Information Extraction from Lumbar Spine MRI Radiology Reports Using GPT4: Accuracy and Benchmarking Against Research-Grade Comprehensive Scoring
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Population and Imaging
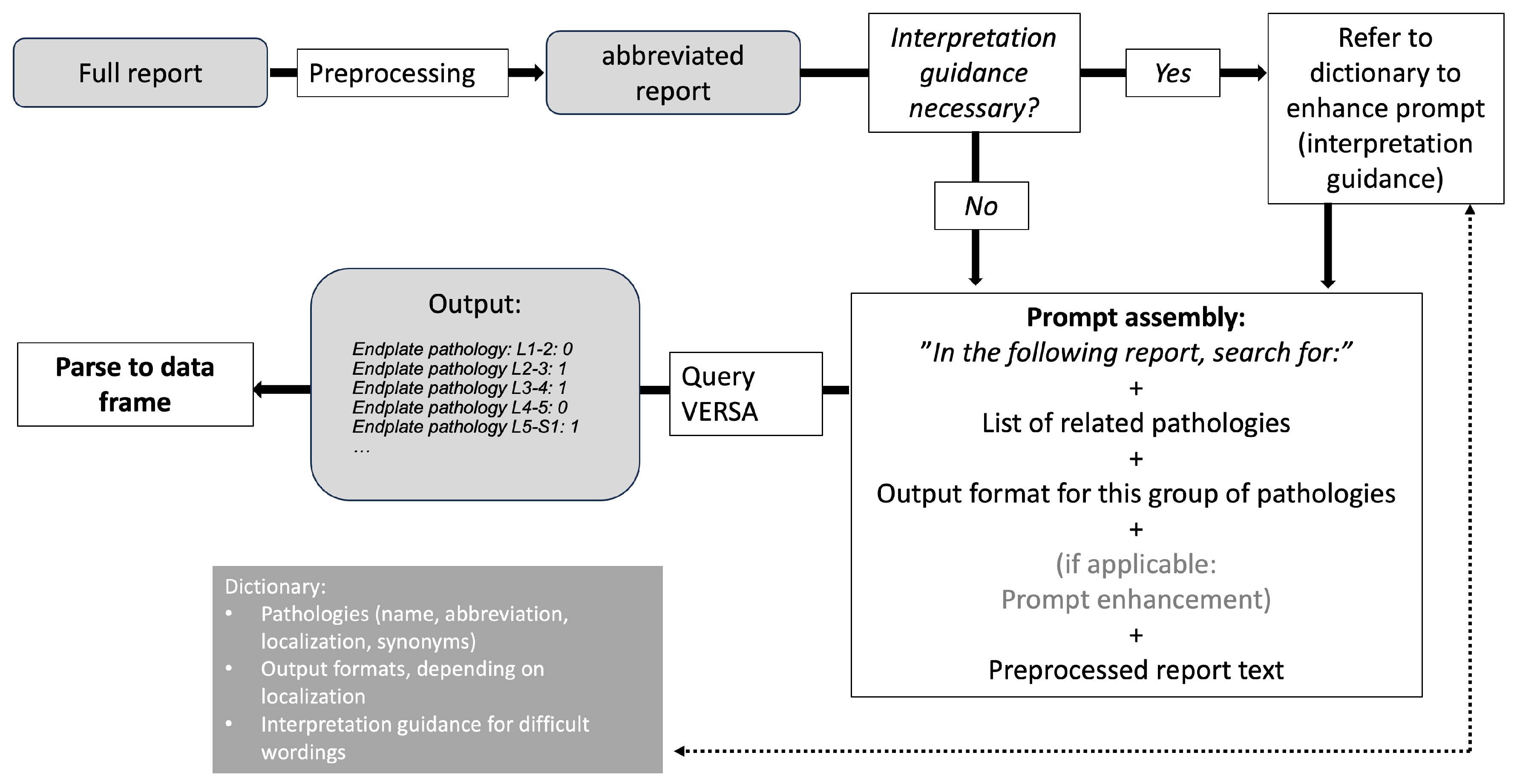
2.2. Prompt Engineering and LLM Pipelines
2.3. Comprehensive Image Scoring and Manual Report Data Extraction
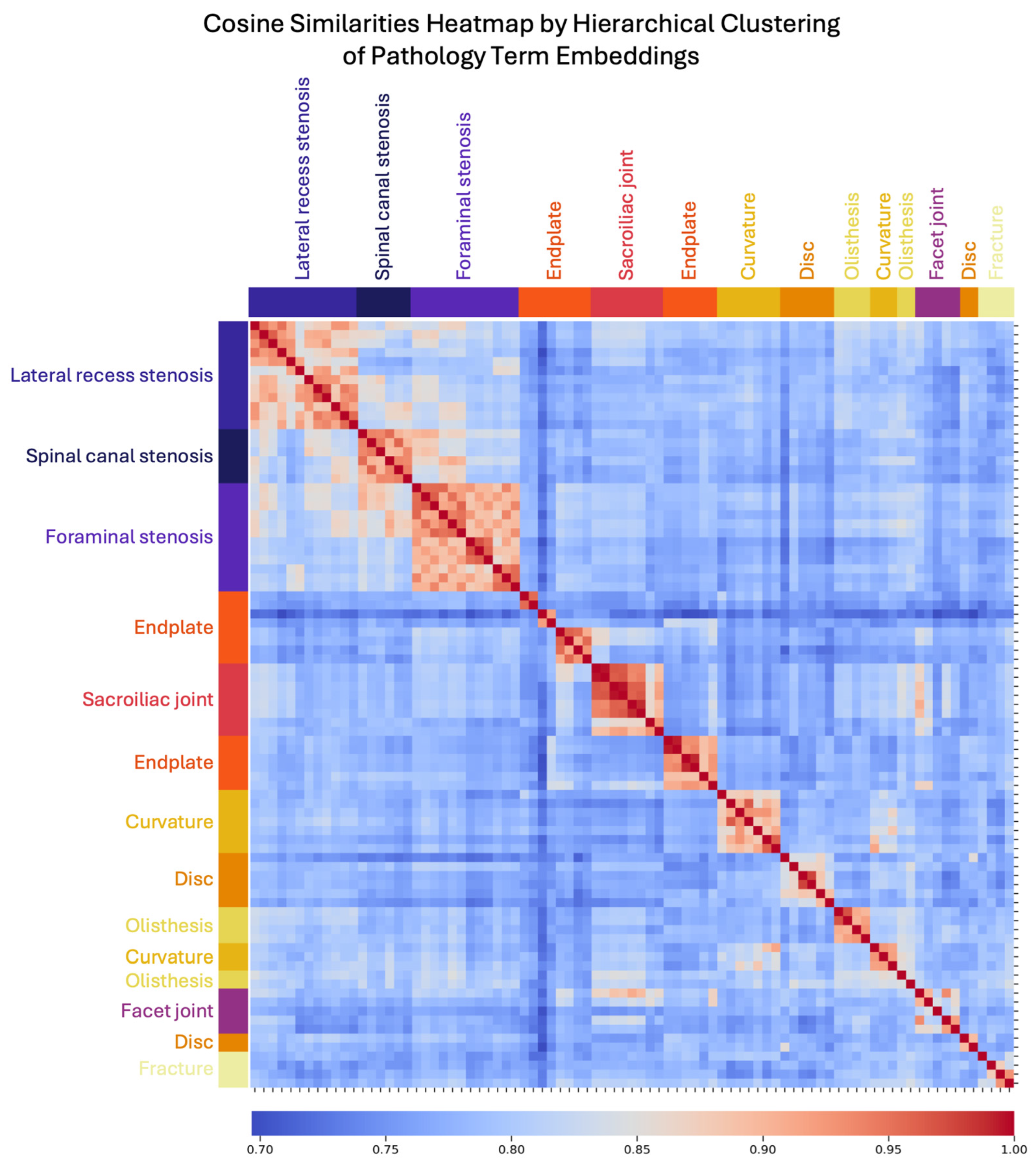
2.4. Semantic Information Representation
2.5. Statistical Analysis
3. Results
3.1. LLM Performance
3.2. LLM Embedding Representation of Semantic Information
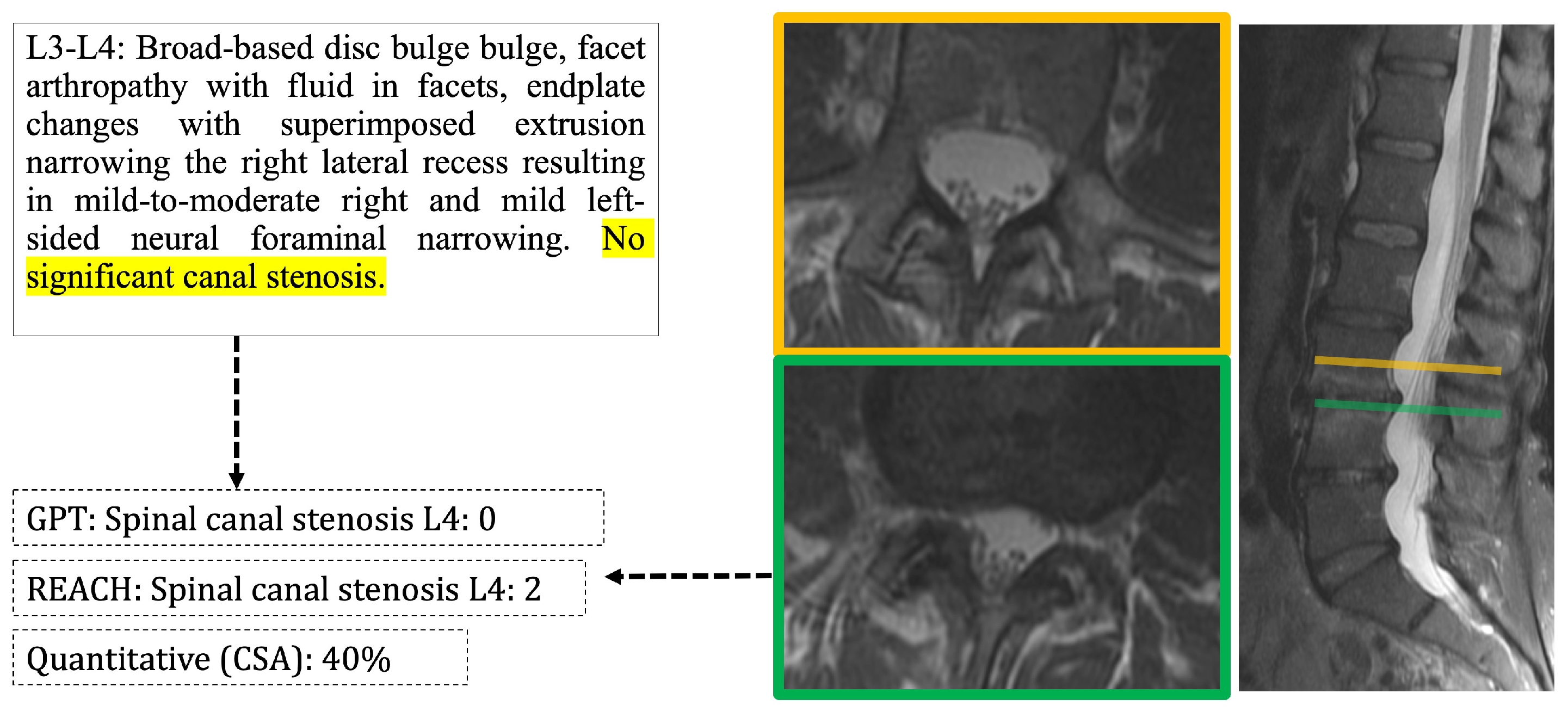
3.3. Agreement of Report-Derived Data with Comprehensive Image Scoring
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| API | Application programming interface |
| cLBP | Chronic low back pain |
| FNR | False-negative rate |
| FPR | False-positive rate |
| LLM | Large language model |
| PHI | Protected health information |
| UMAP | Unsupervised uniform manifold approximation projections |
References
- Hoy, D.; March, L.; Brooks, P.; Woolf, A.; Blyth, F.; Vos, T.; Buchbinder, R. Measuring the Global Burden of Low Back Pain. Best Pract. Res. Clin. Rheumatol. 2010, 24, 155–165. [Google Scholar] [CrossRef] [PubMed]
- Chang, D.; Lui, A.; Matsoyan, A.; Safaee, M.M.; Aryan, H.; Ames, C. Comparative Review of the Socioeconomic Burden of Lower Back Pain in the United States and Globally. Neurospine 2024, 21, 487–501. [Google Scholar] [CrossRef] [PubMed]
- Balagué, F.; Mannion, A.F.; Pellisé, F.; Cedraschi, C. Non-Specific Low Back Pain. Lancet 2012, 379, 482–491. [Google Scholar] [CrossRef]
- Reichenpfader, D.; Müller, H.; Denecke, K. A Scoping Review of Large Language Model Based Approaches for Information Extraction from Radiology Reports. NPJ Digit. Med. 2024, 7, 222. [Google Scholar] [CrossRef]
- Dagdelen, J.; Dunn, A.; Lee, S.; Walker, N.; Rosen, A.S.; Ceder, G.; Persson, K.A.; Jain, A. Structured Information Extraction from Scientific Text with Large Language Models. Nat. Commun. 2024, 15, 1418. [Google Scholar] [CrossRef]
- Ge, J.; Li, M.; Delk, M.B.; Lai, J.C. A Comparison of a Large Language Model vs Manual Chart Review for the Extraction of Data Elements From the Electronic Health Record. Gastroenterology 2024, 166, 707–709.e3. [Google Scholar] [CrossRef]
- Zhang, X.; Zhang, Y.; Zhang, Q.; Ren, Y.; Qiu, T.; Ma, J.; Sun, Q. Extracting Comprehensive Clinical Information for Breast Cancer Using Deep Learning Methods. Int. J. Med. Inf. 2019, 132, 103985. [Google Scholar] [CrossRef]
- Fink, M.A.; Bischoff, A.; Fink, C.A.; Moll, M.; Kroschke, J.; Dulz, L.; Heußel, C.P.; Kauczor, H.-U.; Weber, T.F. Potential of ChatGPT and GPT-4 for Data Mining of Free-Text CT Reports on Lung Cancer. Radiology 2023, 308, e231362. [Google Scholar] [CrossRef]
- Hue, T.F.; Lotz, J.C.; Zheng, P.; Black, D.M.; Bailey, J.; Ewing, S.K.; Fields, A.J.; Mehling, W.; Scheffler, A.; Strigo, I.; et al. Design of the COMEBACK and BACKHOME Studies, Longitudinal Cohorts for Comprehensive Deep Phenotyping of Adults with Chronic Low-Back Pain (cLBP): A Part of the BACPAC Research Program. MedRxiv Prepr. Serv. Health Sci. 2024, 2024.04.09.24305574. [Google Scholar] [CrossRef]
- Mauck, M.C.; Lotz, J.; Psioda, M.A.; Carey, T.S.; Clauw, D.J.; Majumdar, S.; Marras, W.S.; Vo, N.; Aylward, A.; Hoffmeyer, A.; et al. The Back Pain Consortium (BACPAC) Research Program: Structure, Research Priorities, and Methods. Pain Med. Malden Mass 2023, 24, S3–S12. [Google Scholar] [CrossRef]
- Batorsky, A.; Bowden, A.E.; Darwin, J.; Fields, A.J.; Greco, C.M.; Harris, R.E.; Hue, T.F.; Kakyomya, J.; Mehling, W.; O’Neill, C.; et al. The Back Pain Consortium (BACPAC) Research Program Data Harmonization: Rationale for Data Elements and Standards. Pain Med. 2023, 24, S95–S104. [Google Scholar] [CrossRef]
- Sollmann, N.; Fields, A.J.; O’Neill, C.; Nardo, L.; Majumdar, S.; Chin, C.T.; Tosun, D.; Han, M.; Vu, A.T.; Ozhinsky, E.; et al. Magnetic Resonance Imaging of the Lumbar Spine: Recommendations for Acquisition and Image Evaluation from the BACPAC Spine Imaging Working Group. Pain Med. Malden Mass 2023, 24, S81–S94. [Google Scholar] [CrossRef]
- Beutel, G.; Geerits, E.; Kielstein, J.T. Artificial Hallucination: GPT on LSD? Crit. Care 2023, 27, 148. [Google Scholar] [CrossRef] [PubMed]
- McInnes, L.; Healy, J.; Saul, N.; Großberger, L. UMAP: Uniform Manifold Approximation and Projection. J. Open Source Softw. 2018, 3, 861. [Google Scholar] [CrossRef]
- McInnes, L.; Healy, J.; Melville, J. UMAP: Uniform Manifold Approximation and Projection for Dimension Reduction. arXiv 2020, arXiv:1802.03426. [Google Scholar]
- Mabotuwana, T.; Lee, M.C.; Cohen-Solal, E.V. An Ontology-Based Similarity Measure for Biomedical Data—Application to Radiology Reports. J. Biomed. Inform. 2013, 46, 857–868. [Google Scholar] [CrossRef]
- Cohen, J. Weighted Kappa: Nominal Scale Agreement with Provision for Scaled Disagreement or Partial Credit. Psychol. Bull. 1968, 70, 213–220. [Google Scholar] [CrossRef]
- Landis, J.R.; Koch, G.G. The Measurement of Observer Agreement for Categorical Data. Biometrics 1977, 33, 159–174. [Google Scholar] [CrossRef]
- Datta, S.; Roberts, K. Weakly Supervised Spatial Relation Extraction from Radiology Reports. JAMIA Open 2023, 6, ooad027. [Google Scholar] [CrossRef]
- Santos, T.; Kallas, O.N.; Newsome, J.; Rubin, D.; Gichoya, J.W.; Banerjee, I. A Fusion NLP Model for the Inference of Standardized Thyroid Nodule Malignancy Scores from Radiology Report Text. AMIA Annu. Symp. Proc. AMIA Symp. 2021, 2021, 1079–1088. [Google Scholar]
- Tan, W.K.; Hassanpour, S.; Heagerty, P.J.; Rundell, S.D.; Suri, P.; Huhdanpaa, H.T.; James, K.; Carrell, D.S.; Langlotz, C.P.; Organ, N.L.; et al. Comparison of Natural Language Processing Rules-Based and Machine-Learning Systems to Identify Lumbar Spine Imaging Findings Related to Low Back Pain. Acad. Radiol. 2018, 25, 1422–1432. [Google Scholar] [CrossRef] [PubMed]
- Banitalebi, H.; Espeland, A.; Anvar, M.; Hermansen, E.; Hellum, C.; Brox, J.I.; Myklebust, T.Å.; Indrekvam, K.; Brisby, H.; Weber, C.; et al. Reliability of Preoperative MRI Findings in Patients with Lumbar Spinal Stenosis. BMC Musculoskelet. Disord. 2022, 23, 51. [Google Scholar] [CrossRef] [PubMed]
- Pohlner, T.; Deppe, D.; Ziegeler, K.; Proft, F.; Protopopov, M.; Rademacher, J.; Rios Rodriguez, V.; Torgutalp, M.; Braun, J.; Diekhoff, T.; et al. Diagnostic Accuracy in Axial Spondyloarthritis: A Systematic Evaluation of the Role of Clinical Information in the Interpretation of Sacroiliac Joint Imaging. RMD Open 2024, 10, e004044. [Google Scholar] [CrossRef] [PubMed]
- Larson, D.B.; Towbin, A.J.; Pryor, R.M.; Donnelly, L.F. Improving Consistency in Radiology Reporting through the Use of Department-Wide Standardized Structured Reporting. Radiology 2013, 267, 240–250. [Google Scholar] [CrossRef]
- Ashman, C.J.; Yu, J.S.; Wolfman, D. Satisfaction of Search in Osteoradiology. Am. J. Roentgenol. 2000, 175, 541–544. [Google Scholar] [CrossRef]
- Taylor-Phillips, S.; Stinton, C. Fatigue in Radiology: A Fertile Area for Future Research. Br. J. Radiol. 2019, 92, 20190043. [Google Scholar] [CrossRef]
- Pons, E.; Braun, L.M.M.; Hunink, M.G.M.; Kors, J.A. Natural Language Processing in Radiology: A Systematic Review. Radiology 2016, 279, 329–343. [Google Scholar] [CrossRef]
- Adams, L.C.; Truhn, D.; Busch, F.; Kader, A.; Niehues, S.M.; Makowski, M.R.; Bressem, K.K. Leveraging GPT-4 for Post Hoc Transformation of Free-Text Radiology Reports into Structured Reporting: A Multilingual Feasibility Study. Radiology 2023, 307, e230725. [Google Scholar] [CrossRef]
- Doo, F.X.; Savani, D.; Kanhere, A.; Carlos, R.C.; Joshi, A.; Yi, P.H.; Parekh, V.S. Optimal Large Language Model Characteristics to Balance Accuracy and Energy Use for Sustainable Medical Applications. Radiology 2024, 312, e240320. [Google Scholar] [CrossRef]
- Mukherjee, P.; Hou, B.; Lanfredi, R.B.; Summers, R.M. Feasibility of Using the Privacy-Preserving Large Language Model Vicuna for Labeling Radiology Reports. Radiology 2023, 309, e231147. [Google Scholar] [CrossRef]

| Pathology Class | Location | Scale | Included Pathologies |
|---|---|---|---|
| Endplate pathology | level | absent/present | Endplate erosions, Schmorl’s nodes, Modic changes |
| Disc pathology | level | absent/present | Bulging, protrusion, extrusion |
| Facet joint arthropathy (FJ-OA) | level | absent/present | Hypertrophic changes, synovial cysts, increased facet joint fluid |
| Lateral recess stenosis (LRS) | level and side | absent/present | Nerve root contact, compression |
| Spinal canal stenosis (SCS) | level | absent, mild, moderate, severe | Varying grades of stenosis, regardless of cause |
| Foraminal stenosis | level and side | absent, mild, moderate, severe | Varying grades of stenosis, regardless of cause |
| Sacroiliac joint (SIJ) pathology | overall | absent/present | Any observed joint changes, including sclerosis, bone marrow signal changes (edema/fat) |
| Scoliosis | overall | absent/present | Changes in curvature of lumbar spine |
| Pathology | F1 | F1: 95% CI | FPR | FPR: 95% CI | FNR | FNR: 95% CI | Acc. | Acc: 95% CI | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| LL | UL | LL | UL | LL | UL | LL | UL | |||||
| Endplate | 0.949 | 0.939 | 0.958 | 0.039 | 0.033 | 0.047 | 0.136 | 0.089 | 0.172 | 0.948 | 0.933 | 0.959 |
| Disc | 0.938 | 0.930 | 0.948 | 0.168 | 0.111 | 0.243 | 0.015 | 0.009 | 0.023 | 0.951 | 0.937 | 0.962 |
| FJ-OA | 0.957 | 0.938 | 0.974 | 0.006 | 0.006 | 0.006 | 0.035 | 0.035 | 0.035 | 0.981 | 0.971 | 0.987 |
| LRS | 0.970 | 0.957 | 0.981 | 0.004 | 0.004 | 0.004 | 0.000 | 0.000 | 0.000 | 0.929 | 0.917 | 0.939 |
| SCS | 0.921 | 0.898 | 0.950 | 0.109 | 0.064 | 0.155 | 0.018 | 0.002 | 0.042 | 0.923 | 0.906 | 0.937 |
| FS | 0.970 | 0.966 | 0.974 | 0.053 | 0.038 | 0.068 | 0.008 | 0.003 | 0.013 | 0.974 | 0.967 | 0.980 |
| SIJ path. | 0.987 | 0.987 | 0.987 | 0.018 | 0.018 | 0.018 | 0.000 | 0.000 | 0.000 | 0.991 | 0.969 | 0.998 |
| Scoliosis | 0.987 | 0.987 | 0.987 | 0.007 | 0.007 | 0.007 | 0.025 | 0.025 | 0.025 | 0.991 | 0.969 | 0.998 |
| Mean | 0.960 | 0.051 | 0.030 | 0.961 | ||||||||
| Pathology | Kappa | F1: 95% CI | FPR | FPR: 95% CI | FNR | FNR: 95% CI | |||
|---|---|---|---|---|---|---|---|---|---|
| Lower | Upper | Lower | Upper | Lower | Upper | ||||
| Endplate | 0.454 | 0.406 | 0.504 | 0.043 | 0.026 | 0.065 | 0.557 | 0.485 | 0.620 |
| Disc | 0.495 | 0.410 | 0.587 | 0.427 | 0.289 | 0.557 | 0.082 | 0.064 | 0.109 |
| Facet arthropathy | 0.341 | 0.248 | 0.439 | 0.135 | 0.135 | 0.135 | 0.441 | 0.441 | 0.441 |
| Lateral recess stenosis | 0.286 | 0.246 | 0.329 | 0.014 | 0.014 | 0.014 | 0.636 | 0.636 | 0.636 |
| Spinal canal stenosis * | 0.652 | 0.483 | 0.774 | 0.129 | 0.056 | 0.213 | 0.267 | 0.157 | 0.405 |
| Foraminal stenosis * | 0.653 | 0.616 | 0.693 | 0.241 | 0.155 | 0.328 | 0.197 | 0.144 | 0.258 |
| SIJ pathology | 0.234 | 0.234 | 0.234 | 0.215 | 0.215 | 0.215 | 0.556 | 0.556 | 0.556 |
| Scoliosis | 0.278 | 0.278 | 0.278 | 0.284 | 0.284 | 0.284 | 0.192 | 0.192 | 0.192 |
| Mean | 0.424 | 0.186 | 0.366 | ||||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ziegeler, K.; Kreutzinger, V.; Tong, M.W.; Chin, C.T.; Bahroos, E.; Wu, P.-H.; Bonnheim, N.; Fields, A.J.; Lotz, J.C.; Link, T.M.; et al. Information Extraction from Lumbar Spine MRI Radiology Reports Using GPT4: Accuracy and Benchmarking Against Research-Grade Comprehensive Scoring. Diagnostics 2025, 15, 930. https://doi.org/10.3390/diagnostics15070930
Ziegeler K, Kreutzinger V, Tong MW, Chin CT, Bahroos E, Wu P-H, Bonnheim N, Fields AJ, Lotz JC, Link TM, et al. Information Extraction from Lumbar Spine MRI Radiology Reports Using GPT4: Accuracy and Benchmarking Against Research-Grade Comprehensive Scoring. Diagnostics. 2025; 15(7):930. https://doi.org/10.3390/diagnostics15070930
Chicago/Turabian StyleZiegeler, Katharina, Virginie Kreutzinger, Michelle W. Tong, Cynthia T. Chin, Emma Bahroos, Po-Hung Wu, Noah Bonnheim, Aaron J. Fields, Jeffrey C. Lotz, Thomas M. Link, and et al. 2025. "Information Extraction from Lumbar Spine MRI Radiology Reports Using GPT4: Accuracy and Benchmarking Against Research-Grade Comprehensive Scoring" Diagnostics 15, no. 7: 930. https://doi.org/10.3390/diagnostics15070930
APA StyleZiegeler, K., Kreutzinger, V., Tong, M. W., Chin, C. T., Bahroos, E., Wu, P.-H., Bonnheim, N., Fields, A. J., Lotz, J. C., Link, T. M., & Majumdar, S. (2025). Information Extraction from Lumbar Spine MRI Radiology Reports Using GPT4: Accuracy and Benchmarking Against Research-Grade Comprehensive Scoring. Diagnostics, 15(7), 930. https://doi.org/10.3390/diagnostics15070930






