Improving Diagnostic Performance for Head and Neck Tumors with Simple Diffusion Kurtosis Imaging and Machine Learning Bi-Parameter Analysis
Abstract
1. Introduction
2. Materials and Methods
2.1. Patients
2.2. MRI System and DWI Sequence
2.3. Pre-Processing with Smoothing Filter for DWI
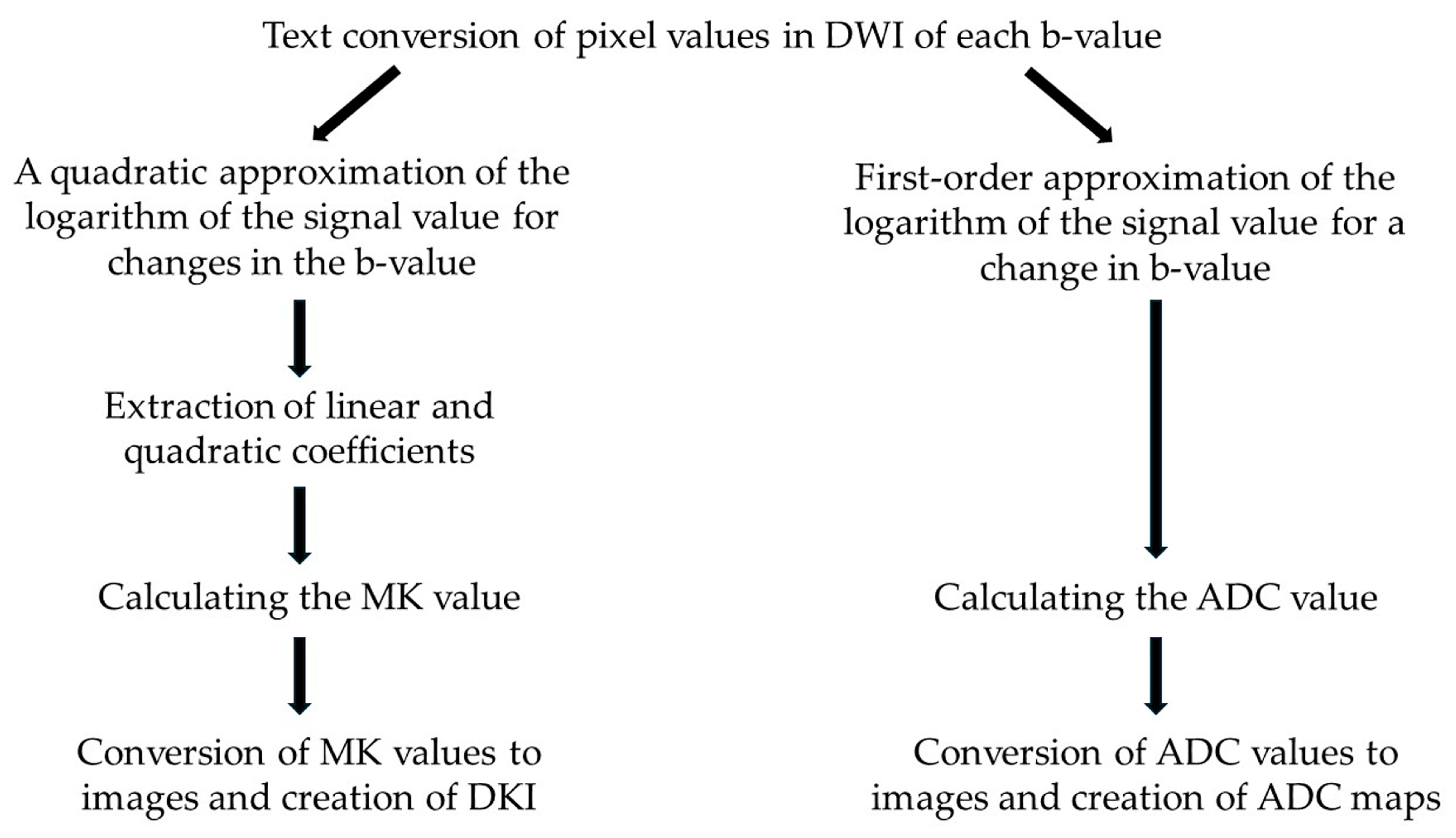
2.4. Creation of DKI and ADC Maps with SDI
2.5. Region of Interest (ROI) Setting and Pixel Count Evaluation
2.6. Evaluation of MK and ADC Values by Tumor Status Histology
2.7. Obtaining AUC Values Using Conventional ROC Analysis for Diagnosis of Tumor Status
2.8. Obtaining AUC Values Using ML ROC Analysis for Diagnosis of Tumor Status
2.8.1. Software and ML Algorithms Used
2.8.2. Data Set (Tables S1 and S2)
2.8.3. Best Modeling and Validation Practices
2.9. Comparison of AUC Values for Diagnosis of Tumor Status
3. Results
3.1. Clinical Case Information
3.2. Comparison of ADC and MK Values in Benign and Malignant Histologic Types
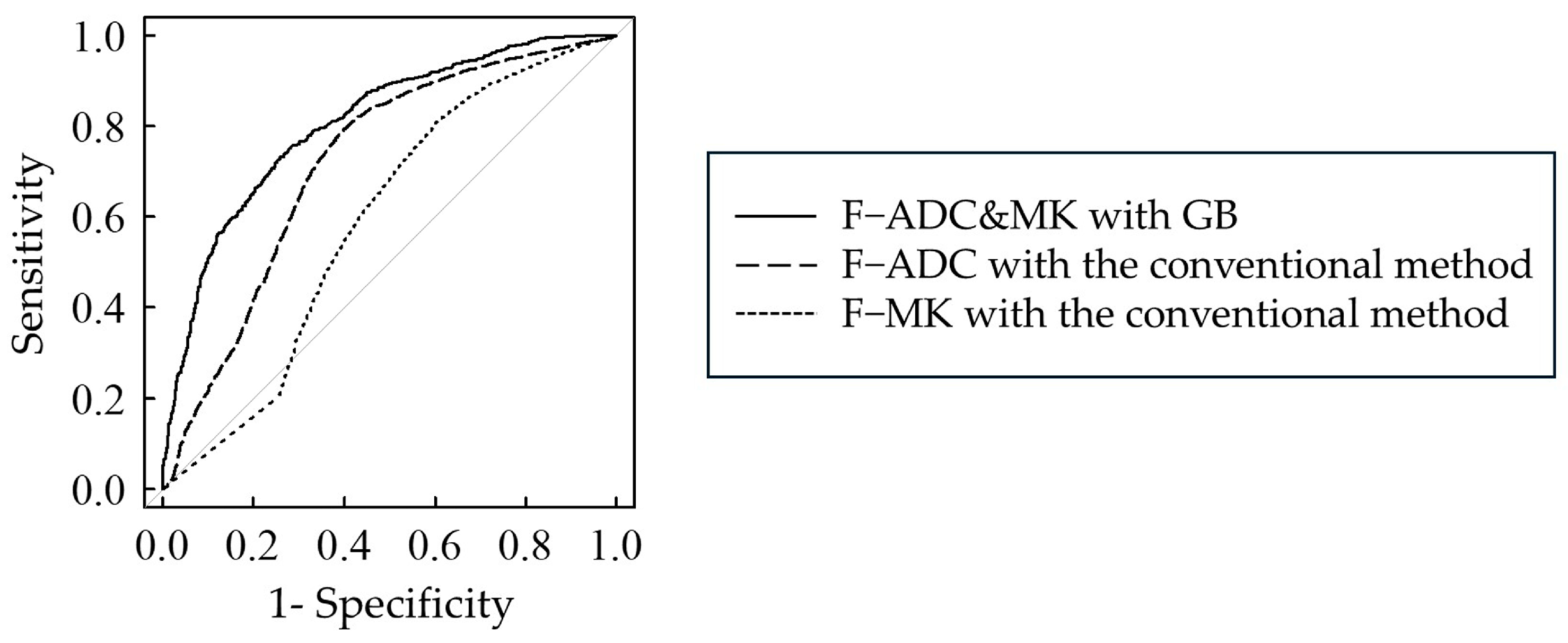
3.3. Comparison of AUC Values Between ML and Conventional Methods
3.4. Comparison of AUC Values Between Bi- and Single-Parameter Analyses
3.5. Influence of Filter Pre-Processing
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Cancer Statistics in Japan-2024. Available online: https://ganjoho.jp/public/qa_links/report/statistics/pdf/cancer_statistics_2024_fig_J.pdf (accessed on 5 March 2025).
- Kokubun, K.; Yamamoto, K.; Nakajima, K.; Akashi, Y.; Chujo, T.; Takano, M.; Katakura, A.; Matsuzaka, K. Frequency of Odontogenic Tumors: A Single Center Study of 1089 Cases in Japan and Literature Review. Head Neck Pathol. 2022, 16, 494–502. [Google Scholar] [CrossRef] [PubMed]
- Cheng, J.; Shao, S.; Chen, W.; Zheng, N. Application of Diffusion Kurtosis Imaging and Dynamic Contrast-Enhanced Magnetic Resonance Imaging in Differentiating Benign and Malignant Head and Neck Lesions. J. Magn. Reason. Imaging 2022, 55, 414–423. [Google Scholar] [CrossRef]
- Ma, G.; Xu, X.Q.; Hu, H.; Su, G.Y.; Shen, J.; Shi, H.B.; Wu, F.Y. Utility of Readout-Segmented Echo-Planar Imaging-Based Diffusion Kurtosis Imaging for Differentiating Malignant from Benign Masses in Head and Neck Region. Korean J. Radiol. 2018, 19, 443–451. [Google Scholar] [CrossRef] [PubMed]
- Karaman, C.Z.; Tanyeri, A.; Özgür, R.; Öztürk, V.S. Parotid gland tumors: Comparison of conventional and diffusion-weighted MRI findings with histopathological results. Dentomaxillofac. Radiol. 2021, 50, 20200391. [Google Scholar] [CrossRef]
- Zhang, Z.; Li, S.; Wang, W.; Zhang, Y.; Wang, K.; Cheng, J.; Wen, B. Synthetic MRI for the quantitative and morphologic assessment of head and neck tumors: A preliminary study. Dentomaxillofac. Radiol. 2023, 52, 20230103. [Google Scholar] [CrossRef] [PubMed]
- Scola, E.; Del Vecchio, G.; Busto, G.; Bianchi, A.; Desideri, I.; Gadda, D.; Mancini, S.; Carlesi, E.; Moretti, M.; Desideri, I.; et al. Conventional and Advanced Magnetic Resonance Imaging Assessment of Non-Enhancing Peritumoral Area in Brain Tumor. Cancers 2023, 15, 2992. [Google Scholar] [CrossRef]
- Honda, M.; Le Bihan, D.; Kataoka, M.; Iima, M. Diffusion kurtosis imaging as a biomarker of breast cancer. BJR Open 2023, 5, 20220038. [Google Scholar] [CrossRef]
- Liu, Y.; Wang, P.; Wang, S.; Zhang, H.; Song, Y.; Yan, X.; Gao, Y. Heterogeneity matching and IDH prediction in adult-type diffuse gliomas: A DKI-based habitat analysis. Front. Oncol. 2023, 13, 1202170. [Google Scholar] [CrossRef]
- Jensen, J.H.; Helpern, J.A.; Ramani, A.; Lu, H.; Kaczynski, K. Diffusional kurtosis imaging: The quantification of non-gaussian water diffusion by means of magnetic resonance imaging. Magn. Reson. Med. 2005, 53, 1432–1440. [Google Scholar] [CrossRef]
- Kuroda, M.; Konishi, K.; Sugimoto, K.; Yoshimura, Y.; Hamada, K.; Khasawnehc, A.; Barham, M.; Tekiki, N.; Sugianto, I.; Bamgbose, B.O.; et al. Evaluation of Fast Diffusion Kurtosis Imaging Using New Software Designed for Widespread Clinical Use. Acta Med. Okayama 2022, 76, 297–305. [Google Scholar]
- Shimizu, Y.; Kuroda, M.; Nakamitsu, Y.; Al-Hammad, W.E.; Yoshida, S.; Fukumura, Y.; Nakamura, Y.; Kuroda, K.; Kamizaki, R.; Imajoh, S.; et al. Usefulness of Simple Diffusion Kurtosis Imaging for Head and Neck Tumors: An Early Clinical Study. Acta Med. Okayama 2023, 77, 273–280. [Google Scholar]
- Fukumura, Y.; Kuroda, M.; Yoshida, S.; Nakamura, Y.; Nakamitsu, Y.; Al-Hammad, W.E.; Kuroda, K.; Kamizaki, R.; Shimizu, Y.; Tanabe, Y.; et al. Characteristic Mean Kurtosis Values in Simple Diffusion Kurtosis Imaging of Dentigerous Cysts. Diagnostics 2023, 13, 3619. [Google Scholar] [CrossRef] [PubMed]
- Nakamitsu, Y.; Kuroda, M.; Shimizu, Y.; Kuroda, K.; Yoshimura, Y.; Yoshida, S.; Nakamura, Y.; Fukumura, Y.; Kamizaki, R.; Al-Hammad, W.E.; et al. Enhancing Diagnostic Precision: Evaluation of Preprocessing Filters in Simple Diffusion Kurtosis Imaging for Head and Neck Tumors. J. Clin. Med. 2024, 13, 1783. [Google Scholar] [CrossRef]
- Chen, Y.; Huang, N.; Zheng, Y.; Wang, F.; Cao, D.; Chen, T. Characterization of parotid gland tumors: Whole-tumor histogram analysis of diffusion weighted imaging, diffusion kurtosis imaging, and intravoxel incoherent motion—A pilot study. Eur. J. Radiol. 2024, 170, 111199. [Google Scholar] [CrossRef]
- Cheng, Q.; Ren, A.; Xu, X.; Meng, Z.; Feng, X.; Pylypenko, D.; Dou, W.; Yu, D. Application of DKI and IVIM imaging in evaluating histologic grades and clinical stages of clear cell renal cell carcinoma. Front. Oncol. 2023, 13, 1203922. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Luo, Y.; Jiang, H.; Meng, N.; Huang, Z.; Feng, P.; Fang, T.; Fu, F.; Li, X.; Bai, Y.; et al. The value of diffusion kurtosis imaging, diffusion weighted imaging and 18F-FDG PET for differentiating benign and malignant solitary pulmonary lesions and predicting pathological grading. Front. Oncol. 2022, 12, 873669. [Google Scholar] [CrossRef]
- Hamada, K.; Kuroda, M.; Yoshimura, Y.; Khasawneh, A.; Barham, M.; Tekiki, N.; Sugianto, I.; Bamgbose, B.O.; Konishi, K.; Sugimoto, K.; et al. Evaluation of the Imaging Process for a Novel Subtraction Method Using Apparent Diffusion Coefficient Values. Acta Med. Okayama 2021, 75, 139–145. [Google Scholar] [PubMed]
- Panyarak, W.; Chikui, T.; Tokumori, K.; Yamashita, Y.; Kamitani, T.; Togao, O.; Yoshiura, K. Utility of a diffusion kurtosis model in the differential diagnosis of orofacial tumours. Clin. Radiol. 2020, 75, 507–519. [Google Scholar] [CrossRef]
- Jiang, J.X.; Tang, Z.H.; Zhong, Y.F.; Qiang, J.W. Diffusion kurtosis imaging for differentiating between the benign and malignant sinonasal lesions. J. Magn. Reason. Imaging 2017, 45, 1446–1454. [Google Scholar] [CrossRef]
- Liu, Z.; Wen, B.; Zhang, Z.; Qu, F.; Wu, Y.; Grimm, R.; Zhang, Y.; Cheng, J.; Zhang, Y. The value of diffusion kurtosis imaging and dynamic contrast-enhanced magnetic resonance imaging in the differential diagnosis of parotid gland tumors. Gland Surg. 2024, 13, 1254–1268. [Google Scholar] [CrossRef]
- Wen, B.; Zhang, Z.; Zhu, J.; Liu, L.; Liu, Z.; Ma, X.; Wang, K.; Xie, L.; Zhang, Y.; Cheng, J. Synthetic MRI plus FSE-PROPELLER DWI for differentiating malignant from benign head and neck tumors: A preliminary study. Front. Oncol. 2023, 13, 1225420. [Google Scholar]
- Xiao, Z.; Tang, Z.; Qiang, J.; Wang, S.; Qian, W.; Zhong, Y.; Wang, R.; Wang, J.; Wu, L.; Tang, W.; et al. Intravoxel Incoherent Motion MR Imaging in the Differentiation of Benign and Malignant Sinonasal Lesions: Comparison with Conventional Diffusion-Weighted MR Imaging. AJNR Am. J. Neuroradiol. 2018, 39, 538–546. [Google Scholar] [CrossRef] [PubMed]
- Wen, B.; Zhang, Z.; Fu, K.; Zhu, J.; Liu, L.; Gao, E.; Qi, J.; Zhang, Y.; Cheng, J.; Qu, F.; et al. Value of pre-/post-contrast-enhanced T1 mapping and readout segmentation of long variable echo-train diffusion-weighted imaging in differentiating parotid gland tumors. Eur. J. Radiol. 2023, 162, 110748. [Google Scholar]
- Xu, Z.; Chen, M.; Zheng, S.; Chen, S.; Xiao, J.; Hu, Z.; Lu, L.; Yang, Z.; Lin, D. Differential diagnosis of parotid gland tumours: Application of SWI combined with DWI and DCE-MRI. Eur. J. Radiol. 2022, 146, 110094. [Google Scholar] [CrossRef]
- Takumi, K.; Nagano, H.; Kikuno, H.; Kumagae, Y.; Fukukura, Y.; Yoshiura, T. Differentiating malignant from benign salivary gland lesions: A multiparametric non-contrast MR imaging approach. Sci. Rep. 2021, 11, 2780. [Google Scholar]
- Takumi, K.; Nakanosono, R.; Nagano, H.; Hakamada, H.; Kanzaki, F.; Kamimura, K.; Nakajo, M.; Eizuru, Y.; Nagano, H.; Yoshiura, T. Multiparametric approach with synthetic MR imaging for diagnosing salivary gland lesions. Jpn. J. Radiol. 2024, 42, 983–992. [Google Scholar] [CrossRef] [PubMed]
- Barchetti, F.; Pranno, N.; Giraldi, G.; Sartori, A.; Gigli, S.; Barchetti, G.; Lo Mele, L.; Marsella, L.T. The role of 3 Tesla diffu-sion-weighted imaging in the differential diagnosis of benign versus malignant cervical lymph nodes in patients with head and neck squamous cell carcinoma. Biomed. Res. Int. 2014, 2014, 532095. [Google Scholar]
- Chen, C.; Lin, Z.; Xiao, Y.; Bai, P.; Yue, Q.; Chen, Y.; Chen, L. Role of diffusion-weighted imaging in the discrimination of benign and metastatic parotid area lymph nodes in patients with nasopharyngeal carcinoma. Sci. Rep. 2018, 8, 281. [Google Scholar]
- Fathi Kazerooni, A.; Nabil, M.; Alviri, M.; Koopaei, S.; Salahshour, F.; Assili, S.; Saligheh Rad, H.; Aghaghazvini, L. Radiomic Analysis of Multi-parametric MR Images (MRI) for Classification of Parotid Tumors. J. Biomed. Phys. Eng. 2022, 12, 599–610. [Google Scholar]
- Malagi, A.V.; Shivaji, S.; Kandasamy, D.; Sharma, R.; Garg, P.; Gupta, S.D.; Gamanagatti, S.; Mehndiratta, A. Pancreatic Mass Characterization Using IVIM-DKI MRI and Machine Learning-Based Multi-Parametric Texture Analysis. Bioengineering 2023, 10, 83. [Google Scholar] [CrossRef]
- Vats, N.; Sengupta, A.; Gupta, R.K.; Patir, R.; Vaishya, S.; Ahlawat, S.; Saini, J.; Agarwal, S.; Singh, A. Differentiation of Pilocytic Astrocytoma from Glioblastoma using a Machine-Learning framework based upon quantitative T1 perfusion MRI. Magn. Reason. Imaging 2023, 98, 76–82. [Google Scholar]
- Lin, C.Y.; Yen, Y.T.; Huang, L.T.; Chen, T.Y.; Liu, Y.S.; Tang, S.Y.; Huang, W.L.; Chen, Y.Y.; Lai, C.H.; Fang, Y.D.; et al. An MRI-Based Clinical-Perfusion Model Predicts Pathological Subtypes of Prevascular Mediastinal Tumors. Diagnostics 2022, 12, 889. [Google Scholar] [CrossRef] [PubMed]
- Tokuc, E.; Eksi, M.; Kayar, R.; Demir, S.; Topaktas, R.; Bastug, Y.; Akyuz, M.; Ozturk, M. Inflammation indexes and machine-learning algorithm in predicting urethroplasty success. Investig. Clin. Urol. 2024, 65, 240–247. [Google Scholar]
- Yang, B.; Gao, Y.; Lu, J.; Wang, Y.; Wu, R.; Shen, J.; Ren, J.; Wu, F.; Xu, H. Quantitative analysis of chest MRI images for benign malignant diagnosis of pulmonary solid nodules. Front. Oncol. 2023, 13, 1212608. [Google Scholar]
- Ding, H.; Fawad, M.; Xu, X.; Hu, B. A framework for identification and classification of liver diseases based on machine learning algorithms. Front. Oncol. 2022, 12, 1048348. [Google Scholar]
- Yin, P.; Mao, N.; Chen, H.; Sun, C.; Wang, S.; Liu, X.; Hong, N. Machine and Deep Learning Based Radiomics Models for Preoperative Prediction of Benign and Malignant Sacral Tumors. Front. Oncol. 2020, 10, 564725. [Google Scholar]
- Baty, F.; Cvetkovic, D.; Boesch, M.; Bauer, F.; Adão Martins, N.R.; Rossi, R.M.; Schoch, O.D.; Annaheim, S.; Brutsche, M.H. Validation of a Textile-Based Wearable Measuring Electrocardiogram and Breathing Frequency for Sleep Apnea Monitoring. Sensors 2024, 24, 6229. [Google Scholar] [CrossRef]


| Type (Sex, Mean Age, Range) | Histological Classification (Differentiation or Type) | Number of Cases | Site * | Number of Pixels ** |
|---|---|---|---|---|
| Malignant (M: 9, F: 8, 69, 37–94) | Squamous cell carcinoma | 11 | Maxilla (4) | 434, 334, 219, 132 |
| (Well: 4 | Tongue (4) | 289, 245, 198, 21 | ||
| Moderately: 2 | Mandible (2) | 310, 63 | ||
| Poorly: 3 Unknown: 2) | Oral floor (1) | 302 | ||
| Adenoid cyst carcinoma | 2 | Maxilla | 412, 59 | |
| Lymphoma (EBV-positive DLBCL: 1, CD5-positive DLBCL: 1) | 2 | Maxilla | 398, 154 | |
| Osteosarcoma | 1 | Mandible | 117 | |
| Acinic cell carcinoma | 1 | Maxilla | 223 | |
| Benign (M: 7, F: 8, 47, 14–80) | Ameloblastoma (Conventional: 3 Unknown: 5) | 8 | Mandible | 1889, 666, 626, 455, 289, 117, 98, 77 |
| Pleomorphic adenoma | 6 | Maxilla (4) | 419, 162, 105, 37 | |
| Submandibular gland (1) | 170 | |||
| Upper lip (1) | 95 | |||
| Dentinogenic ghost cell tumor | 1 | Maxilla | 431 |
| ADC | MK | |||
|---|---|---|---|---|
| Malignant Median (Q1, Q3) | Benign Median (Q1, Q3) | Malignant Median (Q1, Q3) | Benign Median (Q1, Q3) | |
| Without filter | 0.001193 (0.000964, 0.001604) | 0.001631 (0.001361, 0.002070) | 0.93 (0, 1.43) | 0.63 (0.11, 1.04) |
| With filter | 0.001196 (0.000982, 0.001590) * | 0.001623 (0.001384, 0.002044) * | 0.87 (0.06, 1.36) * | 0.65 (0.20, 1.04) |
| Method | Algorithm | Without Filter | With Filter | ||||
|---|---|---|---|---|---|---|---|
| ADC&MK | ADC | MK | ADC&MK | ADC | MK | ||
| Machine learning | Gradient boosting | 0.81 ** | 0.74 | 0.66 ** | 0.80 ** | 0.75 | 0.67 ** |
| Deep neural network | 0.80 ** | 0.73 § | 0.66 ** | 0.79 ** §§ | 0.74 | 0.66 ** | |
| Random forest | 0.80 ** | 0.73 § | 0.65 ** §§ | 0.79 ** § | 0.74 | 0.66 ** | |
| Support vector machine | 0.79 ** § | 0.74 | 0.65 ** § | 0.78 ** §§ | 0.74 | 0.65 ** §§ | |
| Decision tree | 0.78 ** §§§ | 0.73 | 0.66 ** | 0.77 * §§§ | 0.74 §§ | 0.66 ** | |
| Median | 0.80 **1 **2 ☨☨1 ☨☨2 | 0.73 **2 ☨☨2 ☨☨3 | 0.66 ☨☨1 ☨☨3 | 0.79 ☨☨1 ☨☨2 | 0.74 ☨☨2 | 0.66 | |
| Conventional method | 0.71 ***2 ☨1 ☨☨☨2 ☨☨3 | 0.72 ***2 ☨☨☨2 ☨3 | 0.59 ☨☨☨1 ☨2 ☨☨☨3 | 0.74 ☨☨☨2 | 0.73 ☨☨☨2 | 0.57 | |
| Algorithm | Explanatory Variable | F−ADC&MK | F−ADC | F−MK | F+ADC&MK | F+ADC | F+MK |
|---|---|---|---|---|---|---|---|
| Gradient boosting | F−ADC&MK | N/A | |||||
| F−ADC | <0.001 | N/A | |||||
| F−MK | <0.001 | <0.001 | N/A | ||||
| F+ADC&MK | NS | <0.001 | <0.001 | N/A | |||
| F+ADC | <0.001 | NS | <0.001 | <0.001 | N/A | ||
| F+MK | <0.001 | <0.001 | NS | <0.001 | <0.001 | N/A | |
| Deep neural network | F−ADC&MK | N/A | |||||
| F−ADC | <0.001 | N/A | |||||
| F−MK | <0.001 | <0.001 | N/A | ||||
| F+ADC&MK | NS | <0.001 | <0.001 | N/A | |||
| F+ADC | <0.001 | NS | <0.001 | <0.001 | N/A | ||
| F+MK | <0.001 | <0.001 | NS | <0.001 | <0.001 | N/A | |
| Random forest | F−ADC&MK | N/A | |||||
| F−ADC | <0.001 | N/A | |||||
| F−MK | <0.001 | <0.001 | N/A | ||||
| F+ADC&MK | NS | <0.001 | <0.001 | N/A | |||
| F+ADC | <0.001 | NS | <0.001 | <0.001 | N/A | ||
| F+MK | <0.001 | <0.001 | NS | <0.001 | <0.001 | N/A |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yoshida, S.; Kuroda, M.; Nakamura, Y.; Fukumura, Y.; Nakamitsu, Y.; Al-Hammad, W.E.; Kuroda, K.; Shimizu, Y.; Tanabe, Y.; Oita, M.; et al. Improving Diagnostic Performance for Head and Neck Tumors with Simple Diffusion Kurtosis Imaging and Machine Learning Bi-Parameter Analysis. Diagnostics 2025, 15, 790. https://doi.org/10.3390/diagnostics15060790
Yoshida S, Kuroda M, Nakamura Y, Fukumura Y, Nakamitsu Y, Al-Hammad WE, Kuroda K, Shimizu Y, Tanabe Y, Oita M, et al. Improving Diagnostic Performance for Head and Neck Tumors with Simple Diffusion Kurtosis Imaging and Machine Learning Bi-Parameter Analysis. Diagnostics. 2025; 15(6):790. https://doi.org/10.3390/diagnostics15060790
Chicago/Turabian StyleYoshida, Suzuka, Masahiro Kuroda, Yoshihide Nakamura, Yuka Fukumura, Yuki Nakamitsu, Wlla E. Al-Hammad, Kazuhiro Kuroda, Yudai Shimizu, Yoshinori Tanabe, Masataka Oita, and et al. 2025. "Improving Diagnostic Performance for Head and Neck Tumors with Simple Diffusion Kurtosis Imaging and Machine Learning Bi-Parameter Analysis" Diagnostics 15, no. 6: 790. https://doi.org/10.3390/diagnostics15060790
APA StyleYoshida, S., Kuroda, M., Nakamura, Y., Fukumura, Y., Nakamitsu, Y., Al-Hammad, W. E., Kuroda, K., Shimizu, Y., Tanabe, Y., Oita, M., Sugianto, I., Barham, M., Tekiki, N., Kamaruddin, N. N., Hisatomi, M., Yanagi, Y., & Asaumi, J. (2025). Improving Diagnostic Performance for Head and Neck Tumors with Simple Diffusion Kurtosis Imaging and Machine Learning Bi-Parameter Analysis. Diagnostics, 15(6), 790. https://doi.org/10.3390/diagnostics15060790





