Prognostic Significance of DNAJB4 Expression in Gastric Cancer: Correlation with CD31, Caspase-3, and Tumor Progression
Abstract
1. Introduction
2. Materials and Methods
2.1. Patients and Sample Collection
2.2. Tissue Microarray Preparation and Immunohistochemistry
2.3. Antibodies
2.4. Statistical Analysis
3. Results
3.1. Patient Characteristics
3.2. Association of DNAJB4 Expression with Protein and Biomarker Levels
3.3. Factors Associated with DNAJB4 Expression
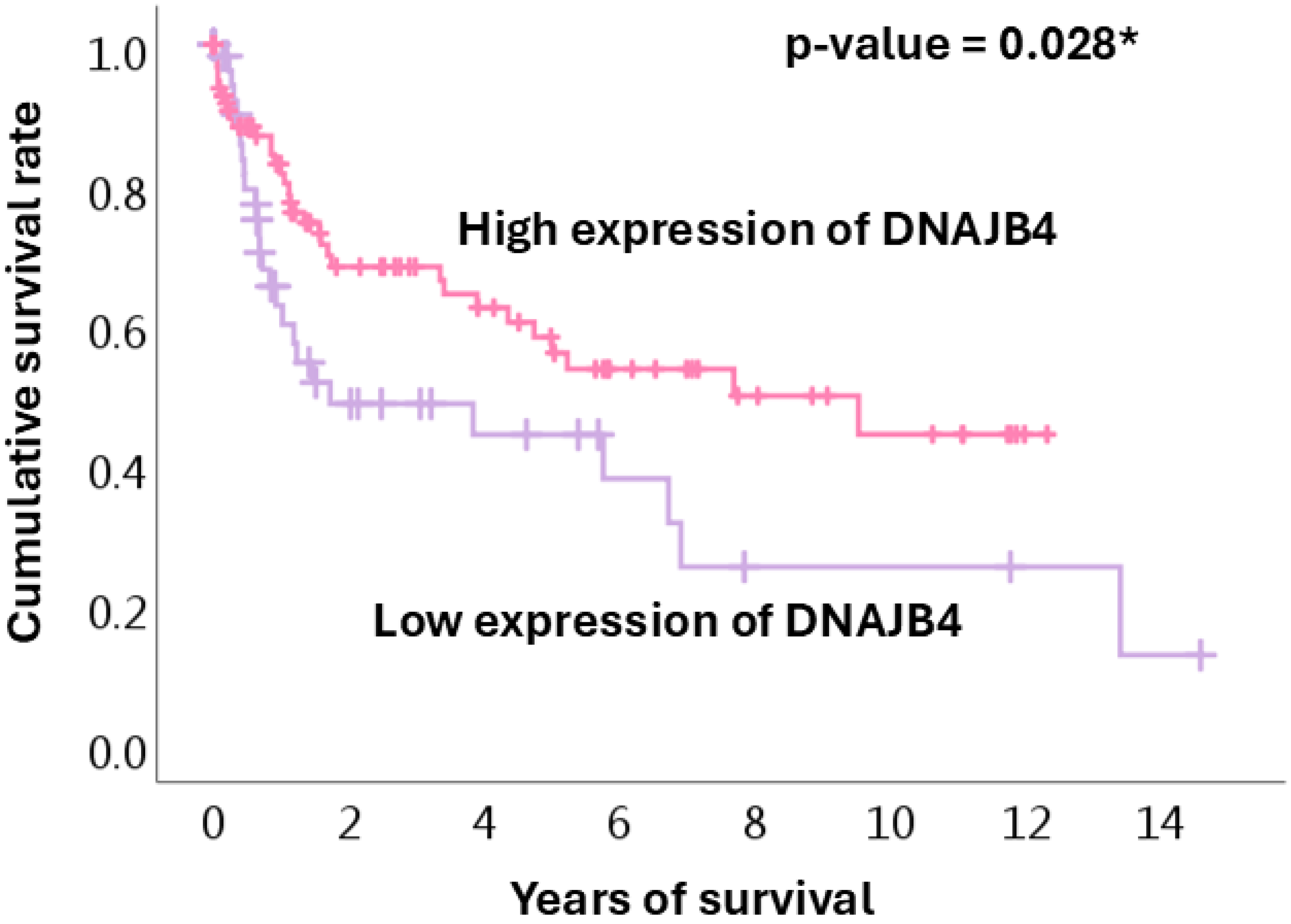
3.4. Survival Analysis
3.5. Role of DNAJB4 in Gastric Cancer by Stage
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Grantham, T.; Ramachandran, R.; Parvataneni, S.; Budh, D.; Gollapalli, S.; Gaduputi, V. Epidemiology of Gastric Cancer: Global Trends, Risk Factors and Premalignant Conditions. J. Community Hosp. Intern. Med. Perspect. 2023, 13, 100–106. [Google Scholar] [CrossRef]
- Inoue, M. Epidemiology of Gastric Cancer-Changing Trends and Global Disparities. Cancers 2024, 16, 2948. [Google Scholar] [CrossRef] [PubMed]
- Joshi, S.S.; Badgwell, B.D. Current treatment and recent progress in gastric cancer. CA Cancer J. Clin. 2021, 71, 264–279. [Google Scholar] [CrossRef]
- Kang, K.; Bagaoisan, M.A.; Zhang, Y. Unveiling the Younger Face of Gastric Cancer: A Comprehensive Review of Epidemiology, Risk Factors, and Prevention Strategies. Cureus 2024, 16, e62826. [Google Scholar] [CrossRef] [PubMed]
- Rocken, C. Predictive biomarkers in gastric cancer. J. Cancer Res. Clin. Oncol. 2023, 149, 467–481. [Google Scholar] [CrossRef]
- Smyth, E.C.; Nilsson, M.; Grabsch, H.I.; van Grieken, N.C.; Lordick, F. Gastric cancer. Lancet 2020, 396, 635–648. [Google Scholar] [CrossRef] [PubMed]
- Lopez, M.J.; Carbajal, J.; Alfaro, A.L.; Saravia, L.G.; Zanabria, D.; Araujo, J.M.; Quispe, L.; Zevallos, A.; Buleje, J.L.; Cho, C.E.; et al. Characteristics of gastric cancer around the world. Crit. Rev. Oncol. Hematol. 2023, 181, 103841. [Google Scholar] [CrossRef]
- Machlowska, J.; Baj, J.; Sitarz, M.; Maciejewski, R.; Sitarz, R. Gastric Cancer: Epidemiology, Risk Factors, Classification, Genomic Characteristics and Treatment Strategies. Int. J. Mol. Sci. 2020, 21, 4012. [Google Scholar] [CrossRef]
- Gomez, D.; Feng, J.J.; Cheok, S.; Shah, I.; Dicharry, H.; Cote, D.J.; Briggs, R.G.; Guerra, G.A.; Peterson, R.; Salhia, B.; et al. Incidence of brain metastasis according to patient race and primary cancer origin: A systematic review. J. Neuro-oncol. 2024, 169, 457–467. [Google Scholar] [CrossRef]
- Lewis, D.; Jimenez, L.; Mansour, M.H.; Horton, S.; Wong, W.W.L. A Systematic Review of Cost-Effectiveness Studies on Gastric Cancer Screening. Cancers 2024, 16, 2353. [Google Scholar] [CrossRef]
- Hoe, K.L.; Won, M.; Chung, K.S.; Jang, Y.J.; Lee, S.B.; Kim, D.U.; Lee, J.W.; Yun, J.H.; Yoo, H.S. Isolation of a new member of DnaJ-like heat shock protein 40 (Hsp40) from human liver. Biochim. Biophys. Acta 1998, 1383, 4–8. [Google Scholar] [CrossRef] [PubMed]
- Ohtsuka, K.; Hata, M. Mammalian HSP40/DNAJ homologs: Cloning of novel cDNAs and a proposal for their classification and nomenclature. Cell Stress Chaperones 2000, 5, 98–112. [Google Scholar] [CrossRef] [PubMed]
- Simoes-Correia, J.; Silva, D.I.; Melo, S.; Figueiredo, J.; Caldeira, J.; Pinto, M.T.; Girao, H.; Pereira, P.; Seruca, R. DNAJB4 molecular chaperone distinguishes WT from mutant E-cadherin, determining their fate in vitro and in vivo. Hum. Mol. Genet. 2014, 23, 2094–2105. [Google Scholar] [CrossRef] [PubMed]
- Mo, L.; Liu, J.; Yang, Z.; Gong, X.; Meng, F.; Zou, R.; Hou, L.; Fang, F. DNAJB4 identified as a potential breast cancer marker: Evidence from bioinformatics analysis and basic experiments. Gland. Surg. 2020, 9, 1955–1972. [Google Scholar] [CrossRef]
- Fang, F.; Mo, L.; Pan, X.; Yang, Z.; Huang, H.; Zhu, L.; Wang, Y.; Jiang, G. DNAJB4 promotes triple-negative breast cancer cell apoptosis via activation of the Hippo signaling pathway. Discov. Oncol. 2023, 14, 40. [Google Scholar] [CrossRef]
- Chen, Y.; Li, J.; Pu, L.; Hu, J.; Fang, L.; Zhou, F.; Zhang, H.; Yang, Y.; Rong, X.; Deng, S.; et al. DNAJB4 suppresses breast cancer progression and promotes tumor immunity by regulating the Hippo signaling pathway. Discov. Oncol. 2023, 14, 144. [Google Scholar] [CrossRef]
- She, K.; Yu, S.; He, S.; Wang, W.; Chen, B. CircRNA 0009043 suppresses non-small-cell lung cancer development via targeting the miR-148a-3p/DNAJB4 axis. Biomark. Res. 2022, 10, 61. [Google Scholar] [CrossRef]
- Park, E.G.; Lee, D.H.; Kim, W.R.; Lee, Y.J.; Bae, W.H.; Kim, J.M.; Shin, H.J.; Ha, H.; Yi, J.M.; Cho, S.G.; et al. Human Endogenous Retrovirus-H-Derived miR-4454 Inhibits the Expression of DNAJB4 and SASH1 in Non-Muscle-Invasive Bladder Cancer. Genes 2023, 14, 1410. [Google Scholar] [CrossRef]
- Fang, S.Q.; Liu, Y.H.; Zhao, K.P.; Zhang, H.X.; Wang, H.W.; Deng, Y.H.; Zhou, Y.X.; Ge, G.B.; Ni, H.M.; Chen, Q.L. Transcriptional profiling and network pharmacology analysis identify the potential biomarkers from Chinese herbal formula Huosu Yangwei Formula treated gastric cancer in vivo. Chin. J. Nat. Med. 2021, 19, 944–953. [Google Scholar] [CrossRef]
- Acun, T.; Doberstein, N.; Habermann, J.K.; Gemoll, T.; Thorns, C.; Oztas, E.; Ried, T. HLJ1 (DNAJB4) Gene Is a Novel Biomarker Candidate in Breast Cancer. OMICS 2017, 21, 257–265. [Google Scholar] [CrossRef]
- Chen, C.H.; Chang, W.H.; Su, K.Y.; Ku, W.H.; Chang, G.C.; Hong, Q.S.; Hsiao, Y.J.; Chen, H.C.; Chen, H.Y.; Wu, R.; et al. HLJ1 is an endogenous Src inhibitor suppressing cancer progression through dual mechanisms. Oncogene 2016, 35, 5674–5685. [Google Scholar] [CrossRef] [PubMed]
- Eskandari, E.; Eaves, C.J. Paradoxical roles of caspase-3 in regulating cell survival, proliferation, and tumorigenesis. J. Cell Biol. 2022, 221, e202201159. [Google Scholar] [CrossRef] [PubMed]
- Thirupathi, A.; Chang, Y.Z. Role of AMPK and its molecular intermediates in subjugating cancer survival mechanism. Life Sci. 2019, 227, 30–38. [Google Scholar] [CrossRef]
- Fan, F.; Mo, H.; Zhang, H.; Dai, Z.; Wang, Z.; Qu, C.; Liu, F.; Zhang, L.; Luo, P.; Zhang, J.; et al. HOXA5: A crucial transcriptional factor in cancer and a potential therapeutic target. Biomed. Pharmacother. 2022, 155, 113800. [Google Scholar] [CrossRef] [PubMed]
- Kloppel, G.; La Rosa, S. Ki67 labeling index: Assessment and prognostic role in gastroenteropancreatic neuroendocrine neoplasms. Virchows Arch. 2018, 472, 341–349. [Google Scholar] [CrossRef]
- Smith, J.P.; Nadella, S.; Osborne, N. Gastrin and Gastric Cancer. Cell Mol. Gastroenterol. Hepatol. 2017, 4, 75–83. [Google Scholar] [CrossRef]
- Ashrafizadeh, M.; Zarrabi, A.; Orouei, S.; Zarrin, V.; Rahmani Moghadam, E.; Zabolian, A.; Mohammadi, S.; Hushmandi, K.; Gharehaghajlou, Y.; Makvandi, P.; et al. STAT3 Pathway in Gastric Cancer: Signaling, Therapeutic Targeting and Future Prospects. Biology 2020, 9, 126. [Google Scholar] [CrossRef]
- Azimi, M.; Totonchi, M.; Ebrahimi, M. Determining The Role of MicroRNAs in Self-Renewal, Metastasis and Resistance to Drugs in Human Gastric Cancer Based on Data Mining Approaches: A Systematic Review. Cell J. 2022, 24, 1–6. [Google Scholar]
- Wang, X.M.; Li, Q.Y.; Ren, L.L.; Liu, Y.M.; Wang, T.S.; Mu, T.C.; Fu, S.; Liu, C.; Xiao, J.Y. Effects of MCRS1 on proliferation, migration, invasion, and epithelial mesenchymal transition of gastric cancer cells by interacting with Pkmyt1 protein kinase. Cell Signal. 2019, 59, 171–181. [Google Scholar] [CrossRef]
- Han, X.; Liu, T.; Zhai, J.; Liu, C.; Wang, W.; Nie, C.; Wang, Q.; Zhu, X.; Zhou, H.; Tian, W. Association between EPHA5 methylation status in peripheral blood leukocytes and the risk and prognosis of gastric cancer. PeerJ 2022, 10, e13774. [Google Scholar] [CrossRef]
- Fresno Vara, J.A.; Casado, E.; de Castro, J.; Cejas, P.; Belda-Iniesta, C.; Gonzalez-Baron, M. PI3K/Akt signalling pathway and cancer. Cancer Treat. Rev. 2004, 30, 193–204. [Google Scholar] [CrossRef]
- Raica, M.; Cimpean, A.M.; Ribatti, D. Angiogenesis in pre-malignant conditions. Eur. J. Cancer 2009, 45, 1924–1934. [Google Scholar] [CrossRef] [PubMed]
- Mirzaei, S.; Saghari, S.; Bassiri, F.; Raesi, R.; Zarrabi, A.; Hushmandi, K.; Sethi, G.; Tergaonkar, V. NF-kappaB as a regulator of cancer metastasis and therapy response: A focus on epithelial-mesenchymal transition. J. Cell. Physiol. 2022, 237, 2770–2795. [Google Scholar] [CrossRef]
- Ouyang, J.; Li, J.; Li, D.; Jiang, J.; Hao, T.; Xia, Y.; Lu, X.; Zhang, C.; He, Y. IGF2BP2 Promotes Epithelial to Mesenchymal Transition and Metastasis through Stabilizing HMGA1 mRNA in Gastric Cancer. Cancers 2022, 14, 5381. [Google Scholar] [CrossRef] [PubMed]
- Blagih, J.; Buck, M.D.; Vousden, K.H. p53, cancer and the immune response. J. Cell Sci. 2020, 133, jcs237453. [Google Scholar] [CrossRef] [PubMed]
- De Re, V.; Alessandrini, L.; Brisotto, G.; Caggiari, L.; De Zorzi, M.; Casarotto, M.; Miolo, G.; Puglisi, F.; Garattini, S.K.; Lonardi, S.; et al. HER2-CDH1 Interaction via Wnt/B-Catenin Is Associated with Patients’ Survival in HER2-Positive Metastatic Gastric Adenocarcinoma. Cancers 2022, 14, 1266. [Google Scholar] [CrossRef]
- Pan, D.; Hao, J.; Wu, T.; Shen, T.; Yu, K.; Li, Q.; Hu, R.; Yang, Z.; Li, Y. Sodium Butyrate Inhibits the Malignant Proliferation of Colon Cancer Cells via the miR-183/DNAJB4 Axis. Biochem. Genet. 2024, 62, 4174–4190. [Google Scholar] [CrossRef]
- Fukita, Y.; Yasuda, I.; Ishibashi, H.; Asaki, T.; Adachi, S.; Toyomizu, M.; Takeda, T.; Suematsu, N. Multifocal angiosarcoma of the gastrointestinal tract: A case report. Nihon Shokakibyo Gakkai Zasshi 2017, 114, 1665–1674. [Google Scholar]
- Kapagan, T.; Bulut, N.; Erdem, G.U.; Yildirim, S.; Erdem, Z.B.; Sahin, H. Synchronous Double Primary Angiosarcoma Originating from the Stomach and Rectum: A Case Report and a Literature Review. Arch. Iran Med. 2024, 27, 168–173. [Google Scholar] [CrossRef]
- Matsuno, K.; Kanazawa, Y.; Kakinuma, D.; Hagiwara, N.; Ando, F.; Masuda, Y.; Fujita, I.; Arai, H.; Nomura, T.; Kato, S.; et al. Preoperatively diagnosed gastric collision tumor with mixed adenocarcinoma and gastrointestinal stromal tumor: A case report and literature review. Clin. J. Gastroenterol. 2021, 14, 494–499. [Google Scholar] [CrossRef]
- Nefedova, N.A.; Kharlova, O.A.; Danilova, N.V.; Malkov, P.G.; Gaifullin, N.M. Markers of angiogenesis in tumor growt. Arkh. Patol. 2016, 78, 55–63. [Google Scholar] [CrossRef] [PubMed]
- Spychala, A.; Nowaczyk, P.; Budnicka, A.; Antoniewicz, E.; Murawa, D. Intramural gastric hematoma imitating a gastrointestinal stromal tumor—Case report and literature review. Pol. Przegl. Chir. 2017, 89, 62–65. [Google Scholar] [CrossRef] [PubMed]
- Tang, D.; Gao, J.; Wang, S.; Ye, N.; Chong, Y.; Huang, Y.; Wang, J.; Li, B.; Yin, W.; Wang, D. Cancer-associated fibroblasts promote angiogenesis in gastric cancer through galectin-1 expression. Tumour Biol. 2016, 37, 1889–1899. [Google Scholar] [CrossRef]
- Blank, S.; Deck, C.; Dreikhausen, L.; Weichert, W.; Giese, N.; Falk, C.; Schmidt, T.; Ott, K. Angiogenic and growth factors in gastric cancer. J. Surg. Res. 2015, 194, 420–429. [Google Scholar] [CrossRef]
- Huang, K.H.; Fang, W.L.; Li, A.F.; Liang, P.H.; Wu, C.W.; Shyr, Y.M.; Yang, M.H. Caspase-3, a key apoptotic protein, as a prognostic marker in gastric cancer after curative surgery. Int. J. Surg. 2018, 52, 258–263. [Google Scholar] [CrossRef]
- Gryko, M.; Lukaszewicz-Zajac, M.; Guzinska-Ustymowicz, K.; Kucharewicz, M.; Mroczko, B.; Algirdas, U. The caspase-8 and procaspase-3 expression in gastric cancer and non-cancer mucosa in relation to clinico-morphological factors and some apoptosis-associated proteins. Adv. Med. Sci. 2023, 68, 94–100. [Google Scholar] [CrossRef] [PubMed]
- Lin, S.Y.; Hsueh, C.M.; Yu, S.L.; Su, C.C.; Shum, W.Y.; Yeh, K.C.; Chang, G.C.; Chen, J.J. HLJ1 is a novel caspase-3 substrate and its expression enhances UV-induced apoptosis in non-small cell lung carcinoma. Nucleic Acids Res. 2010, 38, 6148–6158. [Google Scholar] [CrossRef]
- Hu, Q.; Peng, J.; Liu, W.; He, X.; Cui, L.; Chen, X.; Yang, M.; Liu, H.; Liu, S.; Wang, H. Elevated cleaved caspase-3 is associated with shortened overall survival in several cancer types. Int. J. Clin. Exp. Pathol. 2014, 7, 5057–5070. [Google Scholar]

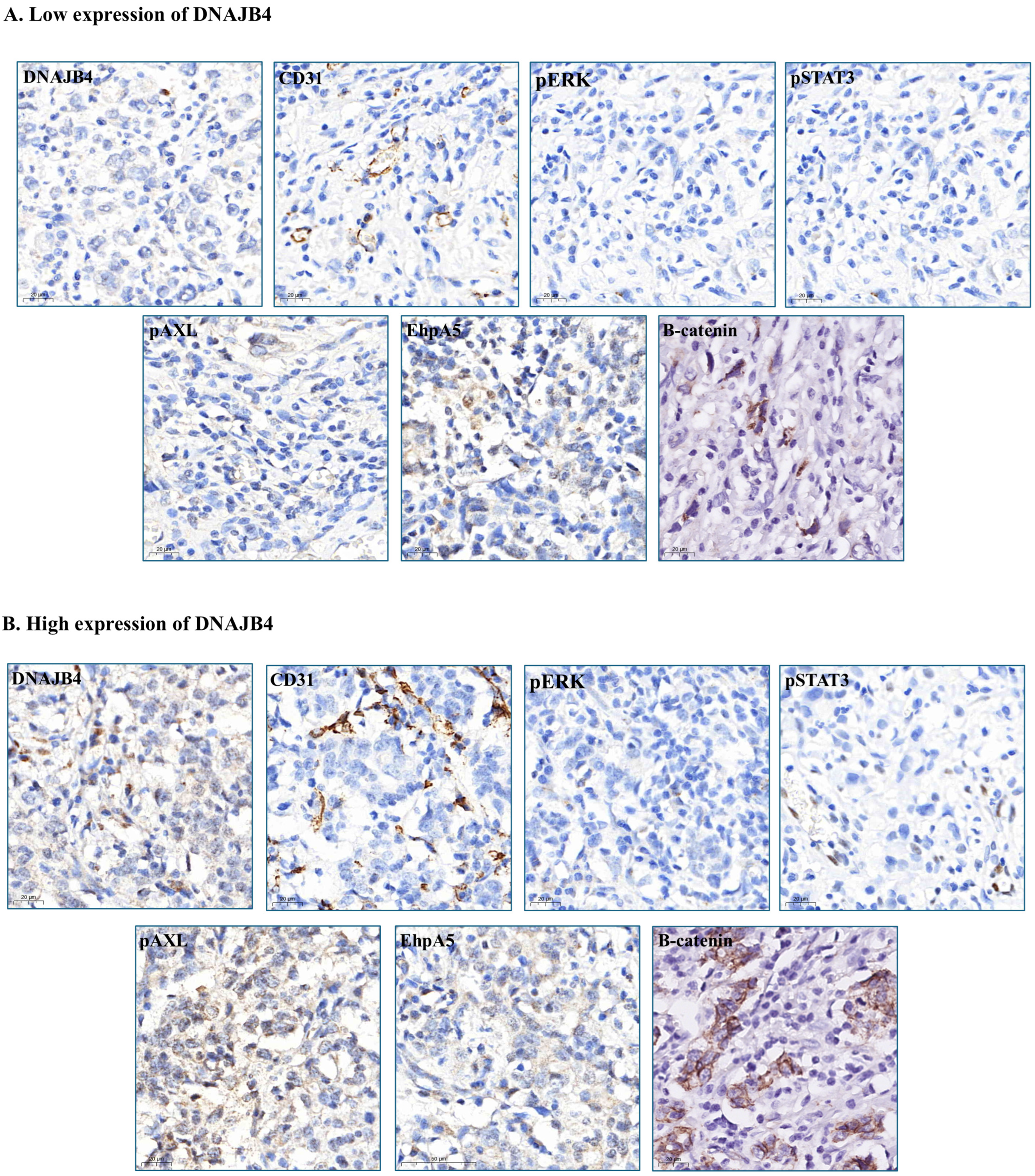
| Characteristic | Low DNAJB4 Expression Group | High DNAJB4 Expression | Total | p-Value |
|---|---|---|---|---|
| Sex | 0.024 * | |||
| Female | 48 (78.7%) | 79 (62.2%) | 127 (67.6%) | |
| Male | 13 (21.3%) | 48 (37.8%) | 61 (32.4%) | |
| Age (years) | 75.0 (69.0–80.0) | 74.0 (60.0–80.0) | 75.0 (63.0–80.0) | 0.299 |
| Differentiation (grade) | 0.911 | |||
| Well | 2 (3.3%) | 5 (4.0%) | 7 (3.8%) | |
| Moderate | 16 (26.7%) | 30 (24.0%) | 46 (24.9%) | |
| Poor | 42 (31.8%) | 90 (72.0%) | 132 (71.4%) | |
| Tumor size (cm) | 5.0 (3.9–7.0) | 4.1 (2.5–6.5) | 4.5 (2.9–7.0) | 0.042 * |
| Cancer stage | 0.009 ** | |||
| I–II | 17 (27.9%) | 61 (48.0%) | 78 (41.5%) | |
| III–IV | 44 (72.1%) | 66 (52.0%) | 110 (58.5%) |
| Biomarker | Low DNAJB4 Expression Group | High DNAJB4 Expression Group | Total | p-Value |
|---|---|---|---|---|
| Caspase-3 | 4.2 (2.8–5.5) | 4.7 (3.2–7.4) | 4.4 (3.2–6.6) | 0.051 |
| Ki67 | 14.8 (4.0–41.3) | 11.9 (3.4–32.7) | 13.61 (3.5–36.0) | 0.375 |
| CD31 | 15.7 (9.3–24.0) | 22.8 (14.8–37.1) | 21.2 (13.1–34.4) | <0.001 *** |
| E-cad | 106.9 (102.2–118.2) | 104.8 (101.4–111.0) | 105.6 (101.8–112.7) | 0.052 |
| N-cad | 5.5 (2.3–13.1) | 7.6 (3.7–11.5) | 6.9 (3.0–11.8) | 0.056 |
| Fibronectin | 36.7 (7.7–97.0) | 38.9 (2.5–104.1) | 37.7 (3.6–101.1) | 0.671 |
| pAkt | 10.0 (3.8–17.8) | 10.3 (3.7–30.4) | 10.1 (3.8–26.4) | 0.279 |
| pErk | 0.3 (0.1–1.1) | 1.6 (0.4–5.8) | 0.9 (0.2–3.3) | <0.001 *** |
| pStat3 | 0.1 (0.0–0.3) | 0.3 (0.0–1.6) | 0.2 (0.0–0.7) | 0.001 ** |
| pAMPK | 4.1 (2.9–7.7) | 3.8 (2.6–5.6) | 3.9 (2.6–6.1) | 0.107 |
| pAXL | 1.7 (1.1–4.1) | 2.2 (1.5–4.4) | 2.2 (1.3–4.3) | 0.046 * |
| HMGA1 | 6.4 (0.7–62.4) | 6.9 (1.0–29.0) | 6.7 (1.0–44.9) | 0.740 |
| HOXA5 | 33.7 (8.9–77.8) | 34.8 (12.1–57.0) | 34.2 (11.2–64.2) | 0.902 |
| IGF2BP1 | 43.8 (14.3–64.1) | 42.0 (19.2–61.4) | 43.0 (19.1–63.6) | 0.856 |
| MCRS1 | 109.7 (71.3–165.9) | 102.8 (69.3–143.9) | 103.5 (70.5–146.9) | 0.295 |
| EhpA5 | 5.2 (2.7–9.6) | 10.3 (3.4–28.2) | 7.7 (3.2–19.8) | 0.001 ** |
| p53 | 1.6 (0.0–14.6) | 2.1 (0.4–11.9) | 2.1 (0.3–12.0) | 0.686 |
| β-catenin | 154.0 (77.4–194.8) | 122.3 (29.8–177.2) | 134.2 (37.7–187.2) | 0.027 * |
| Characteristic | Univariable | p-Value | Multivariable | p-Value |
|---|---|---|---|---|
| Sex | ||||
| Female | Reference | Reference | ||
| Male | 2.24 (1.10, 4.56) | 0.026 * | 2.06 (0.91–4.66) | 0.082 |
| Age | 0.98 (0.95–1.00) | 0.088 | 1.00 (0.97–1.03) | 0.885 |
| Differentiation (grade) | ||||
| Well | Reference | Reference | ||
| Moderate | 0.75 (0.13–4.31) | 0.747 | 2.75 (0.33–22.74) | 0.348 |
| Poor | 0.86 (0.16–4.60) | 0.857 | 3.02 (0.36–25.59) | 0.311 |
| Tumor size | 0.92 (0.83–10.2) | 0.095 | 0.96 (0.84–1.09) | 0.494 |
| Cancer stage | ||||
| I and II | Reference | Reference | ||
| III and IV | 0.42 (0.22–0.81) | 0.009 ** | 0.40 (0.17–0.97) | 0.042 * |
| Marker | ||||
| CD31 | 1.04 (1.01–1.06) | 0.002 ** | 1.03 (1.01–1.06) | 0.015 * |
| pErk | 1.14 (1.03–1.26) | 0.015 * | 1.04 (0.95–1.15) | 0.379 |
| pStat3 | 1.97 (1.10–3.55) | 0.024 * | 1.33 (0.84–2.11) | 0.222 |
| pAXL | 1.08 (0.99–1.18) | 0.070 | ||
| EhpA5 | 1.04 (1.02–1.07) | 0.002 ** | 1.02 (0.99–1.05) | 0.203 |
| β-catenin | 1.00 (0.99–1.00) | 0.056 | ||
| (A) | ||
| Characteristic | Multivariable | p-Value |
| Sex | ||
| Female | Reference | |
| Male | 9.61 (1.09–85.09) | 0.042 * |
| Age | 1.01 (0.95–1.07) | 0.742 |
| Differentiation | ||
| Well | Reference | |
| Moderate | 2.28 (0.22–23.16) | 0.486 |
| Poor | 3.80 (0.32–44.89) | 0.289 |
| Tumor size | 0.79 (0.57–1.10) | 0.164 |
| Biomarker | ||
| N-cad | 1.01 (0.99–1.04) | 0.389 |
| MCRS1 | 0.99 (0.97–1.00) | 0.141 |
| (B) | ||
| Characteristic | Multivariable | p-Value |
| Sex | ||
| Female | Reference | |
| Male | 1.16 (0.40–3.37) | 0.788 |
| Age | 0.97 (0.93–1.01) | 0.518 |
| Differentiation (grade) | ||
| Well | Reference | |
| Moderate | 0.67 (0.16–2.81) | 0.583 |
| Poor | N/A | N/A |
| Tumor size | 0.93 (0.78–1.10) | 0.397 |
| Biomarker | ||
| Caspase-3 | 1.20 (1.04–1.39) | 0.013 * |
| CD31 | 1.06 (1.02–1.10) | 0.004 ** |
| E-cad | 0.93 (0.87–0.98) | 0.011 * |
| EhpA5 | 1.04 (1.00–1.07) | 0.033 * |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cheng, C.-Y.; Chen, Y.-L.; Ho, H.; Huang, C.-Y.; Chu, S.-E.; Liang, Y.-J. Prognostic Significance of DNAJB4 Expression in Gastric Cancer: Correlation with CD31, Caspase-3, and Tumor Progression. Diagnostics 2025, 15, 652. https://doi.org/10.3390/diagnostics15060652
Cheng C-Y, Chen Y-L, Ho H, Huang C-Y, Chu S-E, Liang Y-J. Prognostic Significance of DNAJB4 Expression in Gastric Cancer: Correlation with CD31, Caspase-3, and Tumor Progression. Diagnostics. 2025; 15(6):652. https://doi.org/10.3390/diagnostics15060652
Chicago/Turabian StyleCheng, Chiao-Yin, Yen-Lin Chen, Hua Ho, Chun-Yen Huang, Sheng-En Chu, and Yao-Jen Liang. 2025. "Prognostic Significance of DNAJB4 Expression in Gastric Cancer: Correlation with CD31, Caspase-3, and Tumor Progression" Diagnostics 15, no. 6: 652. https://doi.org/10.3390/diagnostics15060652
APA StyleCheng, C.-Y., Chen, Y.-L., Ho, H., Huang, C.-Y., Chu, S.-E., & Liang, Y.-J. (2025). Prognostic Significance of DNAJB4 Expression in Gastric Cancer: Correlation with CD31, Caspase-3, and Tumor Progression. Diagnostics, 15(6), 652. https://doi.org/10.3390/diagnostics15060652






