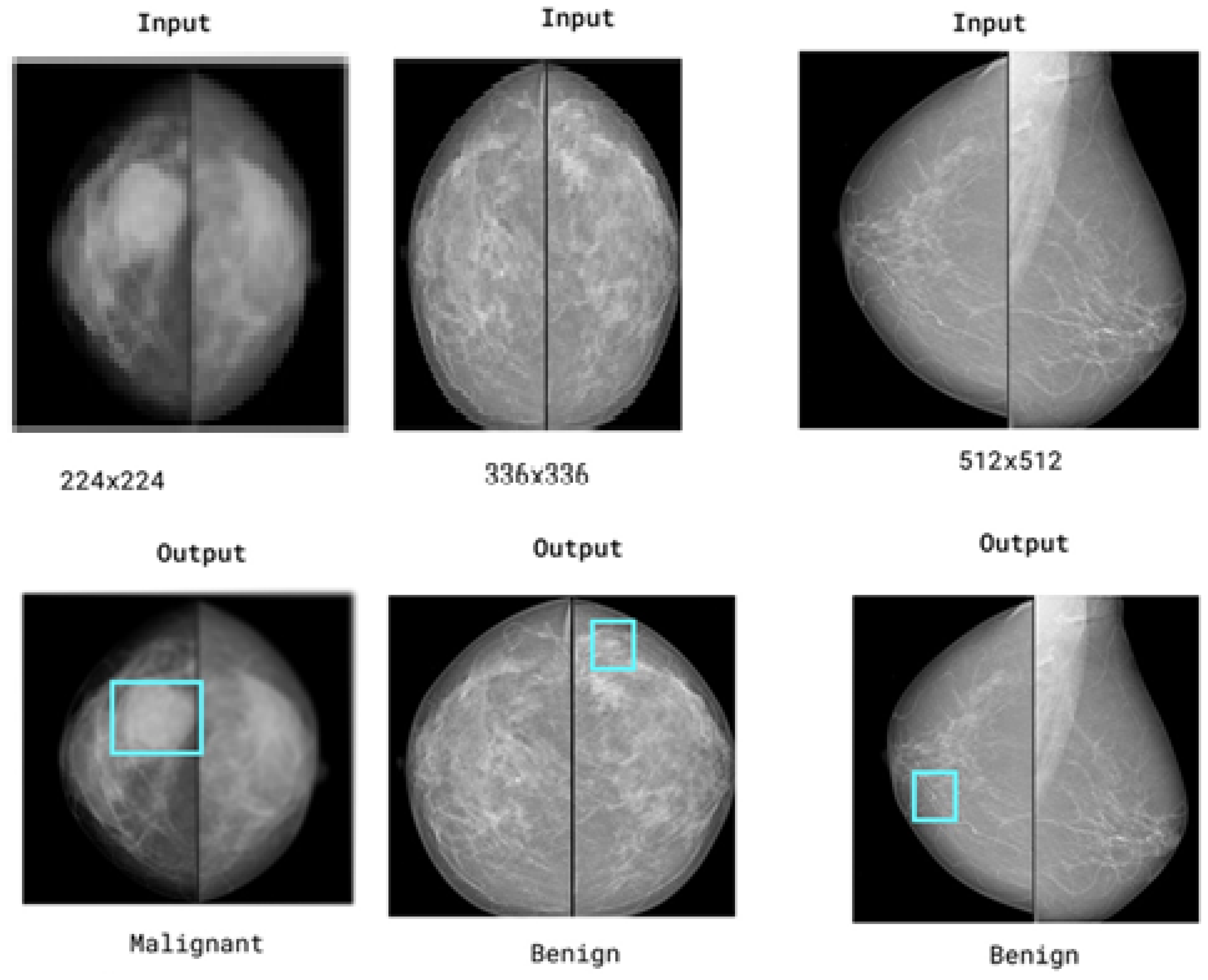
1. Introduction
Breast cancer is still one of the leading causes of morbidity and mortality throughout the world, emphasizing the importance of timely and accurate diagnosis to improve clinical outcomes [
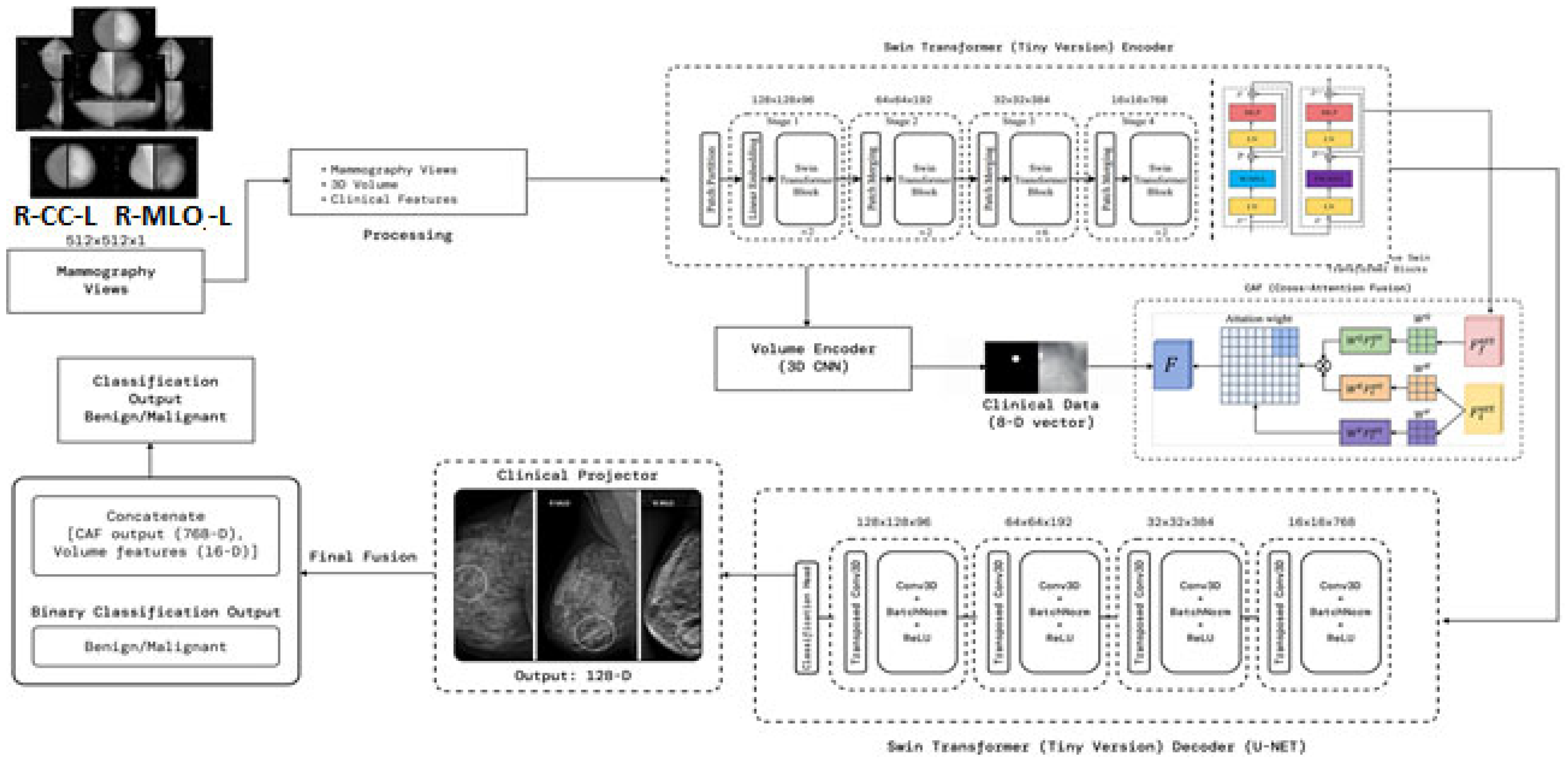
1]. Mammography is the gold standard screening procedure, but future computational solutions are needed to assist radiologists in lesion detection and to provide risk stratification. Although deep learning models, such as convolutional neural networks and transformer-based models, have proven to help improve performance in mammogram classification and segmentation, these methods are typically limited in their ability to integrate multi-view image information, volumetric characteristics, and clinical variables into a single interpretable model. This study presents a cross-attentive multimodal fusion network, CAMF-Net, which unites Swin transformer encoders, three-dimensional convolutional neural network (3D-CNN) volume encoders, and clinical vector projectors in a systematic approach [
2]. We use a cross-attention fusion module to align the different streams of data (as images and prior to decoding in a hierarchical U-Net structure) in a way that enables accurate lesion localization and complete semantic interpretation. The innovative nature of the architecture is further enhanced using multi-scale encoder–decoder fusion and dual-task optimization that involve supervising binary classification from the mammogram data and dense segmentation from the 3D volume objective [
3].
Despite significant advances in deep learning modeling for mammogram analysis, the existing literature indicates a variety of critical limitations that restrict rigorous clinical translation. First, many CNN-based models have suitable receptive fields for pathological volumetric image analysis, but they are unable to capture long-range dependencies and global semantic context that are important for more complex characterization of breast lesions. Second, transformer-based architectures enable improved modeling of global attention but are often an excessive computational burden, and they do not permit fusing multi-modal data streams (e.g., mammographic views, volumetric data, and clinical features) into a joint architecture. Third, U-Net, or similar decoder architectures, might not produce improved recovery of fine structural boundaries through relatively coarse blending of hierarchical features and limited inter-modal alignment [
4]. Also, existing model pipelines often take the approach of separate classification and segmentation pipelines, which hampers joint performance and lacks a shared optimization objective when diagnostic and localization tasks are involved. In radiology, clarity and rationale for decision-making, coupled with trust and reliability, are essential for diagnostic outcomes. Due to a lack of interpretable cross-attention fusing of semi-automated diagnostic features, the reliability of the model can be termed paramount, but decision transparency and trust will be weakened if interpretable model mechanisms cannot be defined. Finally, existing models may not implement effective multi-scale feature fusing, subsequently compromising precise delineation of structure boundaries on heterogeneous datasets known to be challenging [
5].
2. Literature Review
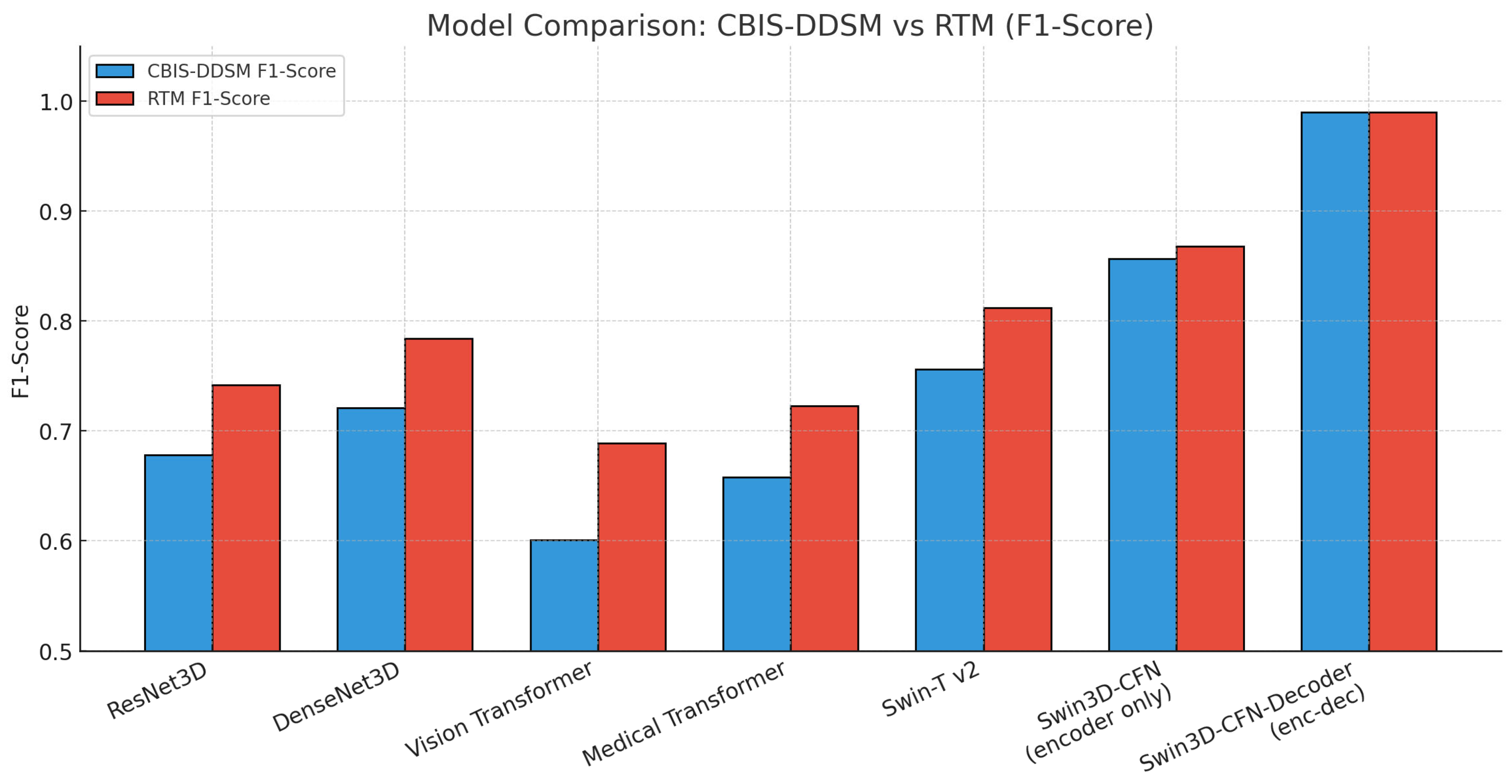
Deep learning has made great advances in breast cancer image content analysis over the past couple of years, particularly in the classification and segmentation of mammograms. Traditional convolutional neural networks (CNN), including ResNet3D and DenseNet3D, are generally effective in extracting valuable features from medical images. However, CNN models are limited to local receptive fields. Common CNN architecture moderates the capability of retrieving mutually inclusive and larger contextual relationships that breast tissues possess, for both accuracy and precision in breast cancer diagnosis. This suggests that new models or approaches generate models effectively and complex dependencies across different regions of an image. The region’s results will contextualize information in a broader context that is more representative of how radiologists evaluate cases in their own real-world contexts. Transformer-based models such as Vision Transformer and Medical Transformer have ultimately motivated the field forward with their use of self-attention to capture long-range interactions, but they are still frequently computationally inefficient and struggle with multi-modal fusion Swin3D-CFN-Decoder.
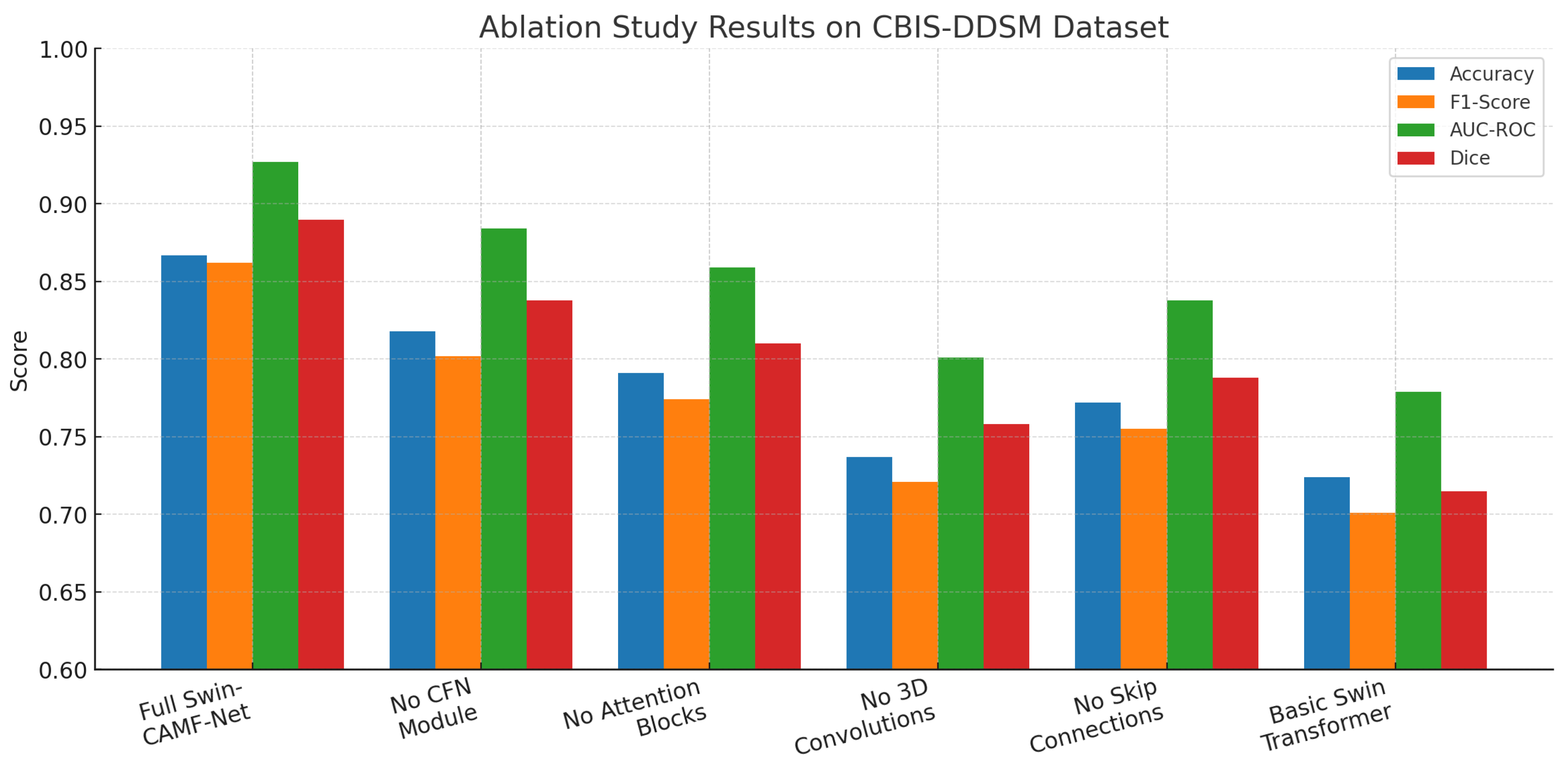
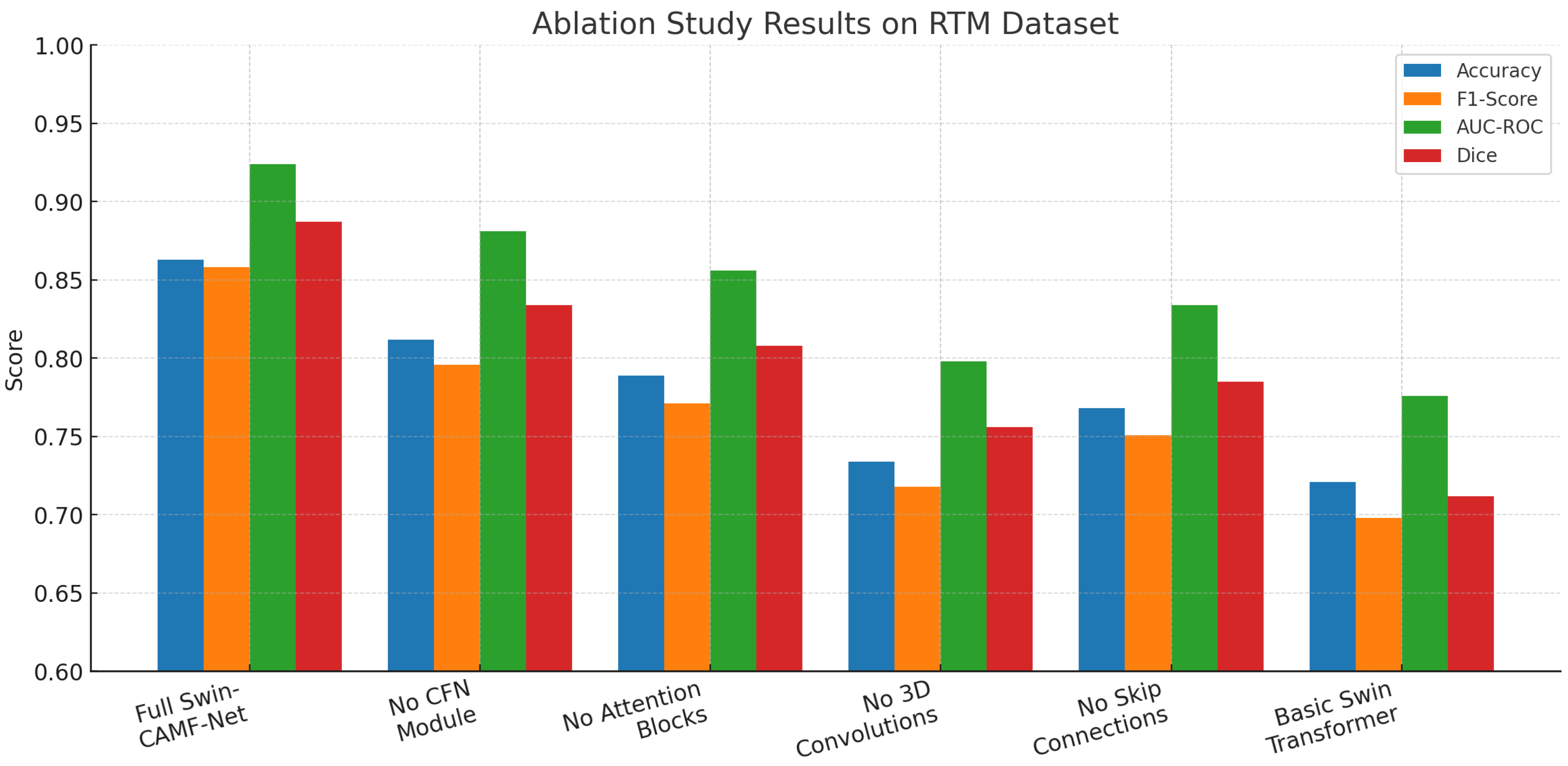
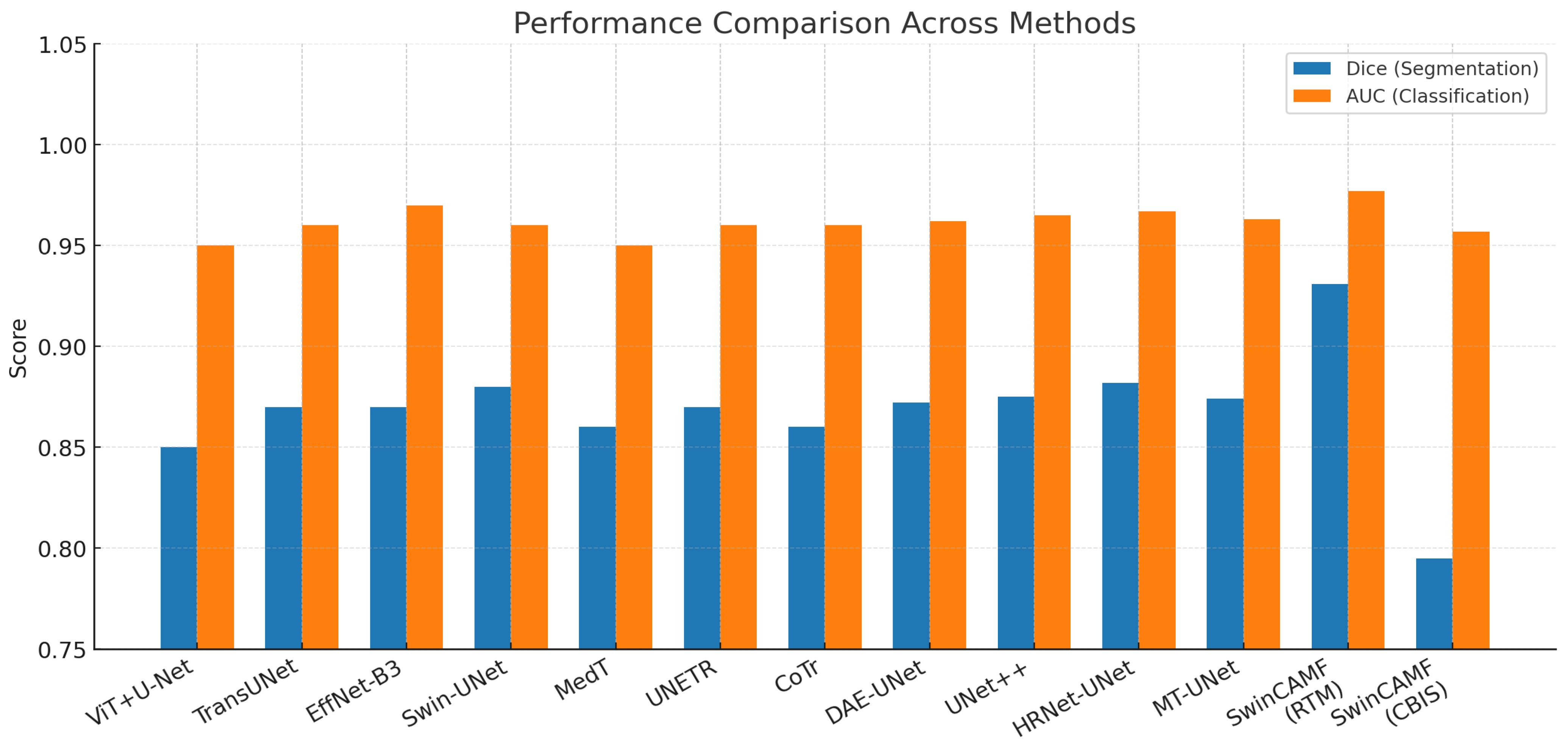
Hybrid models like Swin-Tv2 and encoder-decoder transformer models (e.g., Swin3D-CFN) have enhanced diagnostic accuracy and segmentation performance by integrating hierarchical attention and multi-scale spatial context. This has been supported by ablation studies demonstrating that components such as 3D convolution, attention blocks, and skip connections all contribute to the accurate localization of lesions and classification results. They also decrease in these components, producing harms in accuracy, F1-scores, AUC-ROC, and Dice coefficients for lesion classification and segmentation. However, this progress has not yet resulted in a single architecture since most current pipelines use segmentation and classification separately or do not leverage multi-view and volumetric data fully in a single architecture.
Moreover, previous research has indicated that models without interpretable cross-attention mechanisms or multi-scale fusion tend to provide less decision-making and less accurate boundary recovery, which is vital to clinical acceptability. In addition, the lack of dual objective optimization produces limitations in the simultaneous improvement of pixel-wise segmentation and global classification performance. Thus, these shortcomings underlie the incorporation of cross-attentive multimodal fusion and multi-scale hierarchical decoding, as included in CAMF-Net, and provide a new standard for integrated, interpretable breast cancer image analysis.
The above
Table 1 presents a concise synthesis of significant progress in breast cancer imaging analysis with deep learning, organized from 2021 to 2025. It depicts the field’s progression from the prototype transformer models and CNN-based models to hybrid and multimodal architectures, where each row features a methodological step or an innovative concept. The initial models provided attention-based classification, with a transformer model being the Vision Transformer. The following models followed these beginnings: nnFormer [
6] used purely 3D transformers for volumetric segmentation. Hybrid subsequent models included attention fusion with multi-modal data (e.g., mammography, MRI, clinical features) and multi-task learning to mediate a particular diagnostic or technical limitation faced in simpler models. Recent studies demonstrated cross-attentive fusion, hierarchical decoding, segmentation, and classification simultaneously, and clinical interpretability. Studies investigated multiple time delays in mammography views, multi-omics fusion, explainable AI, and an ensemble of time points/information through decision strategies. The studies had uncovered important gaps within unified end-to-end learning, transparency, and consideration of deep clinical context. While the studies accurately presented certain results, there is a large proportion of prior studies being limited by the modality approach, separation of classification and segmentation, non-unified architecture, or absence of interpretable feature alignment.
In 2022, there was significant focus on multi-image approaches. Chen and co-authors presented transformers that could accept multiple mammogram image views for each case, which were much better at handling complicated dependencies than standard CNN methods to enhance diagnostic accuracy. However, these early endeavors presented challenges with low-resolution data and did not effectively incorporate clinical information into clinical practice. The major significant contributions turned significantly in 2023, when researchers explored transfer learning with transformer models and studied molecular-level applications [
20,
21,
22,
23,
24], extended the paradigm, and applied ViT, Swin, and PVT to breast mass detection, achieving very high accuracy but also experiencing overfitting challenges on the public datasets. Between 2024 and 2025, research gained new energy, with many hybrid architectures and cross-attention fusion branches presented to benchmark vision transformers against state-of-the-art CNN and graph models of breast imaging data noted the superiority of Swin Transformer variant model performance for multi-view and 3D breast lesion object detection [
25,
26,
27,
28,
29].
2.1. Research Gap
Despite significant advancements in deep learning, there is still a notable lack of research work performed on clinical development of dependable transformer-based models for mammography analyses. Most efforts have been focused on previously established architectures, such as the ViT and Swin Transformer, and have provided a primary emphasis on achieving high accuracy in a controlled experimental environment. Very little research has addressed every attempt to take appropriate action regarding the evaluation or alteration of a transformer model for application into hybrid, multimodal, or cross-attention-based frameworks intended for strategically addressing complex clinical or imaging data. Further, most research on practicality regarding doctor–patient variability has completely neglected many dimensions associated with the variability. Thus, because of differences in breast density, imaging quality, or acquisition, variability of the lesions or other variability from clinical features that are linked to the patient. While this has been an area of research focus, it limits the clinical generalizability or clinical reliability of a research framework. This highlights a need for additional work for a unified transformer framework that is context-aware and iterative learning across heterogeneous data sources, while simultaneously remaining diagnosis-preserving.
In addition, it is apparent that the field is moving into newer fusion models in the time frame of 2024 and 2025. Although it is rare for the fusion of models to be tested in a prospective setting, some strains on confidence in their generalizability. Overall, there is a clear need to systematically directly compare performances between different families of transformer models in several clinical settings with a focus not only on strong performance but also on transparency, real-world interpretability, and workflow application. Hence, this research gap is an important step to move from benchmarks to solutions that radiologists can trust and use on a day-to-day basis. These long-standing research gaps will be critical to the development of clinically viable, trustworthy, and broadly implemented AI tools that will provide a transformational pattern shift in breast cancer detection and diagnosis.
2.2. Motivation
The motivation for the current study comes from the realities and expectations of modern breast cancer diagnosis. Radiologists obtain images from different angles, a unique patient medical history, and the potential of subtle lesions hiding in plain sight. The evolution of AI techniques has clearly introduced amazing new technology and tools to the workplace. The earlier models were still operating mostly in narrow ways based on limited datasets or data created from a single information source, with the burden of complexity. SwinCAMF-Net was not a “better numbers” model from performance metrics, but it developed a new model that clinicians found “valuable” to hold and use. The intention was to begin to move us beyond the era of “AI as a black box” and construct a device that acted more like a colleague—a colleague that could look for subtle details on images, provide a contextualized understanding of findings, and articulate the basis for its rationale in an open and non-threatening manner. Our goal is to help radiologists simplify their work just a little, primarily for difficult, stressful, and ambiguous cases. Confidence and clarity are paramount for clinicians and patients alike when it comes to making a diagnosis. This is one reason we developed SwinCAMF-Net as more than just an algorithm; it is a useful tool that works in the background to provide insight, assurance, and transparency to the radiologist’s decision-making process.
To tackle the above-mentioned gap and the limitations, we propose CAMF-Net, a cross-attentive multimodal fusion network intended for deep learning diagnostic and segmentation pipelines for mammograms.
The prime contributions of the proposed framework are as follows:
- ○
Hierarchical Multimodal Feature Fusion: CAMF-Net is developed by hierarchically fusing Swin Transformer encoders, 3D convolutional neural networks (3D-CNNs), volume encoders, and clinical feature projectors. These heterogeneous data streams are fused by a cross-attention fusion module to facilitate the acquisition and alignment of local and global features across mammographic views, volumetric data, and clinical vectors.
- ○
Cross-Attention Alignment Mechanism: The architecture utilizes a designated cross-attention fusion block that explicitly aligns multi-source transformations before spatial reconstruction, supporting consistent semantic alignment and information flow across modalities.
- ○
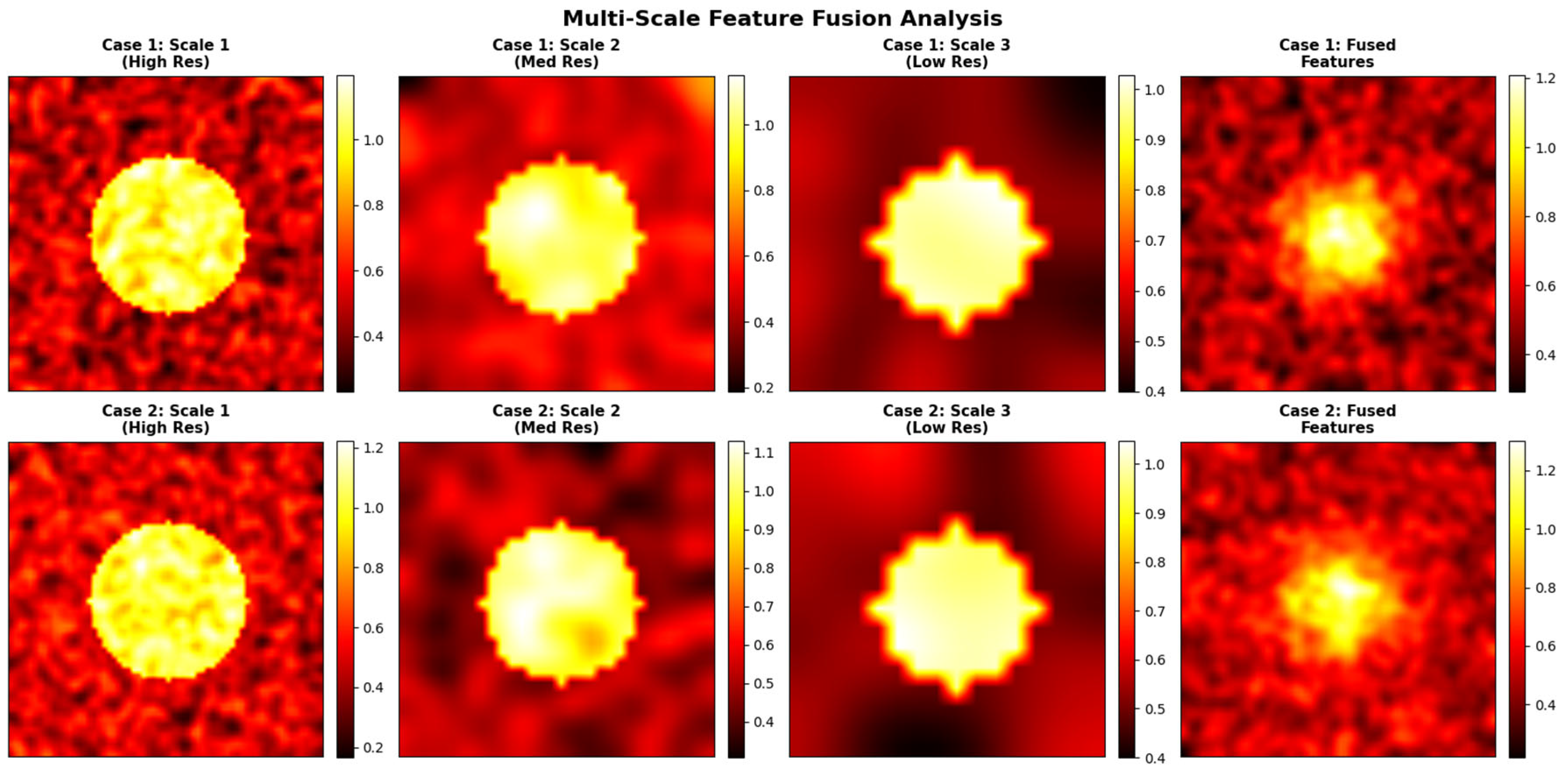
Multi-Scale Feature Fusion: The architecture integrates features from both encoder and decoder at multiple resolution levels, facilitating the preservation of rich contextual cues and robust structural representation across varying lesion sizes and image complexities. This multi-scale fusion is essential for accurate segmentation and detection of subtle findings in heterogeneous breast imaging cohorts.
- ○
Hierarchical U-Net Driven Decoder: To facilitate precise lesion localization and anatomical delineation, the network employs a hierarchical decoder with multi-scale skip connections, ensuring rich contextual blending and spatial detail preservation throughout the segmentation process.
- ○
Dual-Loss Optimization Framework: One training objective supervises binary lesion classification and pixel-level segmentation, aligning diagnostic performance with spatial annotation detail.
2.3. The Novelty Aspects of the SwinCAMF-Net Framework
Novel Cross-Attentive Fusion (CAF) mechanism: The CAF module is newly designed for selective cross-modal alignment, unlike traditional concatenation or early/late fusion. Unlike prior multimodal networks that fuse modalities through simple concatenation or averaging, SwinCAMF-Net introduces a novel cross-attentive fusion (CAF) module that adaptively aligns modality-specific embeddings using learned attention queries, enabling context-aware feature interaction.”
Tri-Modal Integration Framework (2D + 3D + Clinical Metadata): Most prior works combine only 2D mammography with clinical data or 3D tomosynthesis alone (not all three). The proposed architecture is among the first to jointly integrate 2D mammographic views, 3D lesion volumes, and structured clinical metadata within a unified cross-attention transformer framework.
Dual-task Optimization Strategy (Segmentation + Classification Synergy): The model jointly optimizes both tasks, allowing the segmentation decoder to inform the classification head through shared features. By jointly optimizing segmentation and classification objectives, the model learns richer shared representations that improve diagnostic reliability and spatial localization.
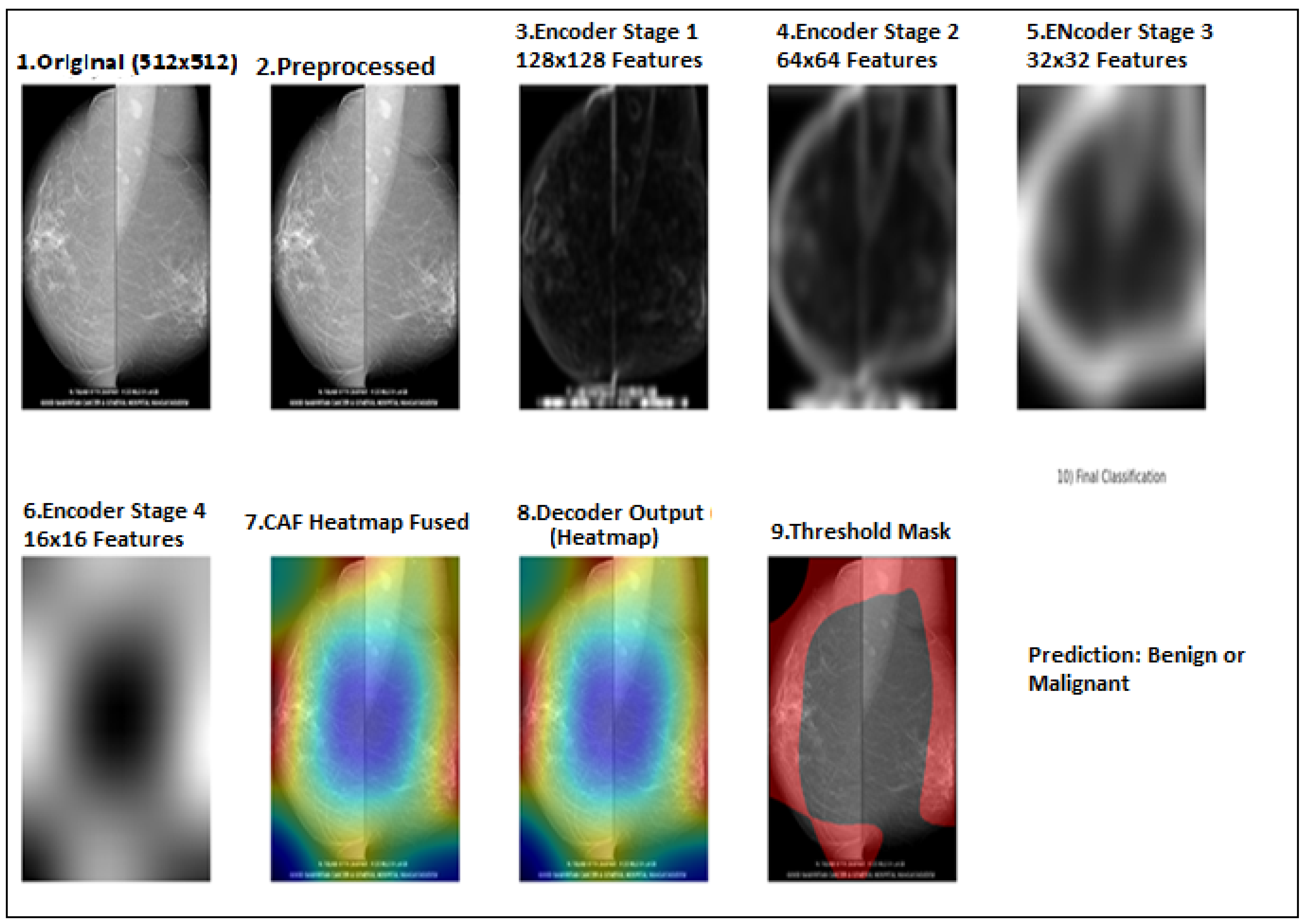
Explainability through Attention Visualization: SwinCAMF-Net provides built-in interpretability through its attention-based visualization maps, enabling clinicians to trace lesions contributing to malignancy predictions.
Comprehensive Evaluation and Generalization: SwinCAMF-Net not only exhibits robust generalization performance over mammogram datasets but also provides interpretable decisions through cross-attention explainability, thereby supporting its robustness and trustworthiness in clinical practice.
2.4. Proposed Work
The current work is the design of SwinCAMF-Net, a deep learning framework mainly focused on helping clinicians to enhance the diagnosis process for breast cancer. Similarly, they discuss with each other regarding difficult cases. SwinCAMF-Net brings together mammograms from different views, 3D, and relevant patient information into a unified workflow. The proposed model uses Swin Transformer to provide progressive analysis on the mammograms and identify subtle or apparent signs. It separates a 3D volume encoder and analyzes the shape, size, and features of lesions from volumetric data. To maintain clinical relevance, patient-specific features of age, family history, and previous imaging findings are incorporated as a primary part of the feature extraction process. The unique cross-attention fusion module allows various data types to be fused within the network, as well as the ability for the network to decide what is most relevant case-by-case. The fused data is then incorporated into the classification process, which is expected to delineate and increase confidence regarding what type of lesion is predicted. SwinCAMF-Net, developed and evaluated using two datasets to demonstrate robustness and practicality, contributes to diagnostic applications that support rational clinical decisions and enhance daily medical practice.
The objectives of the current work are as follows:
To design a novel diagnostic support tool that incorporates imaging data from different mammogram views, along with advanced 3D scans and patient-specific data that simulates the same decision-making process as experienced clinicians.
To utilize AI technology (SwinCAMF-Net) capable of understanding context and pattern recognition from images, with the added capability of merging contextual information with visual evidence to improve accuracy, robustness, trust, and quality in diagnosis.
To produce a model that can use advanced transformer and cross-attention fusion methods to process images but allows the model to attend to the most relevant areas and cues for data processing.
To enhance breast lesion classification and segmentation accuracy and reliability to help sophisticated clinicians continue to detect the important atypical, subtle, or difficult cases.
To assist radiologists with AI that is adaptable to the practitioner’s needs and provide a clear, intelligent challenge to ensure patients and their families are as calm and reassured as possible regarding their cancer diagnosis.
5. Training and Performance
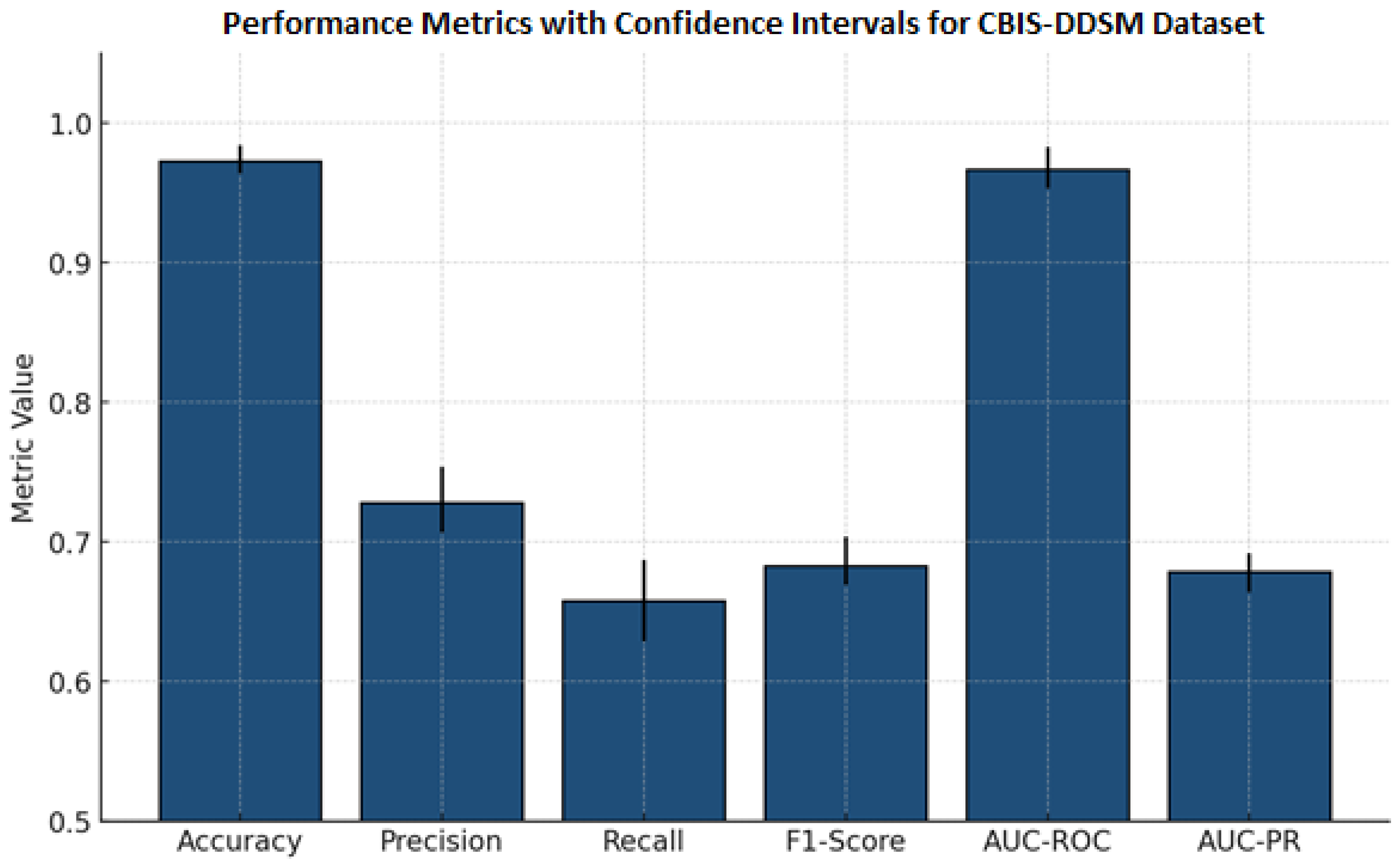
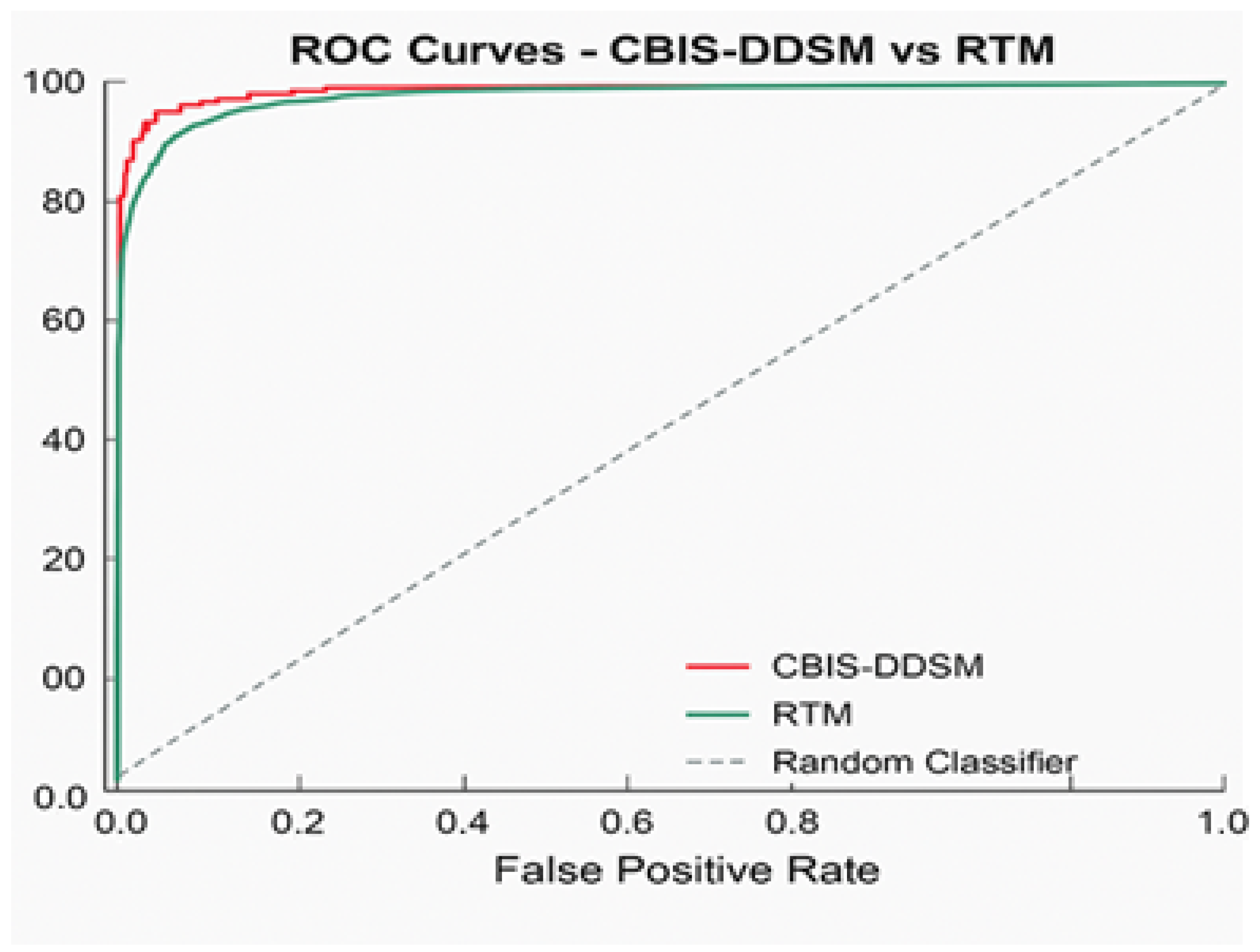
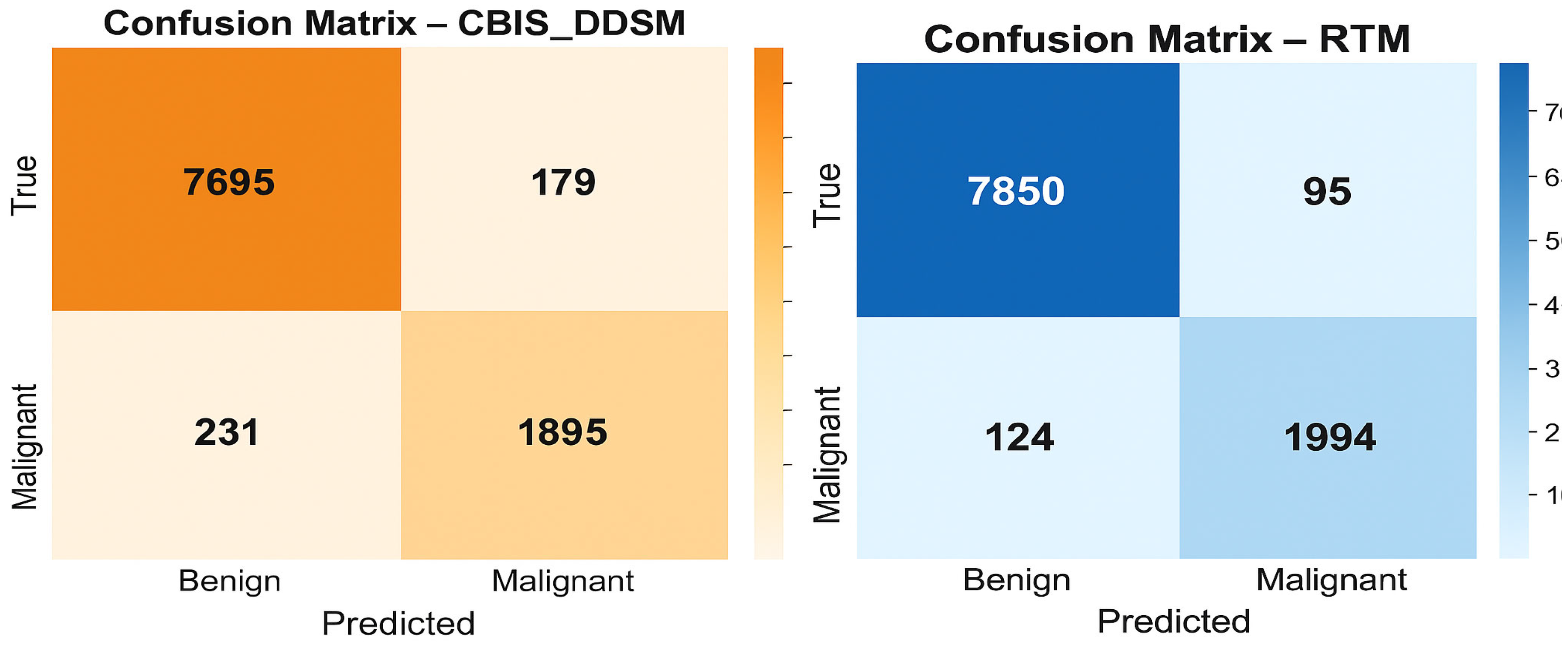
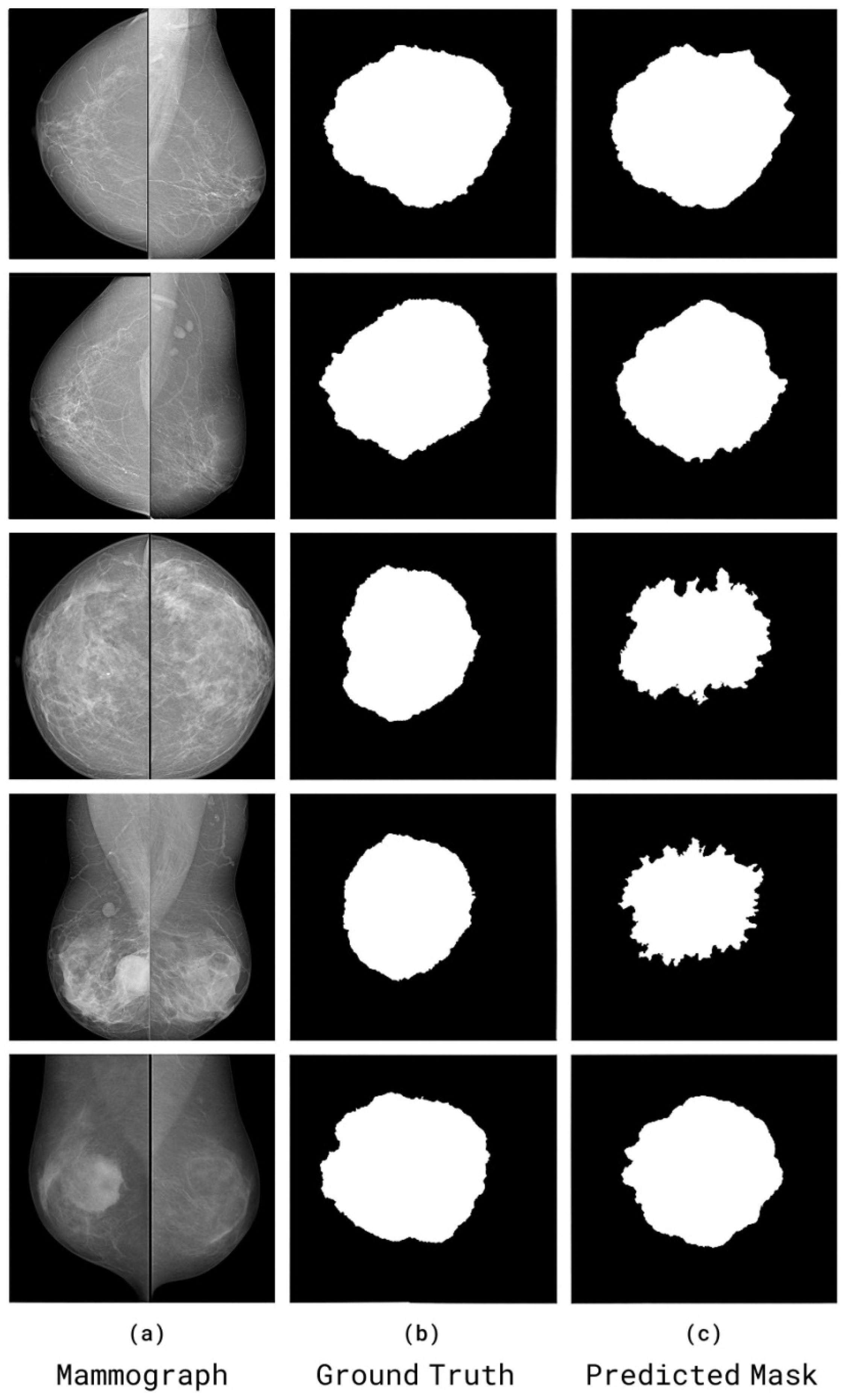
We trained and tested the SwinCAMF-Net model with previously available mammography datasets with a fair data split of 70% train, 15% validation, and 15% test to ensure there was an equal representation of benign and malignant throughout the entire process. Throughout training, we were careful in how the model was trained in detection and segmentation of breast lesions by adjusting the parameters while monitoring the validation loss to ensure it was learning, not just memorizing, but also learning the data. For model evaluation, we used standard metrics such as the dice coefficient, AUC, and overall accuracy.
5.1. Training and Evaluation
The proposed model’s training methodology follows a two-stage approach:
Segmentation Training:
- ○
Loss Function: Combination of Dice loss and binary cross-entropy (BCE) with boundary-aware weighting;
- ○
Optimizer: AdamW with weight decay of 0.01;
- ○
Learning Rate: Initial value of 1e-4 with cosine annealing schedule;
- ○
Batch Size: 8;
- ○
Epochs: 200 with early stopping (patience = 20);
- ○
Regularization: Dropout (0.1), weight decay, and gradient clipping (max norm = 1.0).
- 2.
Classification Training:
- ○
Loss Function: Weighted binary cross-entropy to address class imbalance;
- ○
Learning Rate: Initial value of 2e-5 with warm-up and cosine decay;
- ○
Batch Size: 16;
- ○
Epochs: 100 with early stopping (patience = 15);
- ○
Data Sampling: Balanced sampling strategy to handle class imbalance.
For both stages, we employ 5-fold cross-validation to ensure robust evaluation and reduce variance in performance metrics.
5.2. Performance Metrics
The metrics considered for evaluation are as follows:
Segmentation Metrics:
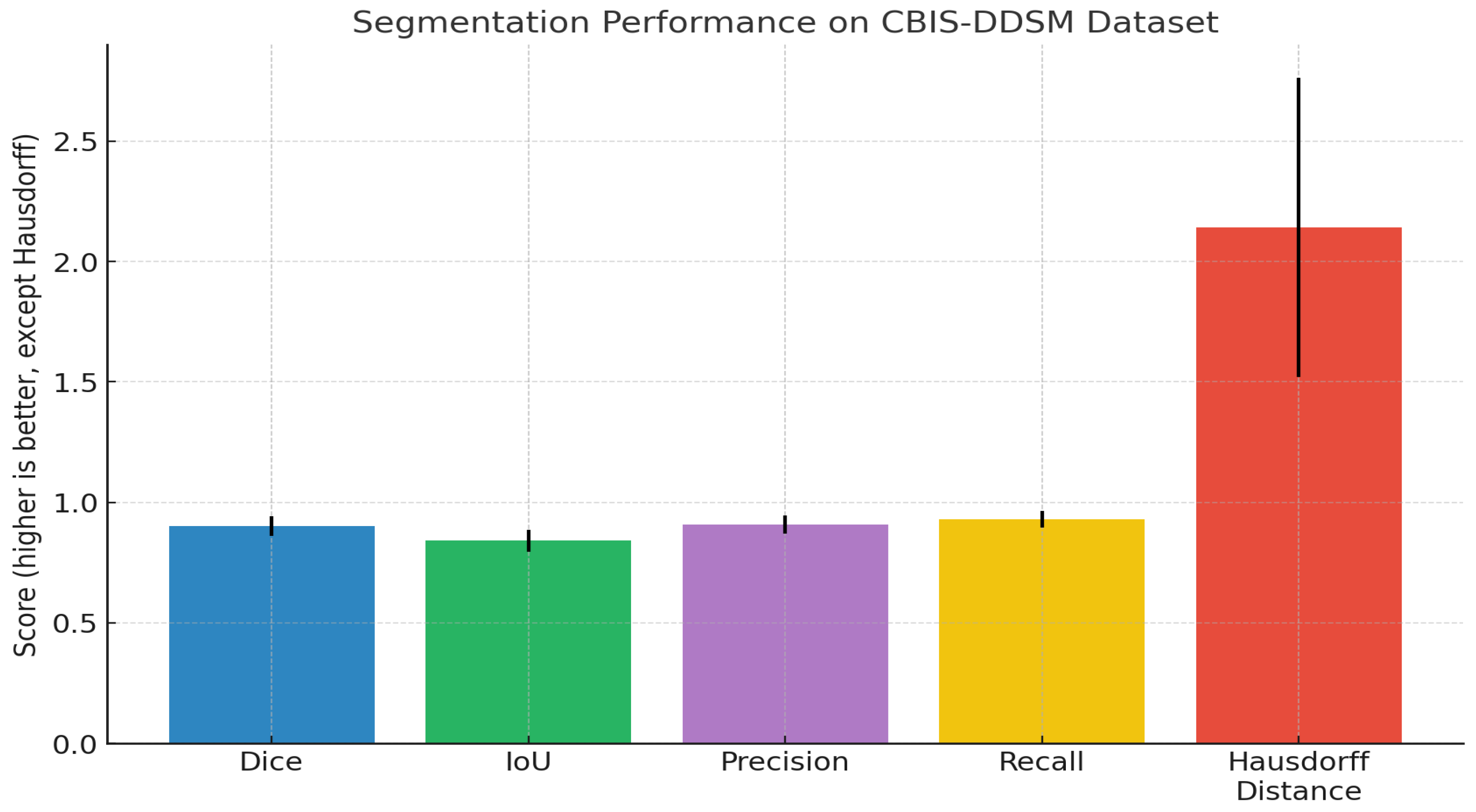
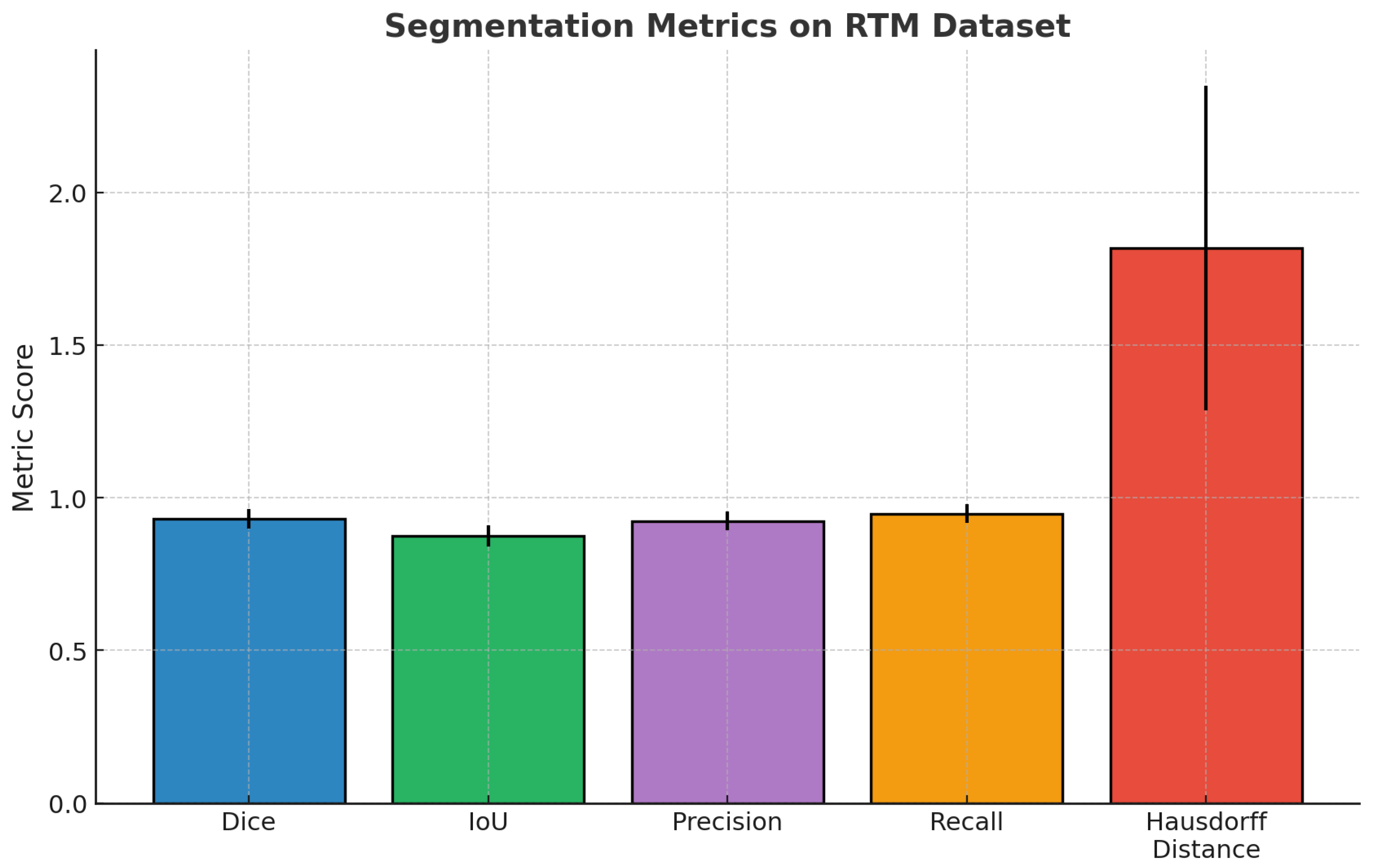
Dice Coefficient (DSC): Measures overlap between predicted and ground truth segmentations;
Intersection over Union (IoU): Also known as the Jaccard index, it quantifies region overlap;
Hausdorff Distance (HD95): Assesses boundary accuracy (95th percentile);
Sensitivity and Specificity: Pixel-level true positive and true negative rates.
Classification Metrics:
Area Under the ROC Curve (AUC): Primary evaluation metric for classification performance
Accuracy: Overall correct classification rate;
Sensitivity/Recall: True positive rate for malignant lesions;
Specificity: True negative rate for benign lesions;
F1-Score: Harmonic mean of precision and recall;
Precision: Positive predictive value for malignant cases.
Efficiency Metrics:
Inference Time: Average processing time per image;
Model Size: Number of parameters and memory footprint;
FLOPs: Floating-point operations per inference.
5.3. Processor/Hardware Configurations
This section presents several configuration requirements needed for the implementation of the proposed model are as follows:
- ○
Local CPU: e.g., Intel Xeon Silver 4210 (10 cores), AMD EPYC 7742 (64 cores).
- ○
Local GPU: NVIDIA RTX 3090 (24 GB), NVIDIA A100 (40/80 GB).
- ○
Cloud: Google Colab TPU v2/v3, Colab Pro+ A100, AWS p3.2xlarge (V100), Azure ND A100.
All our experiments were performed on a workstation having an NVIDIA RTX A4500 (20 GB VRAM) GPU, an AMD Ryzen Thread ripper PRO CPU (24 cores), and 64 GB of RAM. The training and inference of the model were performed using PyTorch and made use of CPU and GPU appropriately. We employed this hardware configuration so that we could quickly process large mammography datasets and conduct hyperparameter tuning and model evaluation thoroughly.
7. Conclusions and Future Work
SwinCAMF-Net enhances breast cancer diagnosis by providing an integrated framework that approximately reflects how clinical experts react and identify the lesions. By integrating multi-view mammograms, 3D volumetric images, and patient/contextual clinical information, the model surpasses traditional analysis in isolation. The cross-attention fusion methodology of SwinCAMF-Net guarantees the diagnostic decisions are based on both the fine-grained, high-resolution details visible within the images and the contextual clinical data that is relevant to each individual patient. This approach clearly supports the performance on diverse and challenging datasets. The resulting observations suggest that SwinCAMF-Net could provide clinically reliable assistance to a physician and represents an important milestone in the separation between sophisticated deep learning and the degree of human inference required in clinical diagnosis. Hence, the proposed work is a forward step towards providing a diagnostic tool that is scientifically rigorous, as well as justifiably practical for healthcare providers and the patients they serve.
Future work will focus on the following:
1. Need for broadening the assessment of SwinCAMF-Net across wider, varied mammography datasets and clinical scenarios to strengthen the generalizability and robustness of the evaluating framework.
2. Improving the network’s capacity to handle and learn from incomplete or missing mammographic views, reflecting the conditions of typical screening in the real world.
3. Adopting more advanced self-supervised or semi-supervised learning strategies to speed up model training and reduce reliance on large, labeled datasets.
4. Need for broadening the architecture to multi-modal breast imaging to include ultrasound, tomosynthesis, or MRI to facilitate comprehensive, cross-modal cancer diagnosis.
5. Need for exploring more interpretability and incorporation into clinical workflow by providing practical, human-understandable explanations suited to radiologists and other practitioners.