Clinicopathologic Analysis of Five Patients with POLE-Mutated Colorectal Cancer in a Single Korean Institute
Abstract
1. Introduction
2. Materials and Methods
2.1. Patients, Inclusion, and Exclusion Criteria
2.2. Molecular Analysis
2.3. Data Presentation
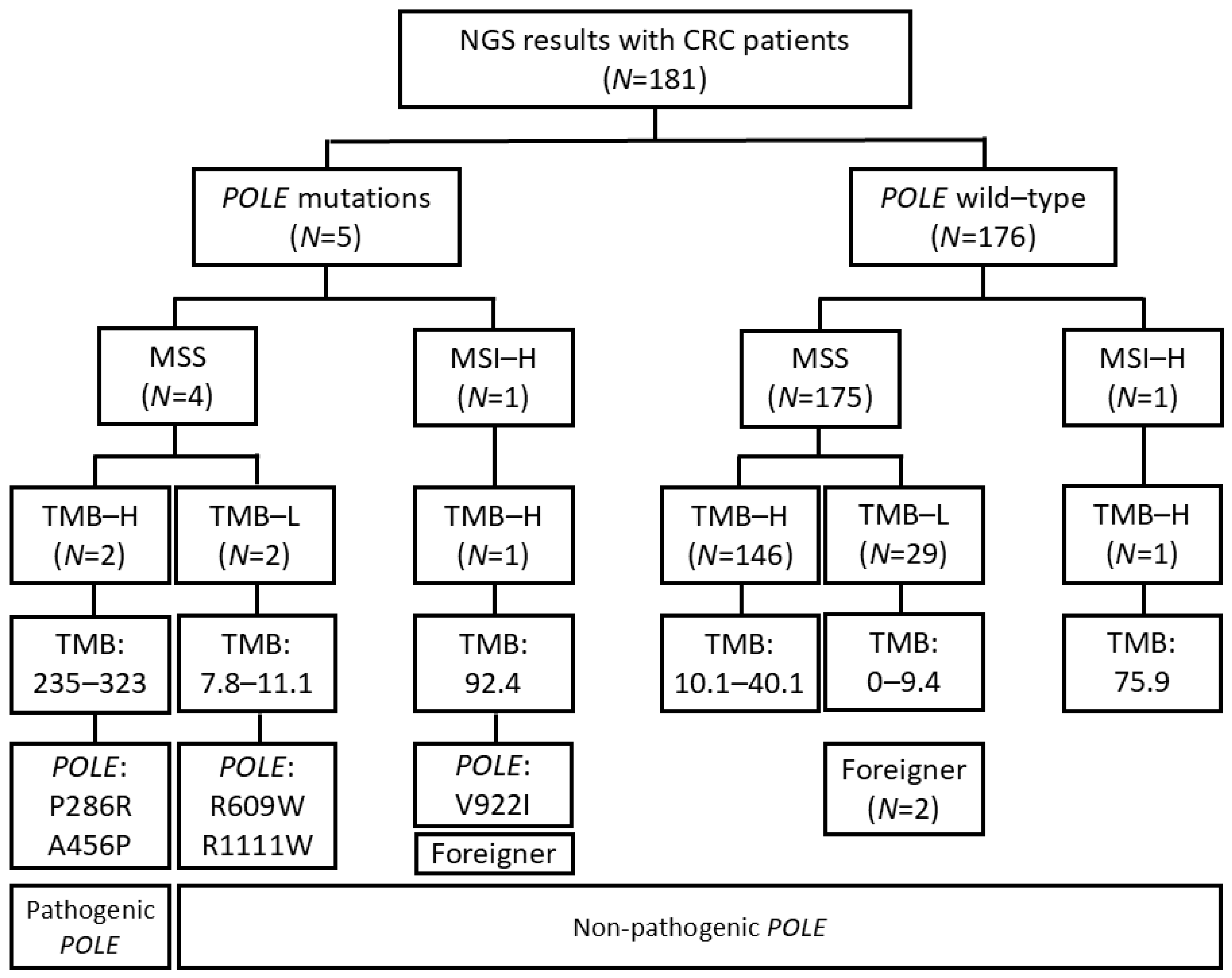
3. Results
3.1. Selection of Patients with Pathogenic POLE Mutation
3.2. Association Between Pathogenic POLE Mutation and Clinicopathological Characteristics
3.3. Clinicopathological Features of Five Patients with POLE Mutations
3.4. Histologic Features of Pathogenic POLE Mutation in CRC
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Loeb, L.A.; Monnat, R.J. DNA polymerases and human disease. Nat. Rev. Genet. 2008, 9, 594–604. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.; Dong, L.; Liu, X.; Ou, K.; Yang, L. POLE/POLD1 mutation and tumor immunotherapy. J. Exp. Clin. Cancer Res. 2022, 41, 216. [Google Scholar] [CrossRef] [PubMed]
- The Cancer Genome Atlas Research Network; Levine, D.A. Integrated genomic characterization of endometrial carcinoma. Nature 2013, 497, 67–73. [Google Scholar] [CrossRef] [PubMed]
- Xing, X.; Jin, N.; Wang, J. Polymerase Epsilon-Associated Ultramutagenesis in Cancer. Cancers 2022, 14, 1467. [Google Scholar] [CrossRef] [PubMed]
- Yarchoan, M.; Hopkins, A.; Jaffee, E.M. Tumor Mutational Burden and Response Rate to PD-1 Inhibition. N. Engl. J. Med. 2017, 377, 2500–2501. [Google Scholar] [CrossRef] [PubMed]
- Benson, A.B.; Venook, A.P.; Adam, M.; Chang, G.; Chen, Y.-J.; Ciombor, K.K.; Cohen, S.A.; Cooper, H.S.; Deming, D.; Garrido-Laguna, I.; et al. Colon Cancer, Version 3.2024, NCCN Clinical Practice Guidelines in Oncology. J. Natl. Compr. Canc. Netw. 2024, 22, e240029. [Google Scholar] [CrossRef] [PubMed]
- Garmezy, B.; Gheeya, J.; Lin, H.Y.; Huang, Y.; Kim, T.; Jiang, X.; Thein, K.Z.; Pilié, P.G.; Zeineddine, F.; Wang, W.; et al. Clinical and Molecular Characterization of POLE Mutations as Predictive Biomarkers of Response to Immune Checkpoint Inhibitors in Advanced Cancers. JCO Precis. Oncol. 2022, 6, e2100267. [Google Scholar] [CrossRef] [PubMed]
- Ambrosini, M.; Rousseau, B.; Manca, P.; Artz, O.; Marabelle, A.; André, T.; Maddalena, G.; Mazzoli, G.; Intini, R.; Cohen, R.; et al. Immune checkpoint inhibitors for POLE or POLD1 proofreading-deficient metastatic colorectal cancer. Ann. Oncol. 2024, 35, 643–655. [Google Scholar] [CrossRef] [PubMed]
- Domingo, E.; Freeman-Mills, L.; Rayner, E.; Glaire, M.; Briggs, S.; Vermeulen, L.; Fessler, E.; Medema, J.P.; Boot, A.; Morreau, H.; et al. Somatic POLE proofreading domain mutation, immune response, and prognosis in colorectal cancer: A retrospective, pooled biomarker study. Lancet Gastroenterol. Hepatol. 2016, 1, 207–216. [Google Scholar] [CrossRef] [PubMed]
- Hino, H.; Shiomi, A.; Kusuhara, M.; Kagawa, H.; Yamakawa, Y.; Hatakeyama, K.; Kawabata, T.; Oishi, T.; Urakami, K.; Nagashima, T.; et al. Clinicopathological and mutational analyses of colorectal cancer with mutations in the POLE gene. Cancer Med. 2019, 8, 4587–4597. [Google Scholar] [CrossRef] [PubMed]
- Amin, M.B.; Edge, S.B.; Greene, F.L.; Byrd, D.R.; Brookland, R.K.; Washington, M.K.; Gershenwald, J.E.; Compton, C.C.; Hess, K.R.; Sullivan, D.C.; et al. (Eds.) AJCC Cancer Staging Manual, 8th ed.; Springer: Cham, Switzerland, 2017; pp. XVII, 1032. [Google Scholar]
- Campbell, B.B.; Light, N.; Fabrizio, D.; Zatzman, M.; Fuligni, F.; de Borja, R.; Davidson, S.; Edwards, M.; Elvin, J.A.; Hodel, K.P.; et al. Comprehensive Analysis of Hypermutation in Human Cancer. Cell 2017, 171, 1042–1056.e1010. [Google Scholar] [CrossRef] [PubMed]
- Kawai, T.; Nyuya, A.; Mori, Y.; Tanaka, T.; Tanioka, H.; Yasui, K.; Toshima, T.; Taniguchi, F.; Shigeyasu, K.; Umeda, Y.; et al. Clinical and epigenetic features of colorectal cancer patients with somatic POLE proofreading mutations. Clin. Epigenetics 2021, 13, 117. [Google Scholar] [CrossRef] [PubMed]
- Ahn, S.-M.; Ansari, A.A.; Kim, J.; Kim, D.; Chun, S.-M.; Kim, J.; Kim, T.W.; Park, I.; Yu, C.-S.; Jang, S.J. The somatic POLE P286R mutation defines a unique subclass of colorectal cancer featuring hypermutation, representing a potential genomic biomarker for immunotherapy. Oncotarget 2016, 7, 68638–68649. [Google Scholar] [CrossRef] [PubMed]
- León-Castillo, A.; Britton, H.; McConechy, M.K.; McAlpine, J.N.; Nout, R.; Kommoss, S.; Brucker, S.Y.; Carlson, J.W.; Epstein, E.; Rau, T.T.; et al. Interpretation of somatic POLE mutations in endometrial carcinoma. J. Pathol. 2020, 250, 323–335. [Google Scholar] [CrossRef] [PubMed]
- Church, D.N.; Briggs, S.E.; Palles, C.; Domingo, E.; Kearsey, S.J.; Grimes, J.M.; Gorman, M.; Martin, L.; Howarth, K.M.; Hodgson, S.V.; et al. DNA polymerase ε and δ exonuclease domain mutations in endometrial cancer. Hum. Mol. Genet. 2013, 22, 2820–2828. [Google Scholar] [CrossRef] [PubMed]
- Church, D.N.; Stelloo, E.; Nout, R.A.; Valtcheva, N.; Depreeuw, J.; ter Haar, N.; Noske, A.; Amant, F.; Tomlinson, I.P.M.; Wild, P.J.; et al. Prognostic significance of POLE proofreading mutations in endometrial cancer. J. Natl. Cancer Inst. 2015, 107, 402. [Google Scholar] [CrossRef] [PubMed]
- McConechy, M.K.; Talhouk, A.; Leung, S.; Chiu, D.; Yang, W.; Senz, J.; Reha-Krantz, L.J.; Lee, C.-H.; Huntsman, D.G.; Gilks, C.B.; et al. Endometrial Carcinomas with POLE Exonuclease Domain Mutations Have a Favorable Prognosis. Clin. Cancer Res. 2016, 22, 2865–2873. [Google Scholar] [CrossRef] [PubMed]
- Crosbie, E.J.; Kitson, S.J.; McAlpine, J.N.; Mukhopadhyay, A.; Powell, M.E.; Singh, N. Endometrial cancer. Lancet 2022, 399, 1412–1428. [Google Scholar] [CrossRef] [PubMed]

| Characteristics | Pathologenic POLE Mutation (n = 2) (%) | Non-Pathogenic POLE Mutation (n = 179) (%) | |
|---|---|---|---|
| Age | year old (mean, SD *) | 49.0 (1.4) | 61.1 (9.6) |
| Sex | Male | 2 (100) | 111 (62.0) |
| Female | 0 (0) | 68 (38.0) | |
| Tumor | Right | 1 (50) | 35 (19.6) |
| location | Left | 0 (0) | 69 (38.5) |
| Rectum | 1 (50) | 75 (41.9) | |
| Histologic | Well | 0 (0) | 9 (5.0) |
| type | Moderately | 1 (50) | 145 (81.0) |
| Poorly | 1 (50) | 14 (7.8) | |
| Others | 0 (0) | 11 (6.1) | |
| 8th AJCC * | I | 0 (0) | 7 (3.9) |
| stage | II | 1 (50) | 18 (10.1) |
| III | 0 (0) | 53 (29.6) | |
| IV | 1 (50) | 101 (56.4) | |
| RAS/RAF mutational status | KRAS mutant | 0 (0) | 101 (56.4) |
| KRAS wild type | 2 (100) | 78 (43.6) | |
| NRAS mutant | 0 (0) | 4 (2.2) | |
| NRAS wild type | 2 (100) | 175 (97.8) | |
| BRAF mutant | 0 (0) | 6 (3.4) | |
| BRAF wild type | 2 (100) | 173 (96.6) | |
| Microsatellite status | MSS * | 2 (100) | 177 (98.9) |
| MSI-H * | 0 (0) | 2 (1.1) | |
| TMB * | mutation/MB * (mean, SD *) | 279.2 (62.0) | 8.1 (9.0) |
| No. | Sex | Age | Tumor Location | Hist * | AJCC Stage * | POLE | POLE Significance | KRAS | NRAS | BRAF | MSI * | TMB * (mut/Mb *) | Tx * | OS * | Alive/ Death |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | M | 48 | Rectum | Poorly * | 4 | A456P | Path * | Wild | Wild | Wild | MSS * | 235.3 | Sx *, Ctx *, ICI * | 16 mo * | Death |
| 2 | M | 50 | Ascending | Mod * | 2 | P286R | Path | Wild | Wild | Wild | MSS | 323 | Sx, Ctx | 44 mo | NED * |
| 3 | F | 62 | Sigmoid | Mod | 4 | R1111W | Non-path | Mut * | Wild | Wild | MSS | 7.8 | Ctx | 22 mo | Alive (mets) * |
| 4 | M | 62 | Sigmoid | Mod | 4 | R609W | Non-path | Mut | Wild | Wild | MSS | 11.1 | Ctx | 9 mo | Alive (mets) |
| 5 | F | 52 | Ascending | Medullary | 2 | V922I | Non-path | Mut | Wild | Wild | MSI-H * | 92.4 | Sx, Ctx | 34 mo | NED |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Oh, H.; Jang, I.; Hwang, J.; Lee, S.; An, J.; Sim, J. Clinicopathologic Analysis of Five Patients with POLE-Mutated Colorectal Cancer in a Single Korean Institute. Diagnostics 2025, 15, 972. https://doi.org/10.3390/diagnostics15080972
Oh H, Jang I, Hwang J, Lee S, An J, Sim J. Clinicopathologic Analysis of Five Patients with POLE-Mutated Colorectal Cancer in a Single Korean Institute. Diagnostics. 2025; 15(8):972. https://doi.org/10.3390/diagnostics15080972
Chicago/Turabian StyleOh, Harim, Inho Jang, Jinha Hwang, Soohyeon Lee, Jungsuk An, and Jongmin Sim. 2025. "Clinicopathologic Analysis of Five Patients with POLE-Mutated Colorectal Cancer in a Single Korean Institute" Diagnostics 15, no. 8: 972. https://doi.org/10.3390/diagnostics15080972
APA StyleOh, H., Jang, I., Hwang, J., Lee, S., An, J., & Sim, J. (2025). Clinicopathologic Analysis of Five Patients with POLE-Mutated Colorectal Cancer in a Single Korean Institute. Diagnostics, 15(8), 972. https://doi.org/10.3390/diagnostics15080972







