Diagnostic Implications of NGS-Based Molecular Profiling in Mature B-Cell Lymphomas with Potential Bone Marrow Involvement
Abstract
1. Introduction
Objective
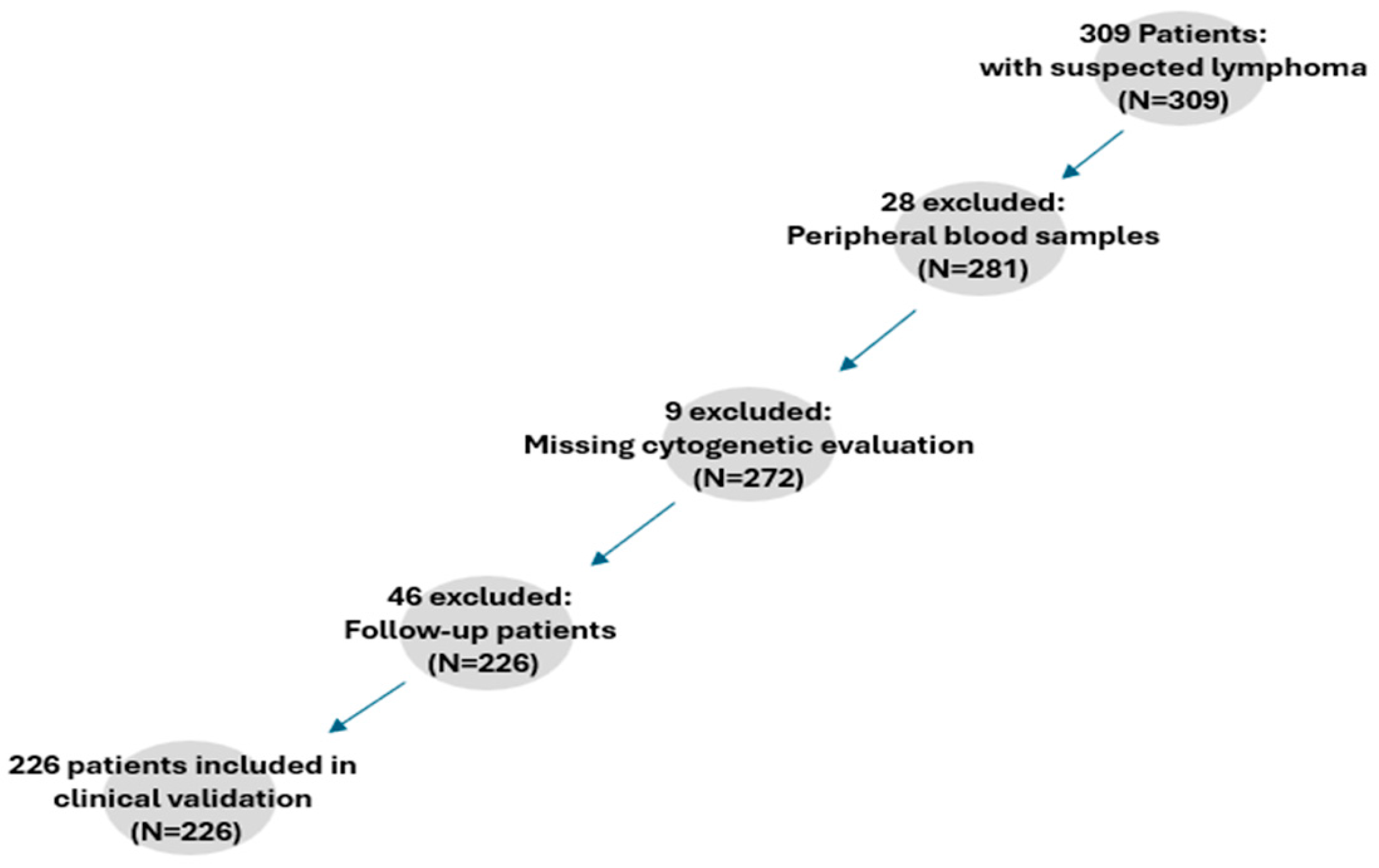
2. Materials and Methods
2.1. Analytical Validation
2.2. Clinical Validation
2.3. Statistical Analysis
3. Results
3.1. Analytical Validation
3.2. NGS Performance in WHO-Relevant B-Cell Lymphomas
3.3. NGS Utility in Lymphoma Exclusion Diagnostics
3.4. NGS Performance in Immunocytologically or Ig-Rearranged Confirmed Lymphomas
3.5. NGS Performance in Cases with Limited or Non-Interpretable Cytogenetics
3.6. NGS Performance in Cases with Limited Morphological Assessment
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Alaggio, R.; Amador, C.; Anagnostopoulos, I.; Attygalle, A.D.; Araujo, I.B.O.; Berti, E.; Bhagat, G.; Borges, A.M.; Boyer, D.; Calaminici, M.; et al. The 5th edition of the World Health Organization Classification of Haematolymphoid Tumours: Lymphoid Neoplasms. Leukemia 2022, 36, 1720–1748. [Google Scholar] [CrossRef] [PubMed]
- Rodenas Quinonero, I.; Marco-Ayala, J.; Chen-Liang, T.H.; de la Cruz-Vicente, F.; Baumann, T.; Navarro, J.T.; Martin Garcia-Sancho, A.; Martin-Santos, T.; Lopez-Jimenez, J.; Andreu, R.; et al. The Value of Bone Marrow Assessment by FDG PET/CT, Biopsy and Aspirate in the Upfront Evaluation of Mantle Cell Lymphoma: A Nationwide Cohort Study. Cancers 2024, 16, 4189. [Google Scholar] [CrossRef] [PubMed]
- Landsburg, D.J. Improving Cure Rates for Patients with Newly Diagnosed Large B-Cell Lymphomas: Targeted Therapies for High-Risk Pathologic Subgroups as Defined by Clinical Laboratory Testing. Cancers 2024, 17, 18. [Google Scholar] [CrossRef] [PubMed]
- Preijers, F.W.; van der Velden, V.H.; Preijers, T.; Brooimans, R.A.; Marijt, E.; Homburg, C.; van Montfort, K.; Gratama, J.W.; on behalf of the Section Immunological and Molecular Cell Diagnostics of the Foundation for Quality Assessment in Medical Laboratories (SKML). Fifteen years of external quality assessment in leukemia/lymphoma immunophenotyping in The Netherlands and Belgium: A way forward. Cytometry B Clin. Cytom. 2016, 90, 267–278. [Google Scholar] [CrossRef]
- Alvarez Flores, M.B.; Sopena Corvinos, M.; Guillen Santos, R.; Cava Valenciano, F. High-Sensitivity Flow Cytometry for the Reliable Detection of Measurable Residual Disease in Hematological Malignancies in Clinical Laboratories. Diseases 2024, 12, 338. [Google Scholar] [CrossRef]
- Win, K.T.; Hsieh, Y.C.; Wu, H.C.; Chuang, S.S. Nodal Low-Grade B-Cell Lymphoma Co-Expressing CD5 and CD10 but Not CD23, IRTA1, or Cyclin D1: The Diagnostic Challenge of a Splenic Marginal Zone Lymphoma. Diagnostics 2024, 14, 640. [Google Scholar] [CrossRef]
- Alfaifi, A.; Refai, M.Y.; Alsaadi, M.; Bahashwan, S.; Malhan, H.; Al-Kahiry, W.; Dammag, E.; Ageel, A.; Mahzary, A.; Albiheyri, R.; et al. Metabolomics: A New Era in the Diagnosis or Prognosis of B-Cell Non-Hodgkin’s Lymphoma. Diagnostics 2023, 13, 861. [Google Scholar] [CrossRef]
- Lopez, C.; Fischer, A.; Rosenwald, A.; Siebert, R.; Ott, G.; Kurz, K.S. Genetic alterations in mature B- and T-cell lymphomas—A practical guide to WHO-HAEM5. Med. Genet. 2024, 36, 59–73. [Google Scholar] [CrossRef]
- Wan Mohamad Zamri, W.N.; Mohd Yunus, N.; Abdul Aziz, A.A.; Zulkipli, N.N.; Sulong, S. Perspectives on the Application of Cytogenomic Approaches in Chronic Lymphocytic Leukaemia. Diagnostics 2023, 13, 964. [Google Scholar] [CrossRef]
- Heimann, P.; Dewispelaere, L. Indications of next-generation sequencing in non-Hodgkin’s lymphoma. Curr. Opin. Oncol. 2020, 32, 391–397. [Google Scholar] [CrossRef]
- Onaindia, A.; Medeiros, L.J.; Patel, K.P. Clinical utility of recently identified diagnostic, prognostic, and predictive molecular biomarkers in mature B-cell neoplasms. Mod. Pathol. 2017, 30, 1338–1366. [Google Scholar] [CrossRef] [PubMed]
- Coupland, S.E.; Du, M.Q.; Ferry, J.A.; de Jong, D.; Khoury, J.D.; Leoncini, L.; Naresh, K.N.; Ott, G.; Siebert, R.; Xerri, L.; et al. The fifth edition of the WHO classification of mature B-cell neoplasms: Open questions for research. J. Pathol. 2024, 262, 255–270. [Google Scholar] [CrossRef] [PubMed]
- Jajosky, A.N.; Havens, N.P.; Sadri, N.; Oduro, K.A.; Moore, E.M.; Beck, R.C.; Meyerson, H.J. Clinical Utility of Targeted Next-Generation Sequencing in the Evaluation of Low-Grade Lymphoproliferative Disorders. Am. J. Clin. Pathol. 2021, 156, 433–444. [Google Scholar] [CrossRef]
- Falini, B.; Martino, G.; Lazzi, S. A comparison of the International Consensus and 5th World Health Organization classifications of mature B-cell lymphomas. Leukemia 2023, 37, 18–34. [Google Scholar] [CrossRef]
- Kim, J.; Park, W.Y.; Kim, N.K.D.; Jang, S.J.; Chun, S.M.; Sung, C.O.; Choi, J.; Ko, Y.H.; Choi, Y.L.; Shim, H.S.; et al. Good Laboratory Standards for Clinical Next-Generation Sequencing Cancer Panel Tests. J. Pathol. Transl. Med. 2017, 51, 191–204. [Google Scholar] [CrossRef]
- Zanelli, M.; Sanguedolce, F.; Zizzo, M.; Ricci, S.; Bisagni, A.; Palicelli, A.; Fragliasso, V.; Donati, B.; Broggi, G.; Boutas, I.; et al. A Diagnostic Approach in Large B-Cell Lymphomas According to the Fifth World Health Organization and International Consensus Classifications and a Practical Algorithm in Routine Practice. Int. J. Mol. Sci. 2024, 25, 13213. [Google Scholar] [CrossRef]
- Hunter, Z.R.; Xu, L.; Yang, G.; Tsakmaklis, N.; Vos, J.M.; Liu, X.; Chen, J.; Manning, R.J.; Chen, J.G.; Brodsky, P.; et al. Transcriptome sequencing reveals a profile that corresponds to genomic variants in Waldenstrom macroglobulinemia. Blood 2016, 128, 827–838. [Google Scholar] [CrossRef]
- Ngo, V.N.; Young, R.M.; Schmitz, R.; Jhavar, S.; Xiao, W.; Lim, K.H.; Kohlhammer, H.; Xu, W.; Yang, Y.; Zhao, H.; et al. Oncogenically active MYD88 mutations in human lymphoma. Nature 2011, 470, 115–119. [Google Scholar] [CrossRef]
- Treon, S.P.; Xu, L.; Guerrera, M.L.; Jimenez, C.; Hunter, Z.R.; Liu, X.; Demos, M.; Gustine, J.; Chan, G.; Munshi, M.; et al. Genomic Landscape of Waldenstrom Macroglobulinemia and Its Impact on Treatment Strategies. J. Clin. Oncol. 2020, 38, 1198–1208. [Google Scholar] [CrossRef]
- Tiacci, E.; Pettirossi, V.; Schiavoni, G.; Falini, B. Genomics of Hairy Cell Leukemia. J. Clin. Oncol. 2017, 35, 1002–1010. [Google Scholar] [CrossRef]
- Oscier, D.; Stamatopoulos, K.; Mirandari, A.; Strefford, J. The Genomics of Hairy Cell Leukaemia and Splenic Diffuse Red Pulp Lymphoma. Cancers 2022, 14, 697. [Google Scholar] [CrossRef]
- Matutes, E. Diagnostic and therapeutic challenges in hairy cell leukemia-variant: Where are we in 2021? Expert Rev. Hematol. 2021, 14, 355–363. [Google Scholar] [CrossRef]
- Xi, L.; Arons, E.; Navarro, W.; Calvo, K.R.; Stetler-Stevenson, M.; Raffeld, M.; Kreitman, R.J. Both variant and IGHV4-34-expressing hairy cell leukemia lack the BRAF V600E mutation. Blood 2012, 119, 3330–3332. [Google Scholar] [CrossRef]
- van der Burg, M.; Poulsen, T.S.; Hunger, S.P.; Beverloo, H.B.; Smit, E.M.; Vang-Nielsen, K.; Langerak, A.W.; van Dongen, J.J. Split-signal FISH for detection of chromosome aberrations in acute lymphoblastic leukemia. Leukemia 2004, 18, 895–908. [Google Scholar] [CrossRef]
- Zhang, Q.; Bai, S.; Vance, G.H. Molecular genetic tests for FLT3, NPM1, and CEBPA in acute myeloid leukemia. Methods Mol. Biol. 2013, 999, 105–121. [Google Scholar] [CrossRef]
- Kayser, S.; Levis, M.J. The clinical impact of the molecular landscape of acute myeloid leukemia. Haematologica 2023, 108, 308–320. [Google Scholar] [CrossRef]
- Morschhauser, F.; Tilly, H.; Chaidos, A.; McKay, P.; Phillips, T.; Assouline, S.; Batlevi, C.L.; Campbell, P.; Ribrag, V.; Damaj, G.L.; et al. Tazemetostat for patients with relapsed or refractory follicular lymphoma: An open-label, single-arm, multicentre, phase 2 trial. Lancet Oncol. 2020, 21, 1433–1442. [Google Scholar] [CrossRef]
- Galimberti, S.; Genuardi, E.; Mazziotta, F.; Iovino, L.; Morabito, F.; Grassi, S.; Ciabatti, E.; Guerrini, F.; Petrini, M. The Minimal Residual Disease in Non-Hodgkin’s Lymphomas: From the Laboratory to the Clinical Practice. Front. Oncol. 2019, 9, 528. [Google Scholar] [CrossRef]
- Santisteban-Espejo, A.; Bernal-Florindo, I.; Perez-Requena, J.; Atienza-Cuevas, L.; Moran-Sanchez, J.; Fernandez-Valle, M.D.C.; Romero-Garcia, R.; Garcia-Rojo, M. The Need for Standardization in Next-Generation Sequencing Studies for Classic Hodgkin Lymphoma: A Systematic Review. Diagnostics 2022, 12, 963. [Google Scholar] [CrossRef]
- Chihara, D.; Nastoupil, L.J.; Flowers, C.R. Clinical approaches for integrating machine learning for patients with lymphoma: Current strategies and future perspectives. Br. J. Haematol. 2023, 202, 219–229. [Google Scholar] [CrossRef]
- Yuan, J.; Zhang, Y.; Wang, X. Application of machine learning in the management of lymphoma: Current practice and future prospects. Digit. Health 2024, 10, 20552076241247963. [Google Scholar] [CrossRef] [PubMed]
- Long, B.; Lai, S.W.; Wu, J.; Bellur, S. Predicting Phase 1 Lymphoma Clinical Trial Durations Using Machine Learning: An In-Depth Analysis and Broad Application Insights. Clin. Pract. 2023, 14, 69–88. [Google Scholar] [CrossRef] [PubMed]


| Diagnosis | Non-Mutated | Mutated | Total | % Non-Mutated | % Mutated |
|---|---|---|---|---|---|
| CLL | 11 | 18 | 29 | 37.93 | 62.07 |
| FL | 11 | 0 | 11 | 100.00 | 0.00 |
| HCL | 0 | 17 | 17 | 0.00 | 100.00 |
| HGBCL | 3 | 3 | 6 | 50.00 | 50.00 |
| LPL | 0 | 12 | 12 | 0.00 | 100.00 |
| MCL | 6 | 4 | 10 | 60.00 | 40.00 |
| MZL | 5 | 6 | 11 | 45.45 | 54.55 |
| SBLPN | 4 | 0 | 4 | 100.00 | 0.00 |
| B-NHL NOS | 3 | 2 | 5 | 60.00 | 40.00 |
| BCOR | BIRC3 | BRAF | BTK | CXCR4 | KRAS | MYD88 | NOTCH1 | NOTCH2 | NRAS | SF3B1 | TP53 | PLCG2 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| B-NOS | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 |
| CLL | 2 | 6 | 1 | 1 | 0 | 0 | 0 | 5 | 0 | 1 | 7 | 6 | 0 |
| HCL | 0 | 0 | 17 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| HGBCL | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 |
| LPL | 0 | 0 | 0 | 0 | 2 | 0 | 11 | 0 | 0 | 0 | 0 | 0 | 0 |
| MCL | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 1 | 0 | 0 | 0 | 3 | 0 |
| MZL | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 3 | 0 | 0 | 3 | 0 |
| 0 | 1 | 2 | 3 | 4 | 6 | 8 | 12 | 17 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Strasser, B.; Mustafa, S.; Seier, J.; Wimmer, E.; Tomasits, J. Diagnostic Implications of NGS-Based Molecular Profiling in Mature B-Cell Lymphomas with Potential Bone Marrow Involvement. Diagnostics 2025, 15, 727. https://doi.org/10.3390/diagnostics15060727
Strasser B, Mustafa S, Seier J, Wimmer E, Tomasits J. Diagnostic Implications of NGS-Based Molecular Profiling in Mature B-Cell Lymphomas with Potential Bone Marrow Involvement. Diagnostics. 2025; 15(6):727. https://doi.org/10.3390/diagnostics15060727
Chicago/Turabian StyleStrasser, Bernhard, Sebastian Mustafa, Josef Seier, Erich Wimmer, and Josef Tomasits. 2025. "Diagnostic Implications of NGS-Based Molecular Profiling in Mature B-Cell Lymphomas with Potential Bone Marrow Involvement" Diagnostics 15, no. 6: 727. https://doi.org/10.3390/diagnostics15060727
APA StyleStrasser, B., Mustafa, S., Seier, J., Wimmer, E., & Tomasits, J. (2025). Diagnostic Implications of NGS-Based Molecular Profiling in Mature B-Cell Lymphomas with Potential Bone Marrow Involvement. Diagnostics, 15(6), 727. https://doi.org/10.3390/diagnostics15060727





