A New Pes Planus Automatic Diagnosis Method: ViT-OELM Hybrid Modeling
Abstract
1. Introduction
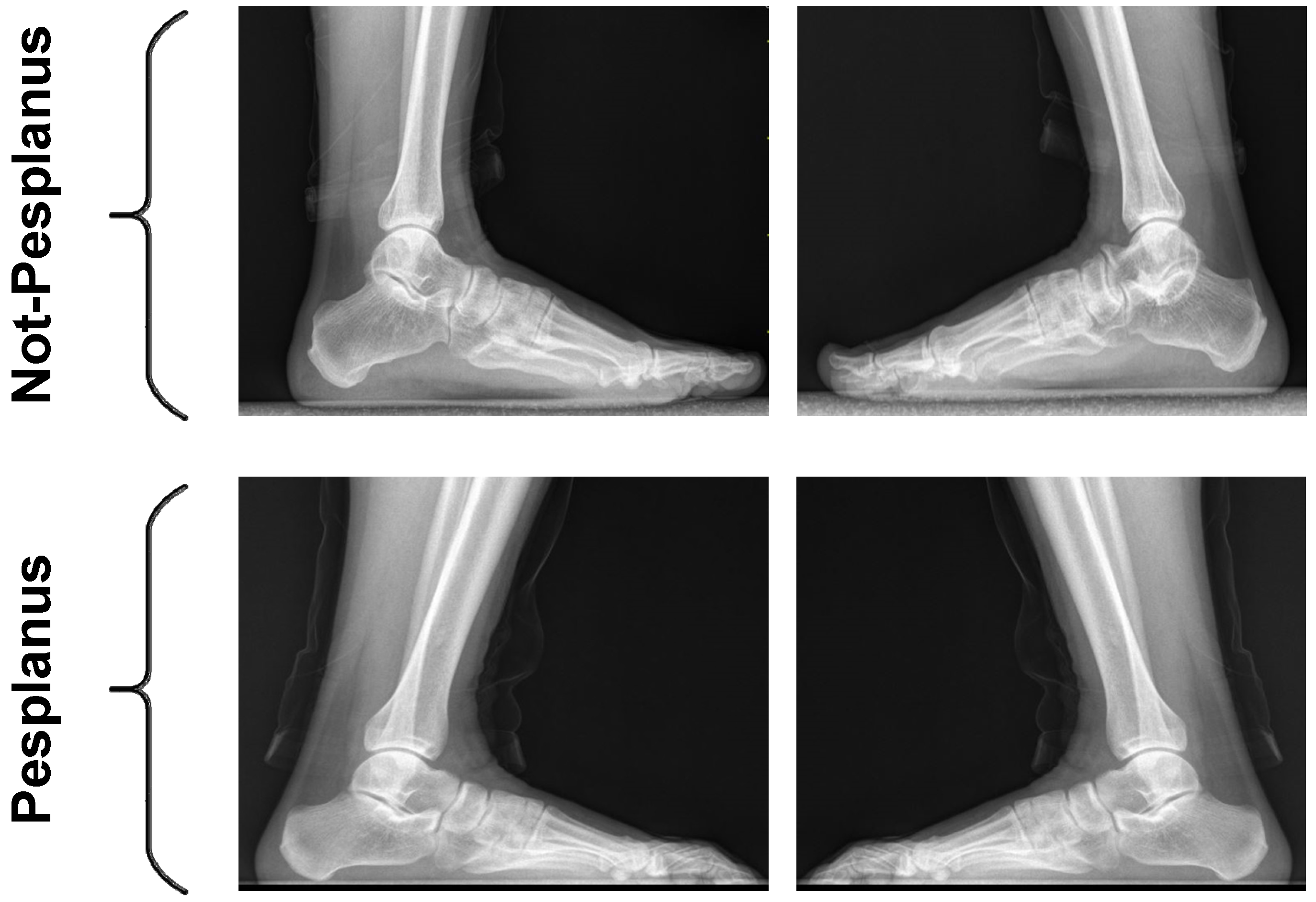
- X-ray Images and Dataset: It is stated in the literature that there are only two studies that share a dataset composed of X-ray pes planus images for automatic flatfoot diagnosis. This indicates the importance of composing X-ray pes planus image datasets for flatfoot diagnosis in this field.
- ViT-OELM Model: In this study, the Optimum Extreme Learning Machine (OELM) head layer was added to the basic ViT architecture model instead of the Multi-Layer Perceptron (MLP)’s head layer being used. The suggested ViT-OELM model comprises 12 encoder blocks. This developed ViT-OELM architecture, with an input size of (224, 224), allows for input images of 224 × 224 pixels. The proposed ViT-OELM model reduces computational load and increases accuracy by using a more comprehensive feature selection method. As a result, a more successful model for flatfoot diagnosis has been developed. The performance results of this proposed ViT-OELM model are superior to other deep learning structures.
- Feature Extraction and Selection: In feature extraction, experiments conducted on raw data show that accuracy increases, and high accuracy rates are achieved. This demonstrates that effective feature extraction increases the high accuracy ratio and contributes to quickly and correctly creating the architecture.
- Classification: By using OELM as classifier in this proposed ViT-OELM hybrid modeling, some classification advantages are obtained. These are the following:
- -
- Faster Training: OELM is faster than fully connected layers and traditional neural networks because it optimizes only the output weights.
- -
- Higher Accuracy: Provides better classification success with optimum weight selection and kernel functions.
- -
- Reduces Over-Learning: The generalization ability of the model is increased with regularization methods.
- Optimization: By using the Adam algorithm as an optimizer in this proposed ViT-OELM hybrid model, some optimization advantages are obtained. These are the following:
- -
- Parameter updates are more balanced thanks to momentum and adaptive learning rate.
- -
- Weights and bias values are optimized to prevent over-learning.
- -
- Faster convergence rates are provided, which is especially useful when working with large datasets.
- Main Contributions: The new ViT-OELM hybrid model using X-ray pes planus images presented in this study can be a flexible instrument for the early diagnosis of flatfoot and can obtain high accuracy success. The ViT-OELM hybrid model can be used in clinical applications thanks to its high classification accuracy.
2. Materials and Methods
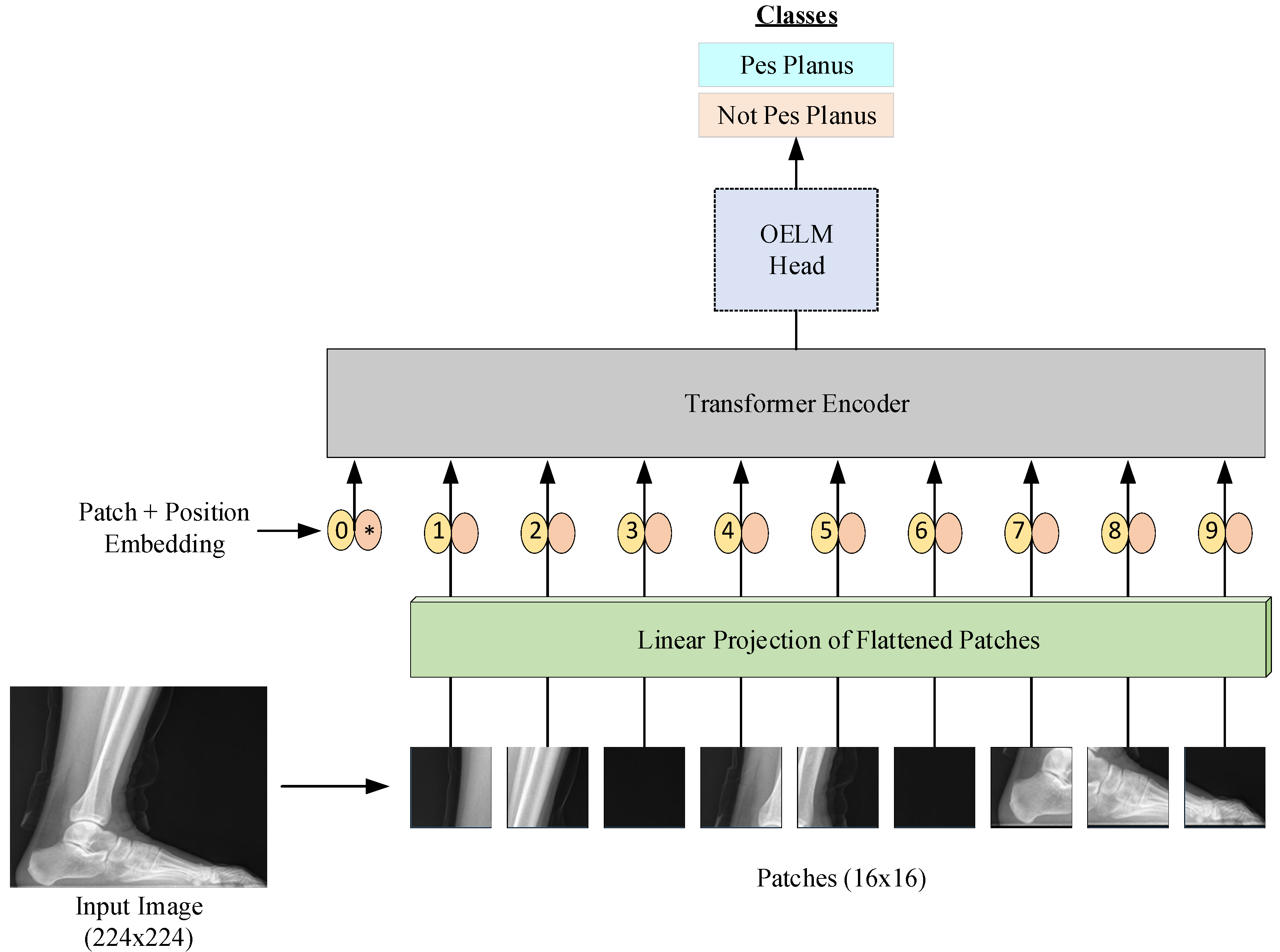
- -
- Input Image (224 × 224 resolution): The process starts with a foot X-ray.
- -
- Patch Extraction (16 × 16 patches): The image is divided into smaller patches of 16 × 16 pixels.
- -
- Linear Projection of Flattened Patches: Each patch is flattened into a vector and projected into a higher dimensional space.
- -
- Patch + Position Embedding Process: Position embeddings are added to the patch embeddings to preserve the positional information.
- -
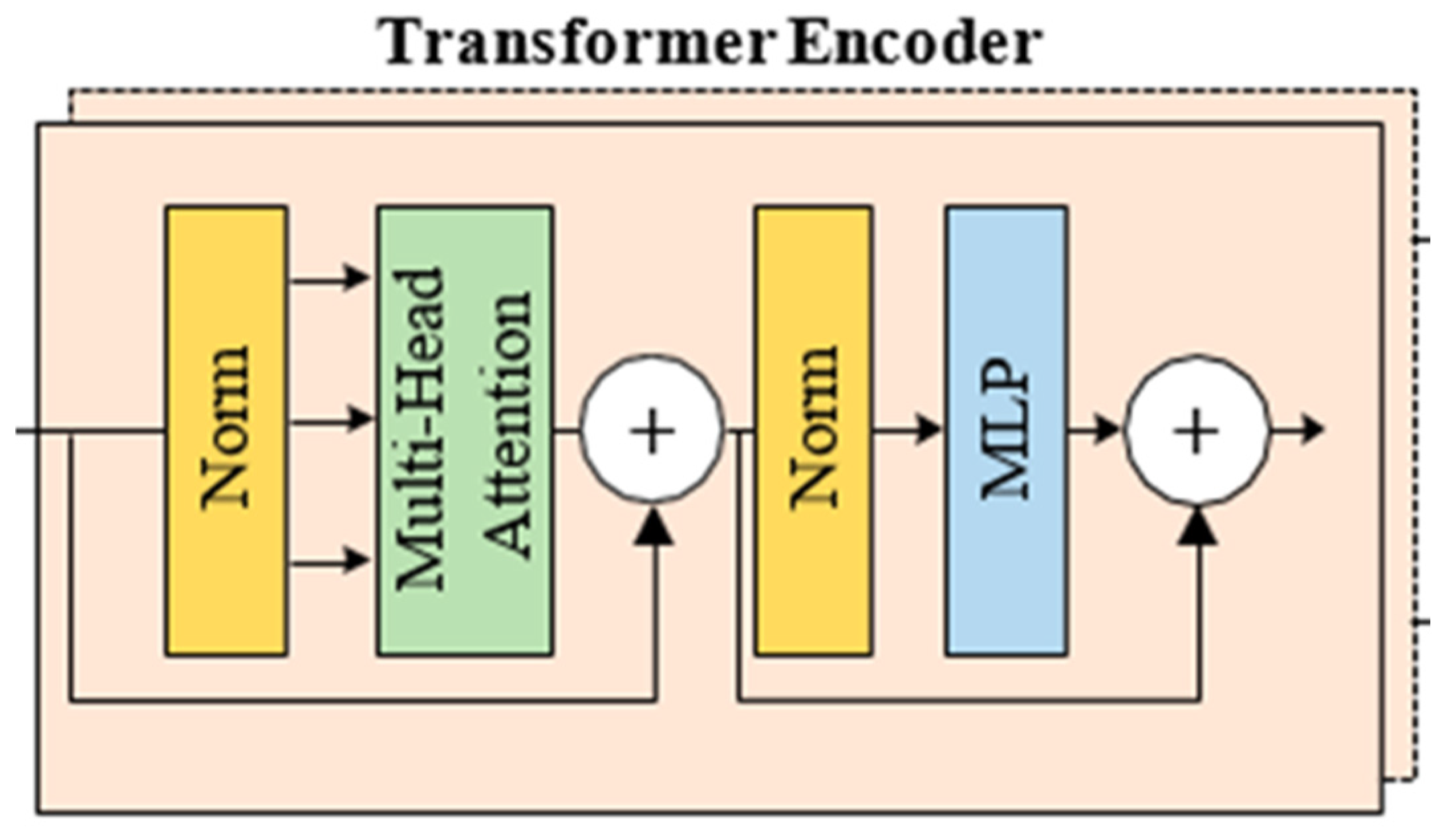
- Transformer Encoder: The embedded patch array is processed by a Transformer encoder.
- -
- OELM Header: The encoded feature representation is transferred to the OELM (Optimized Extreme Learning Machine) header and the classification process is performed.
- -
- Classification Output: The final classification output determines whether the X-ray image is “Pes Planus” (flat foot) or “Not Pes Planus” (not flat foot).
2.1. Used Pes Planus Image Dataset
2.2. Image Classification Method Based on Vision Transformers (ViTs)
- Creating Patching: The image was first divided into small pieces (patches). Each patch has a fixed size of 16 × 16 pixels. These patches were used as the input for the VİT-OELM hybrid model.
- Linear Patch Embeddings: Each patch was transformed into a vector. Here, a linear projection (linear transformation) was used to create a feature vector representing each pixel’s information in the patch.
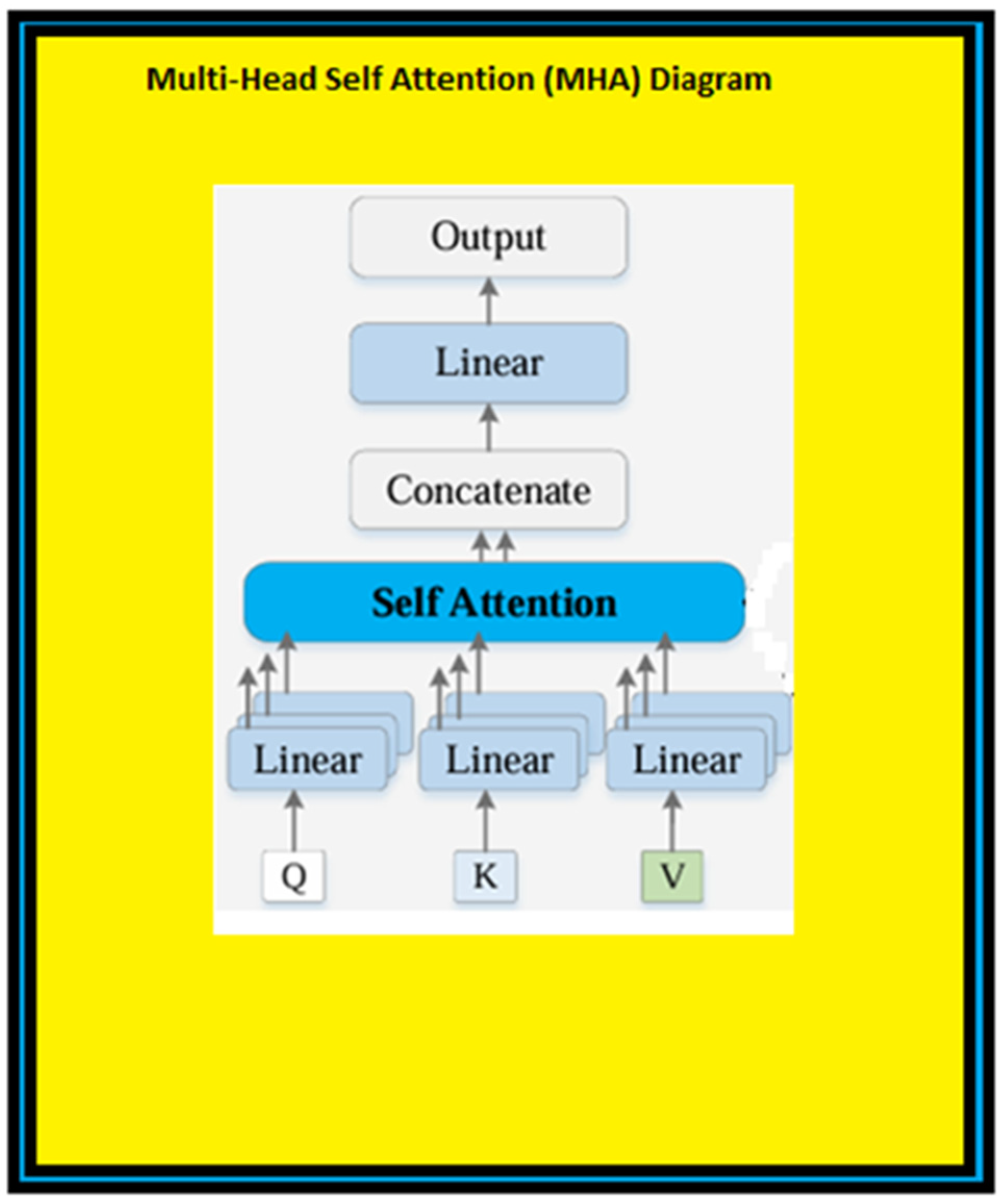
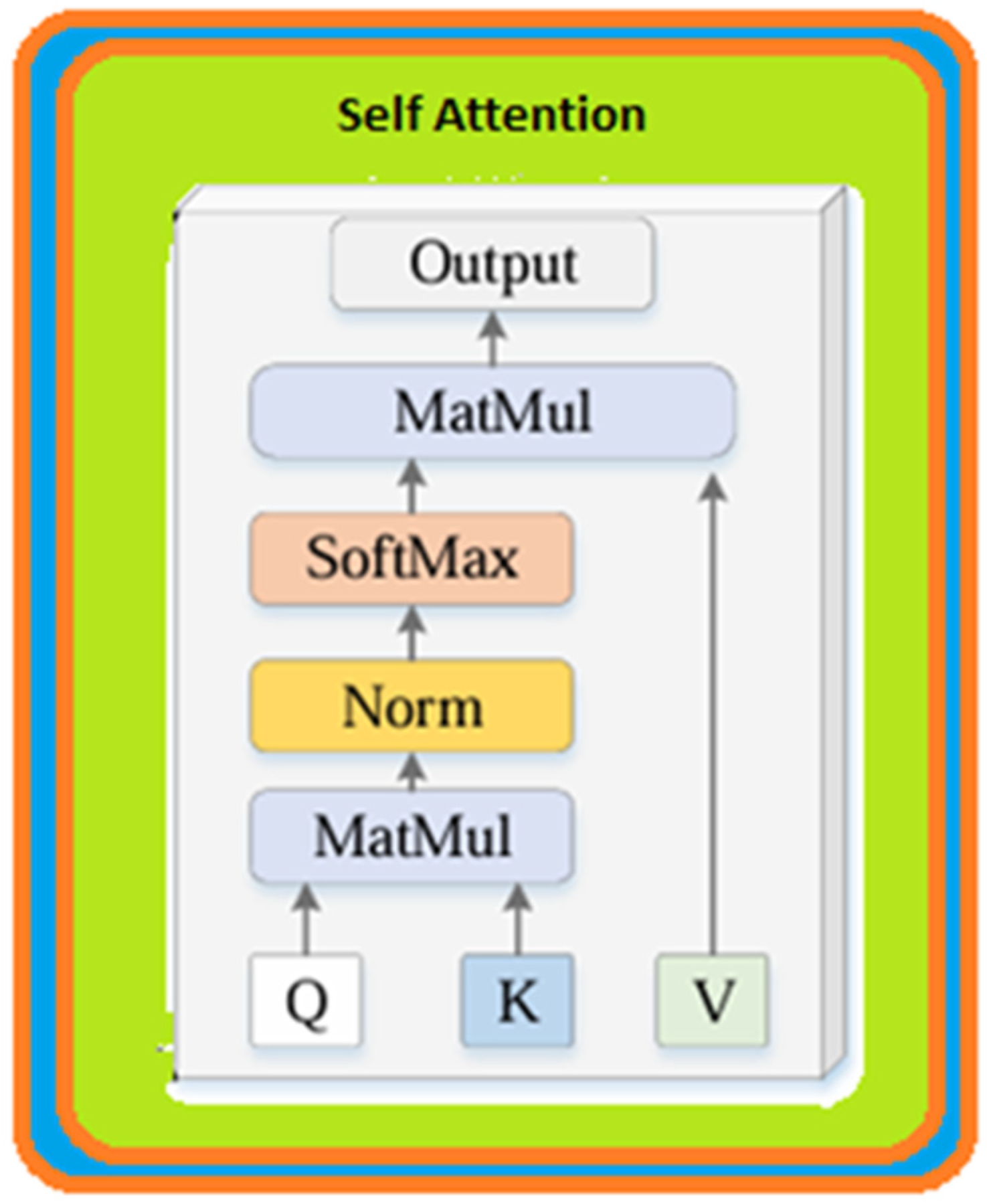
- Transformer Encoder: These features were then passed through Transformer encoder layers. Here, by using the attention (self-attention) mechanism, the model learns the relationship of each patch to the other patches and creates more meaningful feature representations.
- Inference and Classification: Through the Transformer layers, the features of each patch are combined, and the VİT-OELM hybrid model creates a higher-level representation. As the output of the VİT-OELM hybrid model, 1000 features obtained for each image data are set to input for the OELM head classifier.
2.3. Optimum Extreme Learning Machine Classifier
- Speed and Computational Efficiency: OELM is a model that does not require parameter adjustments during its training. This provides faster training times compared to algorithms such as SVM or XGBoost. OELM uses a single-layer artificial neural network, and training this structure is usually faster. Algorithms such as SVM or XGBoost can be more complex in terms of optimizing parameters and the training time of the model.
- Less Parameter Tuning Required: OELM offers a fast training process by assigning random weights only for the input layer. The model generally does not require hyperparameter adjustments, which increases its ease of use. Algorithms such as SVM and XGBoost require more parameter tuning. For example, parameters such as kernel type, C and gamma parameters for SVM, and learning rate and number of trees for XGBoost need to be optimized.
- General Performance: OELM generally has good results on small and medium-sized datasets. Especially on large datasets, methods such as SVM or XGBoost can be more powerful. SVM and XGBoost can generally give better results than OELM in nonlinear classifications, but OELM has great advantages in terms of fast learning and implementation.
- Risk of Overfitting: OELM provides fast learning to reduce the risk of overfitting, and can increase generalization ability by assigning random weights during the training process. SVM and XGBoost may have risks of overfitting if they are not tuned correctly, since they have more parameters.
- Complexity and Flexibility: Since the architecture of OELM is simple, good results can be obtained with less complexity in some cases. However, in more complex and high-accuracy tasks, algorithms such as SVM or XGBoost can be more flexible and powerful.
- Input weights are randomly assigned and not changed.
- Only the weights in the output layer are optimized.
- It provides fast learning and works efficiently on large datasets.
- Randomly determined weights can decrease the accuracy performance of the model.
- It may carry the risk of overfitting.
- It may require sensitive hyperparameter adjustments.
- Optimization of Weights
- Better Generalization Ability
- Fast Learning
- More Stable Results
- Stronger Regularization: Over-learning is prevented by adding L1 or L2 regularization.
- Kernel-Based OELM (K-OELM): It provides better generalization in nonlinear problems by using kernel functions.
- Hyperparameter Optimization: Methods such as Grid Search, Bayesian Optimization or Evolutionary Algorithms can be used to determine the best hyperparameters of the model.
- Higher accuracy: It can generalize better compared to ELM.
- Fast training process: It is faster than traditional neural networks.
- Reduces over-learning: The model becomes more stable with regularization and optimization methods.
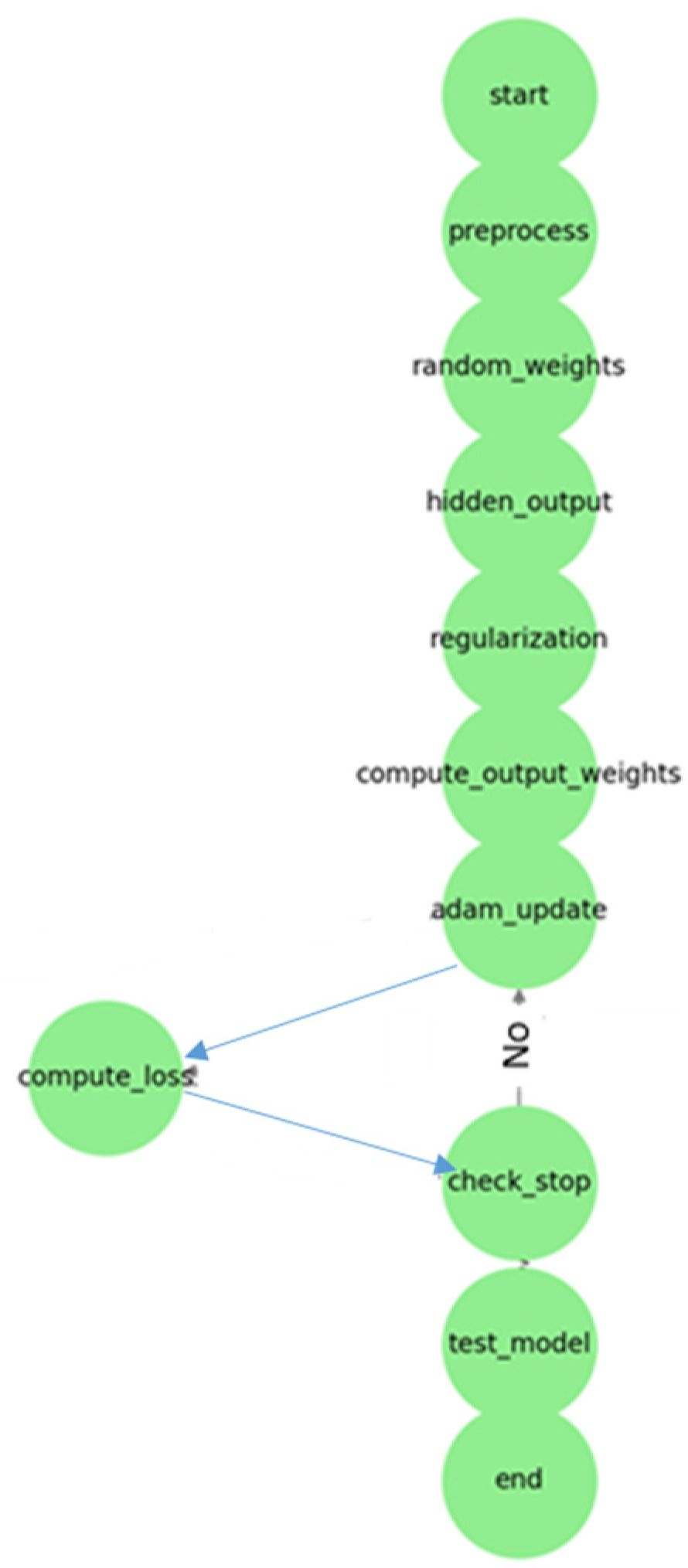
- Flow Steps:
- Start: Input data are prepared. Hyperparameters (lambda, number of hidden layers) are determined.
- Data Preprocessing: Input data are normalized. They are divided into training and test datasets.
- Random Assignment of Hidden Layer Weights: The weights and bias values of the hidden layer are randomly assigned.
- Calculation of Hidden Layer Output: The activation function is applied.
- Regularization and Hyperparameter Optimization: The regularization parameter λ (lambda) is selected. The number of hidden layers is optimized.
- Calculating Output Weights (Adam): Weight updates are performed with Adam optimization instead of the Moore–Penrose Pseudo-Inverse.
- Weight Update with Adam Optimization: The learning rate, beta values, etc., are determined. The first moment (m) and second moment (v) are calculated. Weights are updated.
- Calculating Loss Function: MSE or another error metric is calculated.
- Checking Stopping Criteria: Has the maximum iteration or specified error threshold been reached?Yes: Proceed to testing phase. No: Return to Adam optimization.
- Testing the Model: Model performance is measured with test data.
- Finish: The trained model is saved and the results are reported. With this method, the ViT-OELM hybrid model provided high accuracy and fast calculation advantages in pes planus image classification.
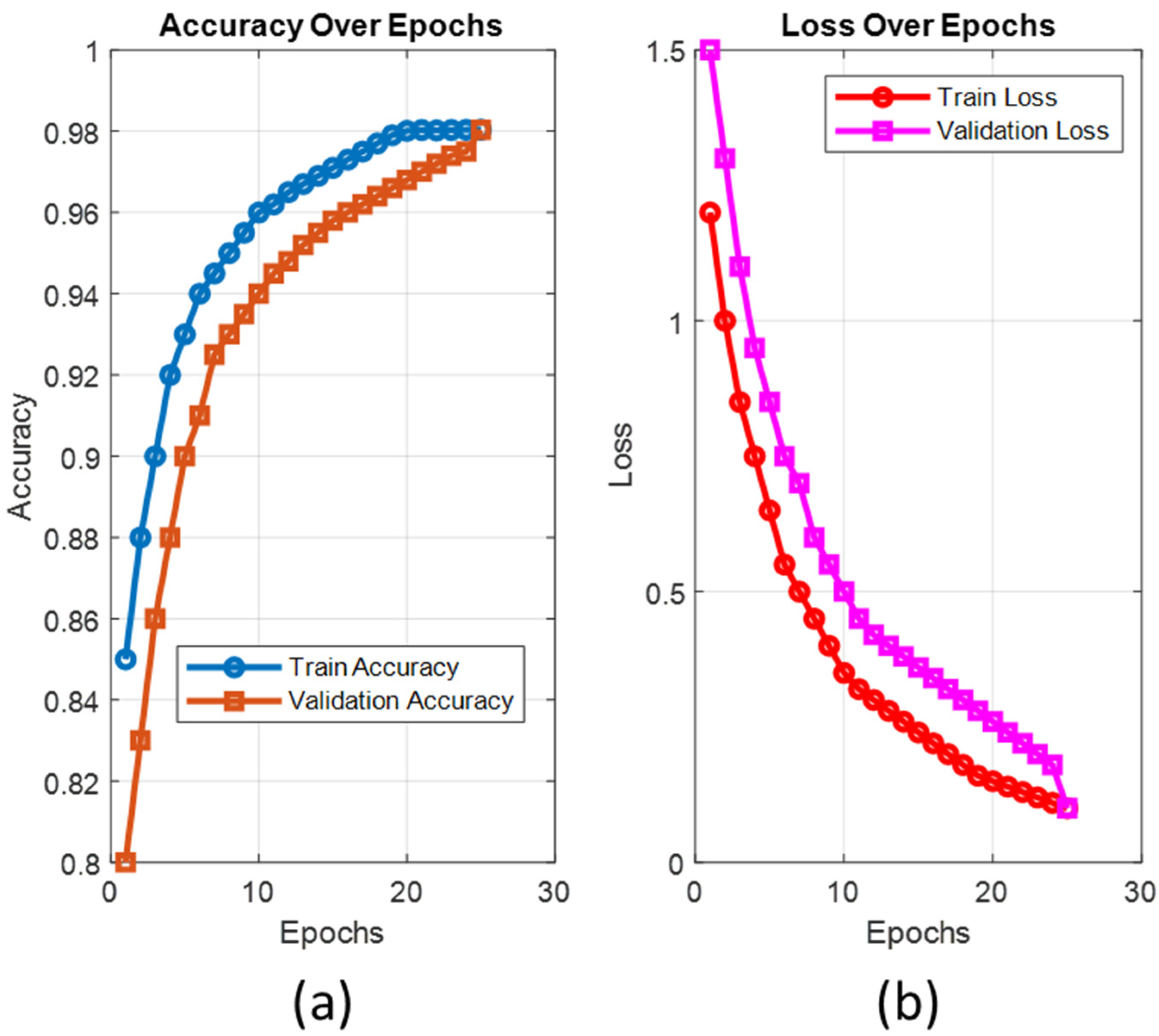
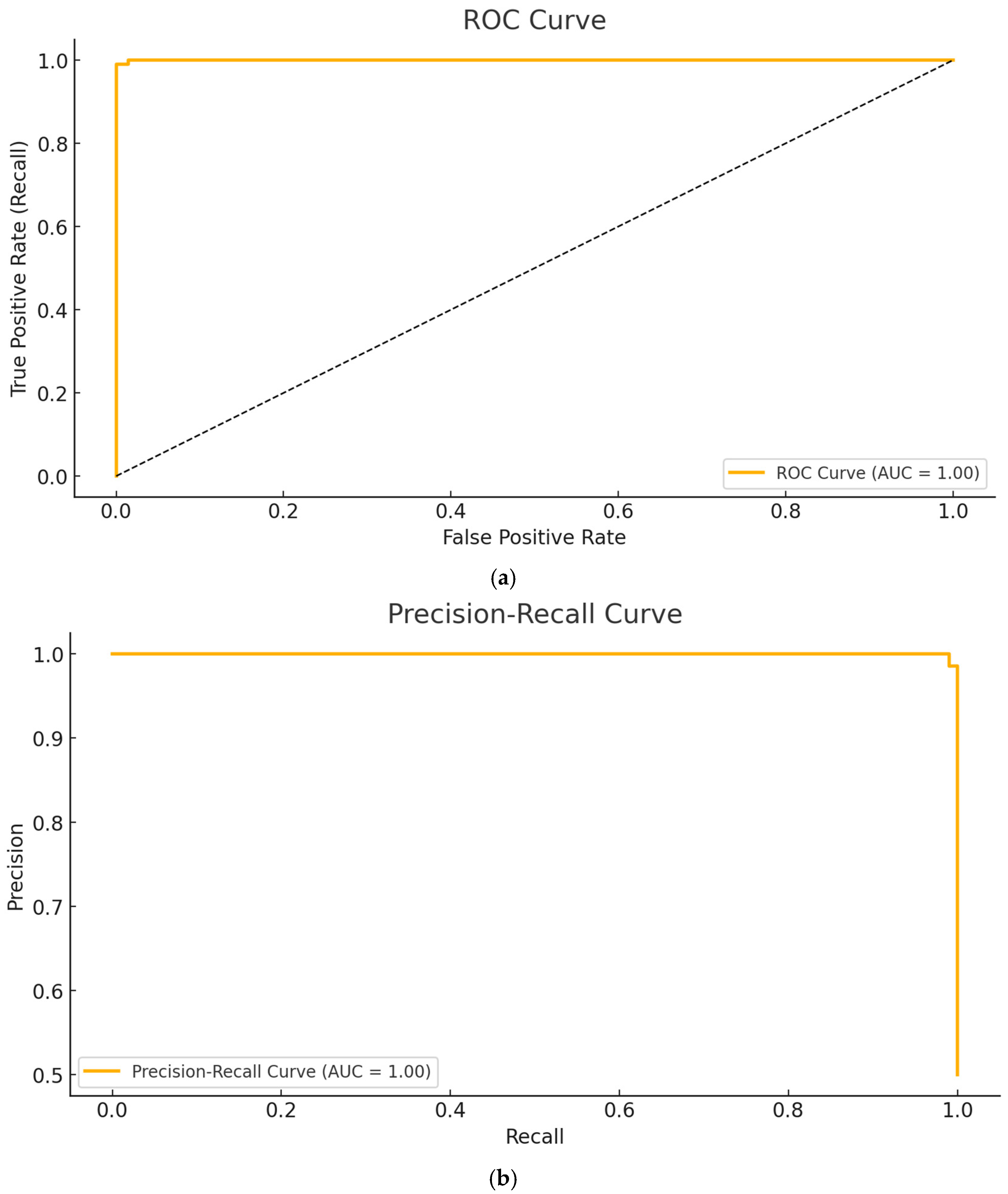
3. Results
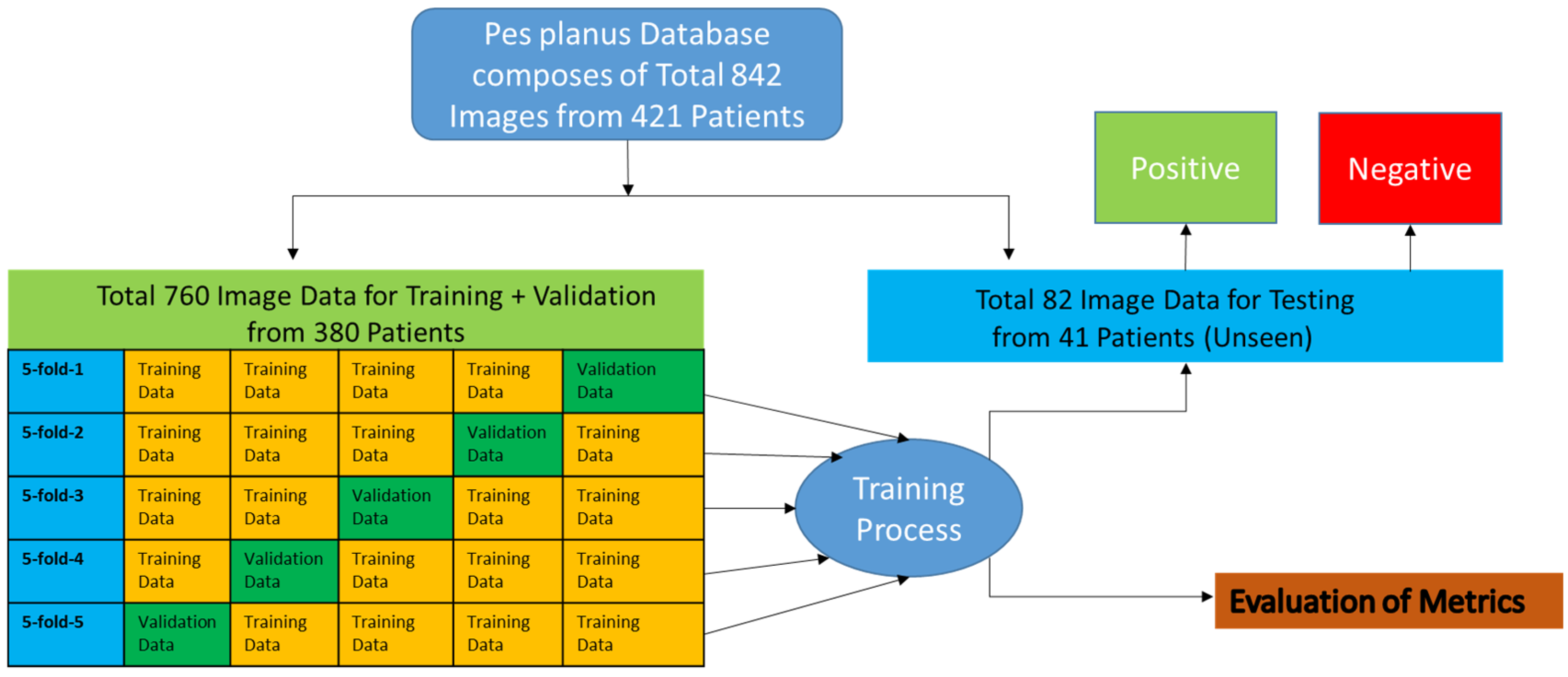
- Definition of Dataset
- 2.
- Dataset Division (K-Fold Cross Validation)
- The dataset was divided into five equal parts.
- In each iteration, one part of the data was used for validation, and the remaining four parts were used for training. Thus, a total of 1520 data were used for training and 380 data were used for validation over five iterations.
- The model was trained from scratch in each layer (fold), and the average performance metrics were reported.
Application Setup and Performance Metrics
- -
- Parameter updates are more balanced, thanks to momentum and the adaptive learning rate.
- -
- Weights and bias values are optimized to prevent over-learning.
- -
- Provides faster convergence, especially useful when working with large datasets.
- Calculated metric values of the proposed ViT-OELM model:
4. Discussion
- The suggested ViT-OELM hybrid model is based on the ViT architecture, which is a popular area in AI. To demonstrate the superiority of the proposed ViT-OELM hybrid model over other deep learning models, which used the same pes planus image database, an application was created here for pes planus classification.
- The ViT-OELM hybrid model was trained on bigger pes planus image datasets; this model achieved successful results in pes planus diagnosis problems with low training costs.
- In this ViT-OELM hybrid model, the ViT and OELM hybrid architecture is different from an end-to-end Transformer structure. In this study, the Optimum Extreme Learning Machine (OELM) head layer was added to basic ViT architecture model instead of Multi-Layer Perceptron (MLP) head layer being used. As shown in Table 2, the performance results of this suggested ViT-OELM hybrid model are superior to other deep learning structures.
- The ViT-OELM hybrid model has the potential to be used in clinical applications thanks to its high classification accuracy.
- By using OELM as classifier in this proposed ViT-OELM hybrid modeling, some classification advantages are obtained. OELM is faster than fully connected layers and traditional neural networks, because it optimizes only the output weights, and it provides better classification success with optimum weight selection and kernel functions. The generalization ability of the model is increased with regularization methods.
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Gül, Y.; Yaman, S.; Avcı, D.; Çilengir, A.H.; Balaban, M.; Güler, H. A novel deep transfer learning-based approach for automated Pes Planus diagnosis using X-ray image. Diagnostics 2023, 13, 1662. [Google Scholar] [CrossRef] [PubMed]
- Danaci, C.; Avci, D.; Tuncer, S.A. Diagnosis of pes planus from X-ray images: Enhanced feature selection with deep learning and machine learning techniques. Biomed. Signal Process. Control. 2025, 106, 107769. [Google Scholar] [CrossRef]
- Gül, Y.; Yaman, S.; Avcı, D.; Çilengir, A.H.; Balaban, M.; Güler, H. Kaggle. 2023. Available online: https://www.kaggle.com/datasets/suleyman32/pesplanus-two-class-dataset (accessed on 10 January 2025).
- Huang, X.; Shan, J.; Vaidya, V. Lung nodule detection in CT using 3D convolutional neural networks. In Proceedings of the 2017 IEEE 14th International Symposium on Biomedical Imaging (ISBI 2017), Melbourne, VIC, Australia, 18–21 April 2017. [Google Scholar]
- Gopatoti, A.; Vijayalakshmi, P. CXGNet: A tri-phase chest X-ray image classification for COVID-19 diagnosis using deep CNN with enhanced grey-wolf optimizer. Biomed. Signal Process. Control. 2022, 77, 103860. [Google Scholar] [CrossRef]
- Litjens, G.; Kooi, T.; Bejnordi, B.E.; Setio, A.A.A.; Ciompi, F.; Ghafoorian, M.; van der Laak, J.A.W.M.; van Ginneken, B.; Sánchez, C.I. A survey on deep learning in medical image analysis. Med. Image Anal. 2017, 42, 60–88. [Google Scholar] [CrossRef] [PubMed]
- Talo, M.; Yildirim, O.; Baloglu, U.B.; Aydin, G.; Acharya, U.R. Convolutional neural networks for multi-class brain disease detection using MRI images. Comput. Med Imaging Graph. 2019, 78, 101673. [Google Scholar] [CrossRef]
- Qin, C.; Yao, D.; Shi, Y.; Song, Z. Computer-aided detection in chest radiography based on artificial intelligence: A survey. Biomed. Eng. Online 2018, 17, 1–23. [Google Scholar] [CrossRef]
- Ozturk, T.; Talo, M.; Yildirim, E.A.; Baloglu, U.B.; Yildirim, O.; Acharya, U.R. Automated detection of COVID-19 cases using deep neural networks with X-ray images. Comput. Biol. Med. 2020, 121, 103792. [Google Scholar] [CrossRef]
- Tang, A.; Tam, R.; Cadrin-Chênevert, A.; Guest, W.; Chong, J.; Barfett, J.; Chepelev, L.; Cairns, R.; Mitchell, J.R.; Cicero, M.D.; et al. Canadian Association of Radiologists white paper on artificial intelligence in radiology. Can. Assoc. Radiol. J. 2018, 69, 120–135. [Google Scholar] [CrossRef]
- Khanna, V.V.; Chadaga, K.; Sampathila, N.; Prabhu, S.; Chadaga, R.; Umakanth, S. Diagnosing COVID-19 using artificial intelligence: A comprehensive review. Netw. Model. Anal. Health Informatics Bioinform. 2022, 11, 25. [Google Scholar] [CrossRef]
- Doğan, K.; Selçuk, T.; Yılmaz, A. A Novel Model Based on CNN–ViT Fusion and Ensemble Learning for the Automatic Detection of Pes Planus. J. Clin. Med. 2024, 13, 4800. [Google Scholar] [CrossRef]
- Zhu, J.; Tan, Y.; Lin, R.; Miao, J.; Fan, X.; Zhu, Y.; Liang, P.; Gong, J.; He, H. Efficient self-attention mechanism and structural distilling model for Alzheimer’s disease diagnosis. Comput. Biol. Med. 2022, 147, 105737. [Google Scholar] [CrossRef]
- Wolf, T.; Debut, L.; Sanh, V.; Chaumond, J.; Delangue, C.; Moi, A.; Cistac, P.; Rault, T.; Louf, R.; Funtowicz, M.; et al. Transformers: State-of-the-art natural language processing. In Proceedings of the 2020 Conference on Empirical Methods in Natural Language Processing: System Demonstrations, Online, 16–20 November 2020. [Google Scholar]
- Guo, M.-H.; Liu, Z.-N.; Mu, T.-J.; Hu, S.-M. Beyond self-attention: External attention using two linear layers for visual tasks. IEEE Trans. Pattern Anal. Mach. Intell. 2022, 45, 5436–5447. [Google Scholar] [CrossRef] [PubMed]
- Qiu, J.; Mitra, J.; Ghose, S.; Dumas, C.; Yang, J.; Sarachan, B.; Judson, M.A. A multichannel CT and radiomics-guided CNN-ViT (RadCT-CNNViT) ensemble network for diagnosis of pulmonary sarcoidosis. Diagnostics 2024, 14, 1049. [Google Scholar] [CrossRef] [PubMed]
- Rahali, A.; Akhloufi, M.A. End-to-end transformer-based models in textual-based NLP. AI 2023, 4, 54–110. [Google Scholar] [CrossRef]
- Bajaj, D.; Bharti, U.; Gupta, I.; Gupta, P.; Yadav, A. GTMicro—Microservice identification approach based on deep NLP transformer model for greenfield developments. Int. J. Inf. Technol. 2024, 16, 2751–2761. [Google Scholar] [CrossRef]
- Zhou, Y. A serial semantic segmentation model based on encoder-decoder architecture. Knowledge-Based Syst. 2024, 295, 111819. [Google Scholar] [CrossRef]
- Al-Fahsi, R.D.H.; Prawirosoenoto, A.N.F.; Nugroho, H.A.; Ardiyanto, I. GIVTED-Net: GhostNet-Mobile Involution ViT Encoder-Decoder Network for Lightweight Medical Image Segmentation. IEEE Access 2024, 12, 81281–81292. [Google Scholar] [CrossRef]
- Parvaiz, A.; Khalid, M.A.; Zafar, R.; Ameer, H.; Ali, M.; Fraz, M.M. Vision Transformers in medical computer vision—A contemplative retrospection. Eng. Appl. Artif. Intell. 2023, 122, 106126. [Google Scholar] [CrossRef]
- Islam, O.; Kumer, K.; Akter, S.; Uddin, M. Multi-Head Self-Attention Mechanisms in Vision Transformers for Retinal Image Classification. In Proceedings of the 2024 IEEE International Conference on Computing, Applications and Systems (COMPAS), Chattogram, Bangladesh, 25–26 September 2024. [Google Scholar]
- Dosovitskiy, A.; Beyer, L.; Kolesnikov, A.; Weissenborn, D.; Zhai, X.; Unterthiner, T.; Dehghani, M.; Minderer, M.; Heigold, G.; Gelly, S.; et al. An image is worth 16x16 words: Transformers for image recognition at scale. arXiv 2020, arXiv:2010.11929. [Google Scholar]
- Zhou, T.; Niu, Y.; Lu, H.; Peng, C.; Guo, Y.; Zhou, H. Vision transformer: To discover the “four secrets” of image patches. Inf. Fusion 2024, 105, 102248. [Google Scholar] [CrossRef]
- Lee, S.H.; Lee, S.; Song, B.C. Vision transformer for small-size datasets. arXiv 2021, arXiv:2112.13492. [Google Scholar]
- Katar, O.; Yildirim, O. An explainable vision transformer model based white blood cells classification and localization. Diagnostics 2023, 13, 2459. [Google Scholar] [CrossRef] [PubMed]
- Diker, A.; Sönmez, Y.; Özyurt, F.; Avcı, E.; Avcı, D. Examination of the ECG signal classification technique DEA-ELM using deep convolutional neural network features. Multimedia Tools Appl. 2021, 80, 24777–24800. [Google Scholar] [CrossRef]
- Diker, A.; Avci, D.; Avci, E.; Gedikpinar, M. A new technique for ECG signal classification genetic algorithm Wavelet Kernel extreme learning machine. Optik 2018, 180, 46–55. [Google Scholar] [CrossRef]
- Reyad, M.; Sarhan, A.M.; Arafa, M. A modified Adam algorithm for deep neural network optimization. Neural Comput. Appl. 2023, 35, 17095–17112. [Google Scholar] [CrossRef]
- Lambora, A.; Gupta, K.; Chopra, K. Genetic algorithm—A literature review. In Proceedings of the 2019 international conference on Machine Learning, Big Data, Cloud and Parallel Computing (COMITCon), Faridabad, India, 14–16 February 2019. [Google Scholar]
- Fang, J.; Liu, W.; Chen, L.; Lauria, S.; Miron, A.; Liu, X. A survey of algorithms, applications and trends for particle swarm optimization. Int. J. Netw. Dyn. Intell. 2023, 24–50. [Google Scholar] [CrossRef]
- Nogay, H.S.; Adeli, H. Diagnostic of autism spectrum disorder based on structural brain MRI images using, grid search optimization, and convolutional neural networks. Biomed. Signal Process. Control. 2023, 79, 104234. [Google Scholar]
| Number of 5-Fold | Accuarcy (%) | Sensitivity (%) | Specificity (%) | Precision (%) | F-1 Score (%) |
|---|---|---|---|---|---|
| 5-fold-1 | 96.34 | 95.12 | 97.56 | 97.50 | 96.29 |
| 5-fold-2 | 98.78 | 100 | 98.78 | 97.61 | 98.79 |
| 5-fold-3 | 98.78 | 100 | 98.78 | 97.61 | 98.79 |
| 5-fold-4 | 97.56 | 97.56 | 97.56 | 97.56 | 97.55 |
| 5-fold-5 | 98.78 | 97.56 | 100 | 100 | 98.76 |
| Mean ± Std | 98.04 ± 0.976 | 98.04 ± 1.826 | 98.53 ± 0.913 | 98.05 ± 0.973 | 98.03 ± 0.995 |
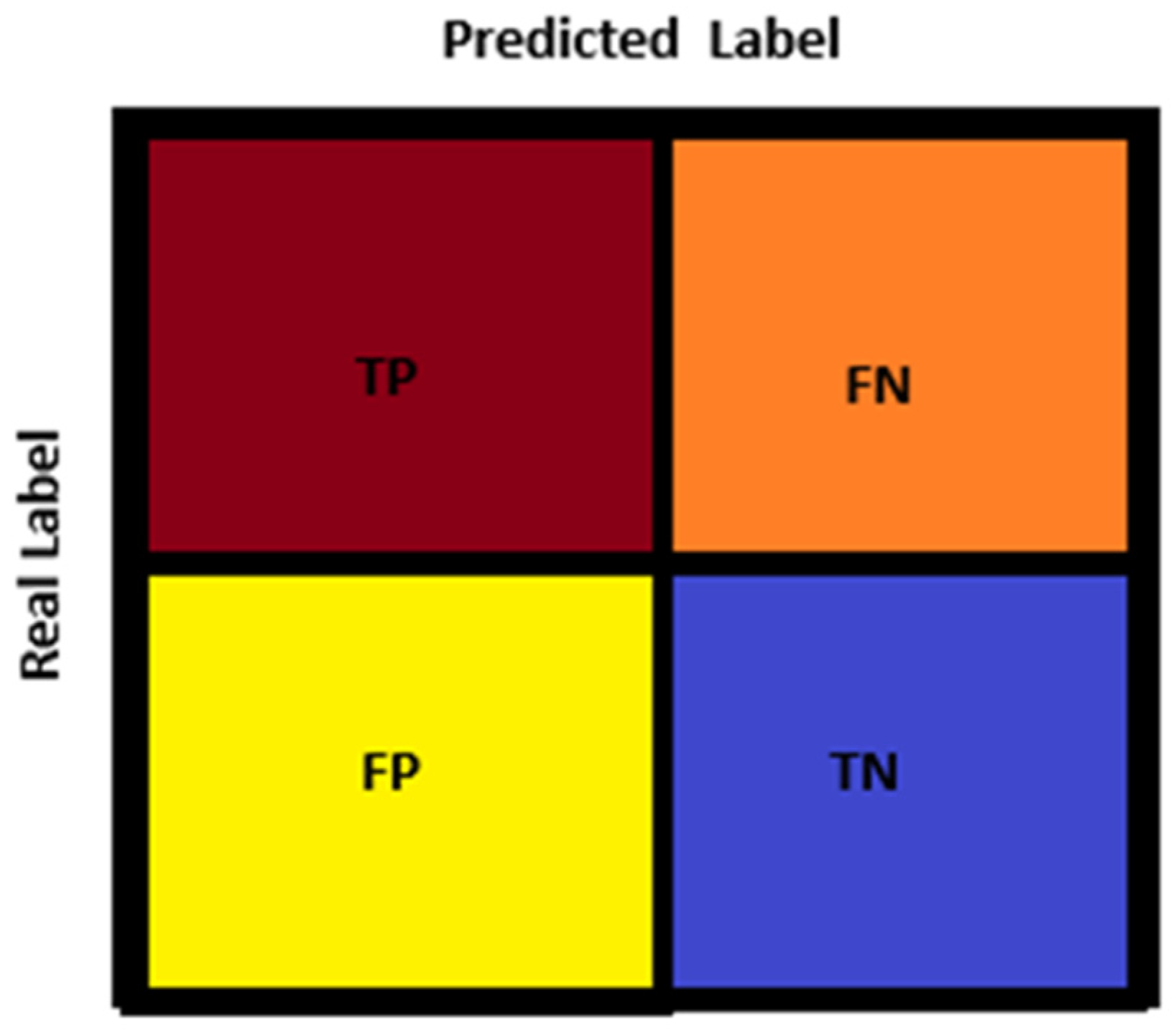
| 5-Fold-1 | 5-Fold-2 | 5-Fold-3 | 5-Fold-4 | 5-Fold-5 | |||||
|---|---|---|---|---|---|---|---|---|---|
| TP: 39 | FN: 2 | TP: 41 | FN: 0 | TP: 41 | FN: 0 | TP: 40 | FN: 1 | TP: 40 | FN: 1 |
| FP: 1 | TN: 40 | FP: 1 | TN: 40 | FP: 1 | TN: 40 | FP: 1 | TN: 40 | FP: 0 | TN: 41 |
| Metrics of Total 5-Fold Testing Data | |
|---|---|
| TP: 201 | FN: 4 |
| FP: 4 | TN: 201 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Avcı, D. A New Pes Planus Automatic Diagnosis Method: ViT-OELM Hybrid Modeling. Diagnostics 2025, 15, 867. https://doi.org/10.3390/diagnostics15070867
Avcı D. A New Pes Planus Automatic Diagnosis Method: ViT-OELM Hybrid Modeling. Diagnostics. 2025; 15(7):867. https://doi.org/10.3390/diagnostics15070867
Chicago/Turabian StyleAvcı, Derya. 2025. "A New Pes Planus Automatic Diagnosis Method: ViT-OELM Hybrid Modeling" Diagnostics 15, no. 7: 867. https://doi.org/10.3390/diagnostics15070867
APA StyleAvcı, D. (2025). A New Pes Planus Automatic Diagnosis Method: ViT-OELM Hybrid Modeling. Diagnostics, 15(7), 867. https://doi.org/10.3390/diagnostics15070867






