 3111 H&E-stained WSIs from 1086 female and 12 male breast cancer patients. Includes matched gene expression data and clinical information. | Widely used to create derivative datasets like MoNuSeg, BCSS, LYSTO, and TIGER. | https://portal.gdc.cancer.gov/projects/TCGA-BRCA (accessed on 18 July 2024) |
| MITOS-ATYPIA 14 | 2013 | 2400 high-power field (HPF) images from 11 breast cancer patients, scanned by two devices: 1200 at 1539 × 1376 pixels and 1200 at 1663 × 1485 pixels, all at 40× magnification. | Provides a comprehensive external test set for evaluating the robustness of mitosis-detection models across different imaging conditions. | https://mitos-atypia-14.grand-challenge.org/Description/ (accessed on 18 July 2024) |
| AMIDA13 [78] | 2013 | 606 HPF images (311 training, 295 test) from 23 subjects, each 2000 × 2000 pixels (0.25 mm2) | Established a benchmark for mitosis-detection algorithms, later integrated into larger datasets for broader impact. | - |
| BreakHis [79] (accessed on 18 July 2024) | 2016 | 9109 breast tissue images from 82 patients at various magnifications (40×–400×). Includes 2480 benign and 5429 malignant samples, all 700 × 460 pixel RGB images. Samples collected via partial mastectomy. | Provides a diverse, well-annotated dataset for developing and evaluating breast cancer classification algorithms. Enables research on automated diagnosis across different magnifications and tumor types, potentially improving clinical diagnostic accuracy. | https://web.inf.ufpr.br/vri/databases/breast-cancer-histopathological-database-breakhis/ (accessed on 18 July 2024) |
| TUPAC-auxiliary [78] | 2016 | 73 breast cancer WSIs from 3 pathology centers, scanned by 2 types of scanners at 40× magnification. | Integrates multi-center data, enhancing the scope of mitosis-detection research in breast cancer. | https://tupac.grand-challenge.org/ (accessed on 18 July 2024) |
| HER2 Challenge Contest [80] | 2016 | High-resolution breast cancer histology dataset. Comprises 100 gigapixel WSIs. Annotated with expert pathologist HER2 scores and percentage assessments. | Pioneering benchmark for automated HER2 quantification in digital pathology. Facilitates the development of AI tools to enhance diagnostic consistency and efficiency in breast cancer assessment. | https://warwick.ac.uk/fac/cross_fac/tia/data/her2contest/ (accessed on 18 July 2024) |
| BACH [81] | 2018 | Includes 400 H&E-stained patches (2048 × 1536 resolution) and 30 WSIs with pixel-level annotations | Useful for training models with pixel-level cancer type annotations | https://www.kaggle.com/datasets/truthisneverlinear/bach-breast-cancer-histology-images (accessed on 18 July 2024) |
| MoNuSeg [82] | 2018 | WSIs from 30 organs, creating 1000 × 1000 pixel sub-images with nuclear annotations. | Ensures variation in nuclear appearance, enhancing model training on diverse tissue samples. | https://monuseg.grand-challenge.org/Data/ (accessed on 18 July 2024) |
| BCSS [83] | 2019 | 151 WSIs with representative regions of interest (ROI) selected, contributing to the TIGER dataset. | Helps in understanding tumor-infiltrating lymphocytes in HER2+ and triple-negative breast cancers. | https://bcsegmentation.grand-challenge.org/ (accessed on 18 July 2024) |
| LYSTO [47] | 2019 | LYSTO data set comprises 20,000 images from 43 patients with breast, colon, and prostate cancers. It’s structured with a patient-level split: 19 for training, 9 for validation, and 6 for testing, ensuring diverse representation across cancer types and stages. | Enables development of cross-cancer lymphocyte-assessment algorithms. Patient-level data division supports robust AI model validation, advancing generalized lymphocyte-detection tools. | https://lysto.grand-challenge.org/ (accessed on 18 July 2024) |
| PanNuke [84] | 2020 | Contains 200,000 nuclei categorized into five clinically significant classes, with high-resolution patches scanned at 20× or 40× magnification | Supports nuanced classification of different tissue types in breast cancer research | https://sites.google.com/view/panoptils (accessed on 18 July 2024) |
| NuClick [85] | 2020 | NuClick dataset comprises 871 images derived from 440 WSIs, strategically partitioned into 471 training, 99 validation, and 300 testing images. The dataset encompasses various cancer types, with careful consideration given to maintaining separation between patient samples across different sets. | Supports AI model development for cancer diagnostics with real-world applicability. Thoughtful data division prevents patient-level leakage, enhancing model reliability and clinical relevance. | https://github.com/navidstuv/NuClick (accessed on 18 July 2024) |
| IMPRESS [86] | 2020 | Large-scale linguistic dataset comprising over 25,000 sentence pairs. Utilizes a rich vocabulary of 3000+ lexical items with grammatical annotations. Generated using advanced natural language processing techniques, following established Natural language inference dataset formats. | Advances pragmatic inference research by providing a comprehensive benchmark for evaluating NLI models’ understanding of presuppositions and implicatures. Facilitates the development of more nuanced language understanding systems capable of grasping subtle linguistic phenomena. | https://github.com/facebookresearch/Imppres?tab=readme-ov-file (accessed on 18 July 2024) |
| BRACS [87] | 2021 | 4539 ROIs from 547 H&E WSIs, meticulously categorized into different lesion types. | Facilitates detailed subtyping in breast cancer research. | https://www.bracs.icar.cnr.it/ (accessed on 18 July 2024) |
| SHIDC-BC-Ki-67 [38] | 2021 | 2357 tru-cut biopsy images of invasive ductal carcinoma, collected from 2017 to 2020. Comprises 1656 training and 701 test samples, all expertly annotated for Ki-67 markers. | Addresses the scarcity of comprehensive Ki-67 marked datasets in breast cancer research. Facilitates development of deep learning models for accurate Ki-67 assessment, potentially enhancing diagnostic precision and treatment planning for invasive ductal carcinoma. | https://shiraz-hidc.com/ki-67-dataset/ (accessed on 18 July 2024) |
| MIDOG21 [87] | 2021 | 280 breast cancer WSIs at 8000 × 8000 pixels, scanned by 4 different devices. | Addresses scanner variability in mitosis detection, promoting robust algorithm development across diverse imaging equipment. | https://midog2021.grand-challenge.org/ (accessed on 18 July 2024) |
| TIGER [88] | 2022 | Includes H&E-stained WSIs of Her2 positive and Triple Negative breast cancer. TIGER comprises cases from clinical routine and a phase 3 clinical trial. Includes annotations for lymphocytes, plasma cells, invasive tumors, and stroma. | First challenge for fully automated assessment of TILs in breast cancer. Aims to validate AI-based TIL scores for clinical use. | https://tiger.grand-challenge.org/ (accessed on 18 July 2024) |
| MIDOG22 [89] | 2022 | 50 canine mast cell tumor cases, scanned by a single device type. | Expands mitosis detection to veterinary pathology, offering cross-species validation for detection algorithms. | https://midog.deepmicroscopy.org/download-dataset/ (accessed on 18 July 2024) |
| PanopTILs [65] | 2023 | Includes annotations for 814,886 nuclei from 151 patients, with WSIs scanned at 20× magnification. | Enhances understanding of the role of TILs in breast cancer detection. | https://sites.google.com/view/panoptils (accessed on 18 July 2024) |
| AI-TUMOR [73] | 2024 | 2500 WSIs with pixel-level annotations for tumor regions, normal tissue, and surrounding stroma. | Focuses on reducing biases in AI models, ensuring better generalizability. | https://github.com/nickryd/ATLAS/blob/main/README.md (accessed on 18 July 2024) |
| Challenge | Description | Impact on Breast Cancer Cell Detection | Source |
|---|---|---|---|
| Size and Complexity of WSIs | WSIs can be several gigabytes in size, containing millions of pixels that need to be processed. | Requires substantial computational resources, high-performance GPUs, and large memory capacities; storage and management of large datasets are logistically challenging. | [28,29,30] |
| Variability in Image Quality and Resolution | Inconsistencies in image quality and resolution across different scanners and datasets. | Leads to variations in training data quality, reducing model generalization and accuracy across different scanners. | [27,28,31,39,42,45,51,54,63,64] |
| Annotation Challenges | High-quality annotations are labor-intensive, time-consuming, and prone to variability. | Inconsistent annotations can introduce biases, affecting model robustness and generalizability. | [29,40,47,57,60,61] |
| Feature Extraction Difficulties | Complex and subtle patterns in breast cancer tissues are challenging to capture accurately. | Traditional methods often fall short, necessitating advanced deep learning techniques for accurate detection. | [29,30,32,33,34,35,36,37,38,40,41,43,44,45,46,47,48,49,50,51,52,53,54,55,56,57,58,59,60,62,63,65] |
| Factor | Description | Impact on Accuracy and Reliability |
|---|---|---|
| Resolution of WSIs | High-resolution images provide detailed information, crucial for detecting subtle differences between normal and cancerous tissues, especially in early-stage cancers. | Enhances detection accuracy but increases computational requirements. |
| Quality and Consistency | Variations in staining, lighting, and scanner types can introduce noise and artifacts, affecting model predictions. | Inconsistent quality can lead to inaccuracies; standardization techniques are vital. |
| Availability of Annotated WSIs | High-quality annotations by expert pathologists are essential for training accurate models. Obtaining such annotations is resource-intensive. | Poor annotations reduce model accuracy; improving annotation quality is key. |
| Diversity and Generalizability of Training | Models trained on diverse WSIs, covering a range of tissue types and patient demographics, are more likely to generalize well across different clinical settings. | Enhances generalization and reliability, but lack of standardization can hinder performance. |
| Criteria | Baseline Models | Optimization and Improvement Strategies |
|---|---|---|
| Accuracy and Performance Metrics | CNN, GAN, U-Net, Transformer, MIL, MLP |
|
| Robustness and Generalizability | CNN, U-Net, Transformer |
|
| Interpretability and Explainability | CNN, MIL |
|
| Computational Efficiency | CNN, U-Net, YOLO |
|
| Annotation Quality and Requirements | CNN, U-Net |
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xu, Q.; Adam, A.; Abdullah, A.; Bariyah, N. Advanced Deep Learning Approaches in Detection Technologies for Comprehensive Breast Cancer Assessment Based on WSIs: A Systematic Literature Review. Diagnostics 2025, 15, 1150. https://doi.org/10.3390/diagnostics15091150
Xu Q, Adam A, Abdullah A, Bariyah N. Advanced Deep Learning Approaches in Detection Technologies for Comprehensive Breast Cancer Assessment Based on WSIs: A Systematic Literature Review. Diagnostics. 2025; 15(9):1150. https://doi.org/10.3390/diagnostics15091150
Chicago/Turabian StyleXu, Qiaoyi, Afzan Adam, Azizi Abdullah, and Nurkhairul Bariyah. 2025. "Advanced Deep Learning Approaches in Detection Technologies for Comprehensive Breast Cancer Assessment Based on WSIs: A Systematic Literature Review" Diagnostics 15, no. 9: 1150. https://doi.org/10.3390/diagnostics15091150
APA StyleXu, Q., Adam, A., Abdullah, A., & Bariyah, N. (2025). Advanced Deep Learning Approaches in Detection Technologies for Comprehensive Breast Cancer Assessment Based on WSIs: A Systematic Literature Review. Diagnostics, 15(9), 1150. https://doi.org/10.3390/diagnostics15091150






