A Novel Deep Learning Framework for Liver Fibrosis Staging and Etiology Diagnosis Using Integrated Liver–Spleen Elastography
Abstract
1. Introduction
2. Methods
2.1. Patients
2.2. Pathological Examination
2.3. Serological Examinations
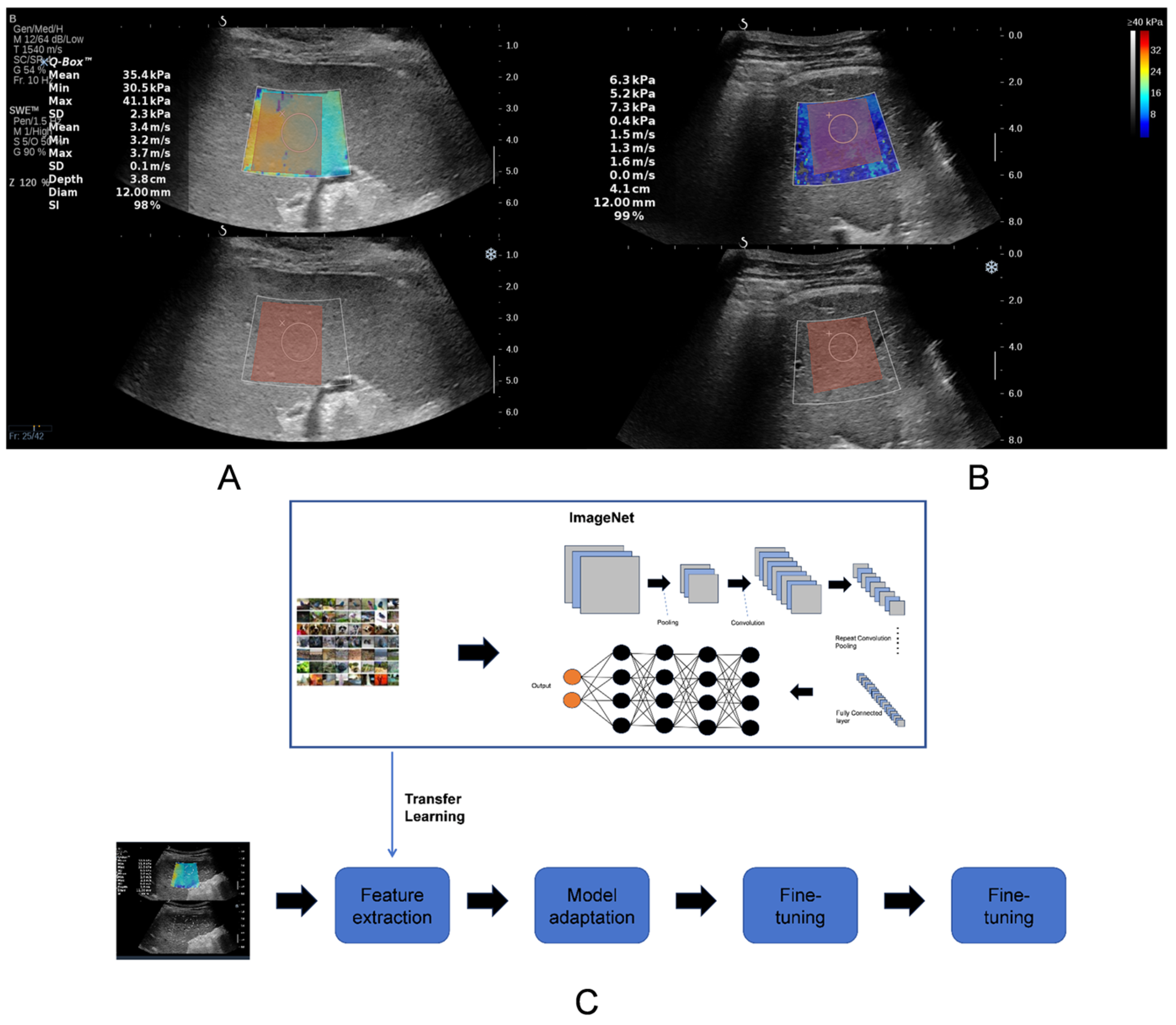
2.4. Two-Dimensional Shear Wave Elastography
2.5. Region-of-Interest Segmentation
2.6. Machine Learning
2.7. Transfer Learning
2.8. Model Construction and Validation
2.9. Statistical Power and Multinomial Logistic Regression
2.10. Statistical Analysis
3. Results
3.1. Baseline Characters
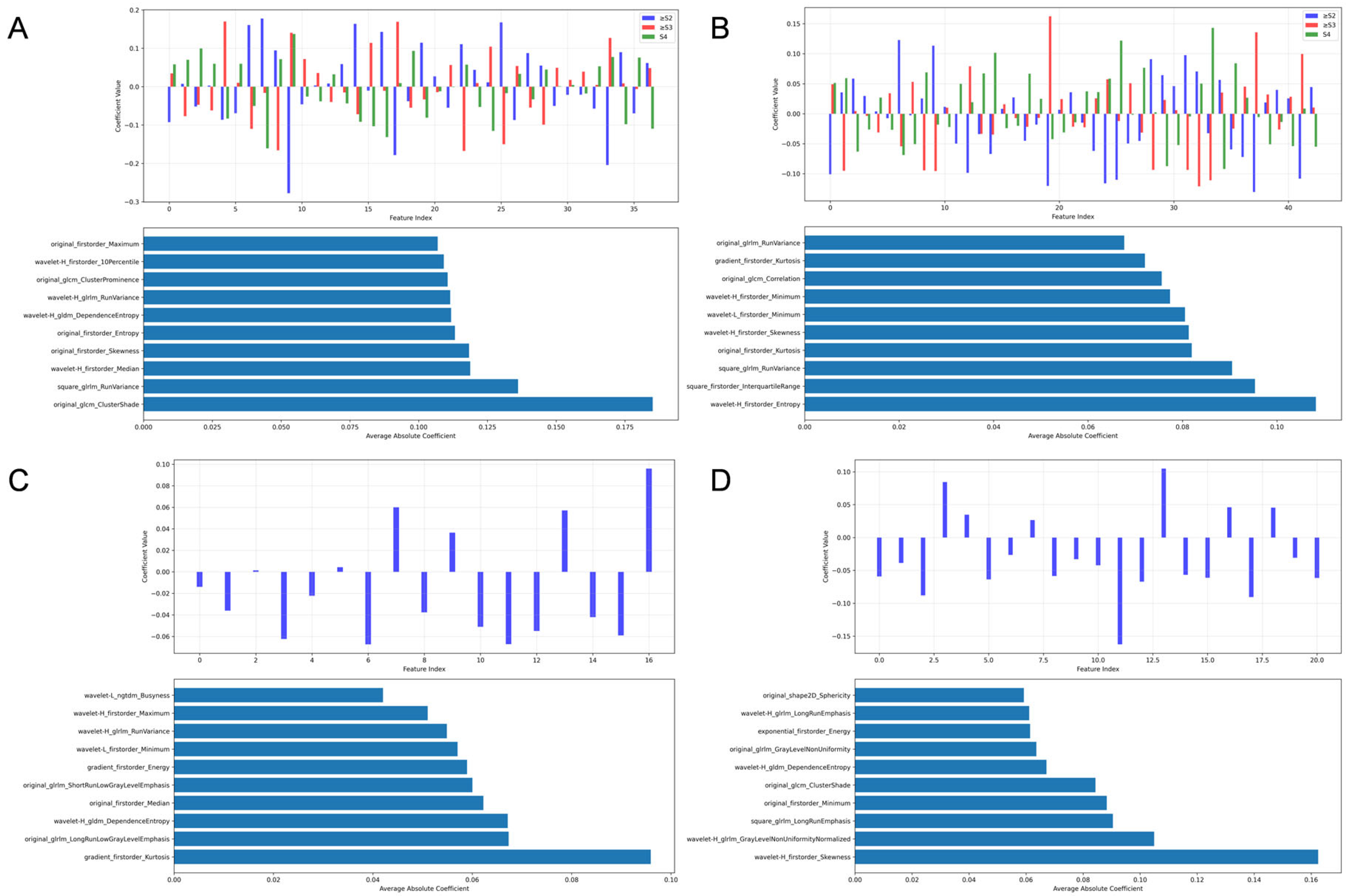
3.2. Radiomic Feature Selection
- 1.
- For liver fibrosis staging (Figure 3):
- 1.1
- Liver grayscale images prioritized Gray-Level Run-Length Matrix (GLRLM) texture and wavelet-transformed first-order statistics.
- 1.2
- Liver 2D-SWE emphasized Gray-Level Co-occurrence Matrix (GLCM) texture and first-order statistics.
- 1.3
- Spleen grayscale imaging prioritized shape features (original_shape2D_Major-AxisLength and original_shape2D_Sphericity).
- 1.4
- Spleen 2D-SWE focused on wavelet-transformed first-order statistics and Gray-Level Dependence Matrix (GLDM) texture.
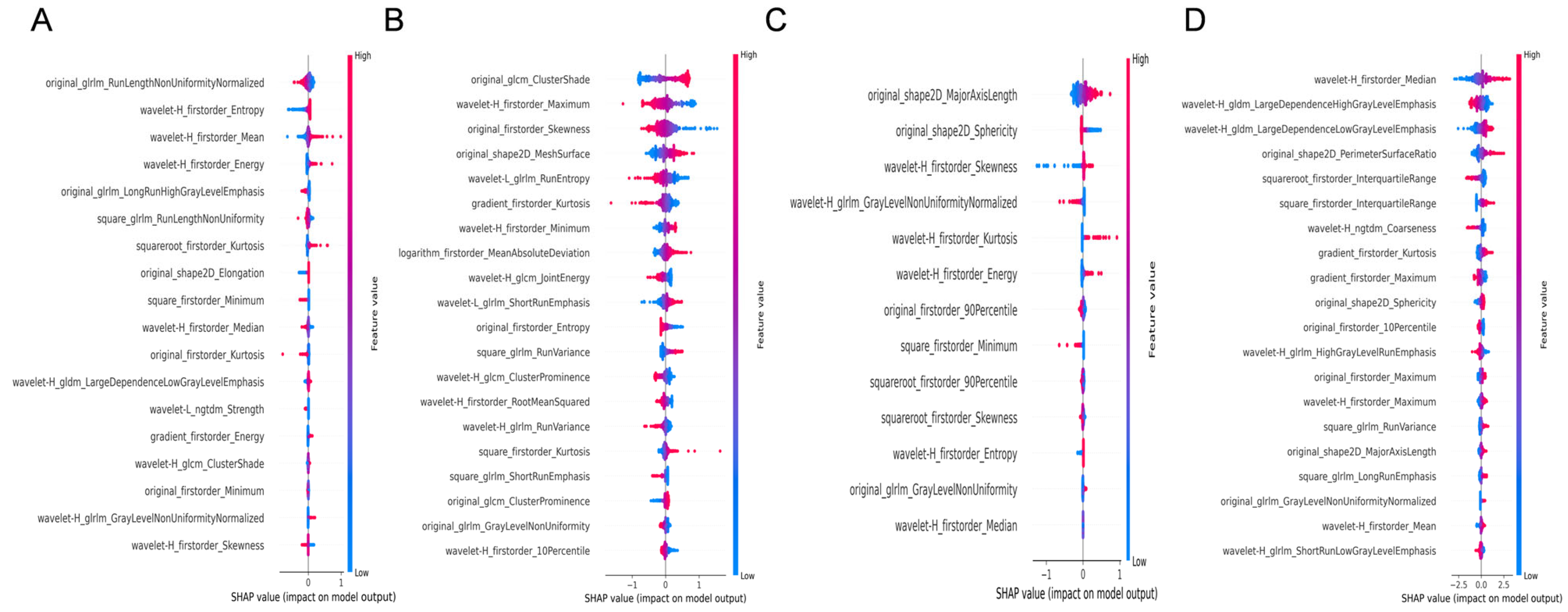
- 2.
- For etiology diagnosis (Figure 4):
- 2.1
- Liver grayscale images prioritized distributional statistics and texture heterogeneity, logarithm_firstorder_InterquartileRange and squareroot_firstorder_90Perce-ntile capturing asymmetric or focal damage patterns.
- 2.2
- Liver 2D-SWE emphasized stiffness distribution and complexity (original_firstorder_Median and wavelet-H_gldm_DependenceEntropy), distinguishing etiologies by stiffness homogeneity/heterogeneity.
- 2.3
- Spleen grayscale imaging integrated morphological and distributional features (original_shape2D_MajorAxisLength and squareroot_firstorder_Maximum).
- 2.4
- Spleen 2D-SWE focused on asymmetric stiffness and local texture (wavelet-H_firstorder_Skewness and square_glrlm_LongRunEmphasis).

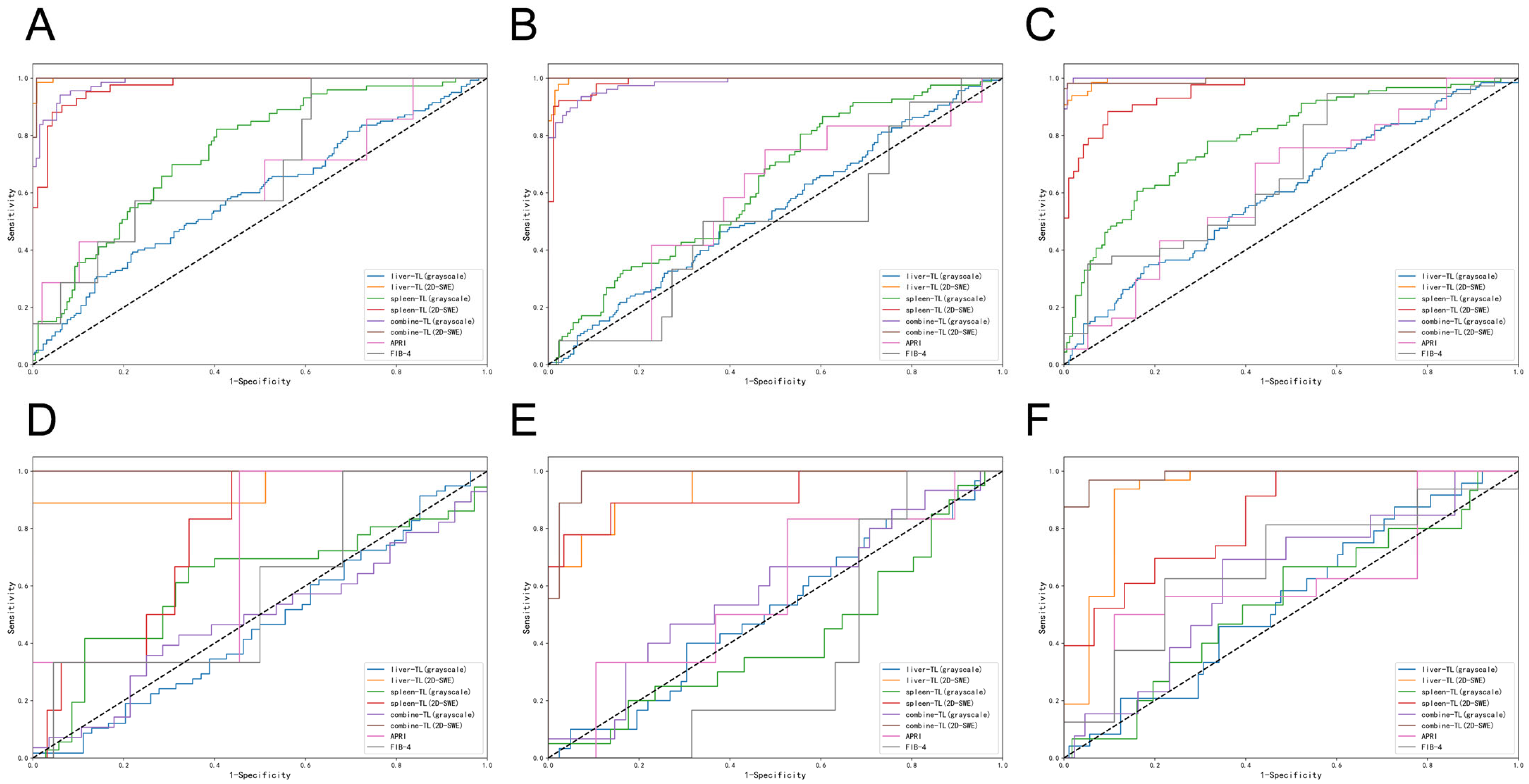
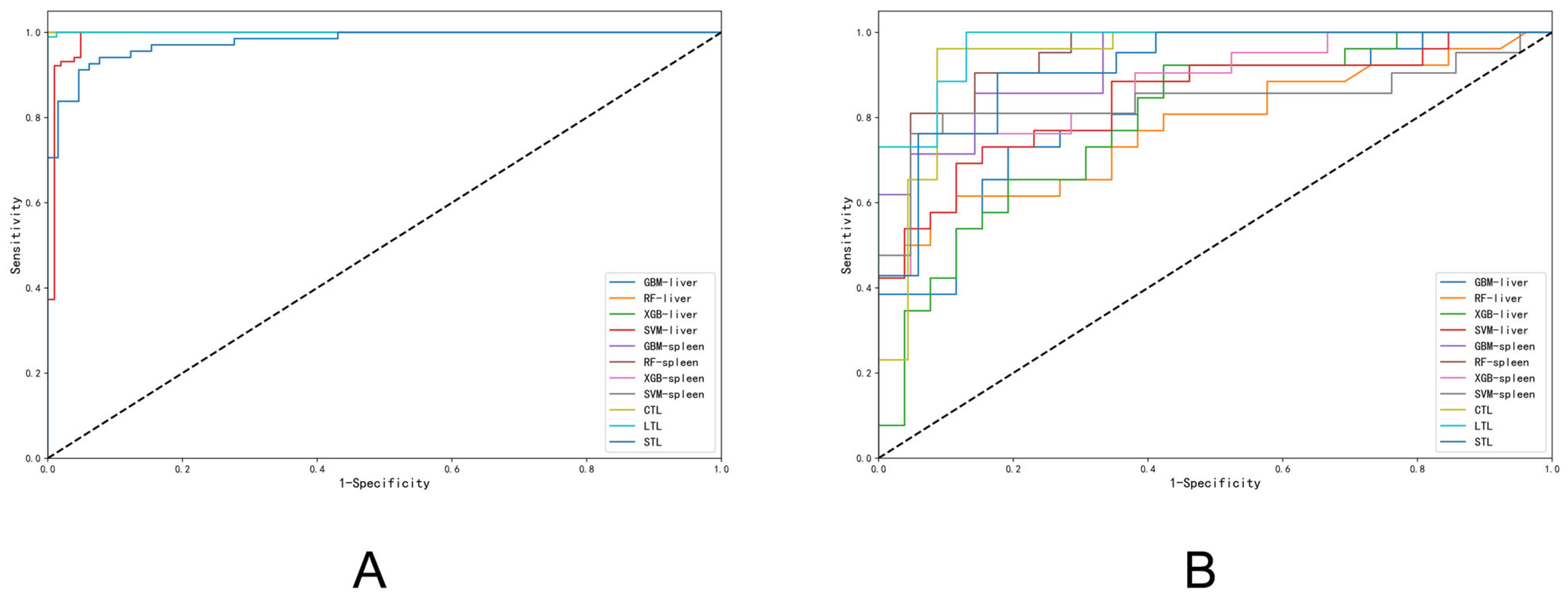
3.3. Diagnostic Accuracy of Liver Fibrosis Staging
3.4. Diagnostic Accuracy of Liver Fibrosis Etiology
3.5. Assessment of Statistical Power and Multinomial Regression Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Asrani, S.K.; Devarbhavi, H.; Eaton, J.; Kamath, P.S. Burden of liver diseases in the world. J. Hepatol. 2019, 70, 151–171. [Google Scholar] [CrossRef] [PubMed]
- Terrault, N.A.; Lok, A.S.F.; McMahon, B.J.; Chang, K.M.; Hwang, J.P.; Jonas, M.M.; Brown, R.S., Jr.; Bzowej, N.H.; Wong, J.B. Update on prevention, diagnosis, and treatment of chronic hepatitis B: AASLD 2018 hepatitis B guidance. Hepatology 2018, 67, 1560–1599. [Google Scholar] [CrossRef] [PubMed]
- Bravo, A.A.; Sheth, S.G.; Chopra, S. Liver biopsy. N. Engl. J. Med. 2001, 344, 495–500. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.N.; Han, J.W.; An, J.; Kim, B.K.; Jin, Y.J.; Kim, S.S.; Lee, M.; Lee, H.A.; Cho, Y.; Kim, H.Y.; et al. KASL clinical practice guidelines for noninvasive tests to assess liver fibrosis in chronic liver disease. Clin. Mol. Hepatol. 2024, 30, S5–S105. [Google Scholar] [CrossRef]
- Cassinotto, C.; Lapuyade, B.; Mouries, A.; Hiriart, J.B.; Vergniol, J.; Gaye, D.; Castain, C.; Le Bail, B.; Chermak, F.; Foucher, J.; et al. Non-invasive assessment of liver fibrosis with impulse elastography: Comparison of Supersonic Shear Imaging with ARFI and FibroScan®. J. Hepatol. 2014, 61, 550–557. [Google Scholar] [CrossRef]
- Dietrich, C.F.; Bamber, J.; Berzigotti, A.; Bota, S.; Cantisani, V.; Castera, L.; Cosgrove, D.; Ferraioli, G.; Friedrich-Rust, M.; Gilja, O.H.; et al. EFSUMB Guidelines and Recommendations on the Clinical Use of Liver Ultrasound Elastography, Update 2017 (Long Version). Ultraschall. Med. 2017, 38, e16–e47. [Google Scholar] [CrossRef]
- Sigrist, R.M.S.; Liau, J.; Kaffas, A.E.; Chammas, M.C.; Willmann, J.K. Ultrasound Elastography: Review of Techniques and Clinical Applications. Theranostics 2017, 7, 1303–1329. [Google Scholar] [CrossRef]
- Herrmann, E.; de Lédinghen, V.; Cassinotto, C.; Chu, W.C.; Leung, V.Y.; Ferraioli, G.; Filice, C.; Castera, L.; Vilgrain, V.; Ronot, M.; et al. Assessment of biopsy-proven liver fibrosis by two-dimensional shear wave elastography: An individual patient data-based meta-analysis. Hepatology 2018, 67, 260–272. [Google Scholar] [CrossRef]
- Gao, Y.; Zheng, J.; Liang, P.; Tong, M.; Wang, J.; Wu, C.; He, X.; Liu, C.; Zhang, S.; Huang, L.; et al. Liver Fibrosis with Two-dimensional US Shear-Wave Elastography in Participants with Chronic Hepatitis B: A Prospective Multicenter Study. Radiology 2018, 289, 407–415. [Google Scholar] [CrossRef]
- Dajti, E.; Huber, A.T.; Ferraioli, G.; Berzigotti, A. Advances in imaging-Elastography. Hepatology 2025. [Google Scholar] [CrossRef]
- Dong, B.T.; Huang, S.; Lyu, G.R.; Qin, R.; Gu, J.H. Assessment of liver fibrosis with liver and spleen stiffness measured by sound touch elastography, serum fibrosis markers in patients with chronic hepatitis B. J. Dig. Dis. 2021, 22, 342–350. [Google Scholar] [CrossRef]
- Wang, X.P.; Wang, Y.; Ma, H.; Wang, H.; Yang, D.W.; Zhao, X.Y.; Jin, E.H.; Yang, Z.H. Assessment of liver fibrosis with liver and spleen magnetic resonance elastography, serum markers in chronic liver disease. Quant. Imaging Med. Surg. 2020, 10, 1208–1222. [Google Scholar] [CrossRef] [PubMed]
- Samir, A.E.; Dhyani, M.; Vij, A.; Bhan, A.K.; Halpern, E.F.; Méndez-Navarro, J.; Corey, K.E.; Chung, R.T. Shear-wave elastography for the estimation of liver fibrosis in chronic liver disease: Determining accuracy and ideal site for measurement. Radiology 2015, 274, 888–896. [Google Scholar] [CrossRef] [PubMed]
- Zhuang, Y.; Ding, H.; Zhang, Y.; Sun, H.; Xu, C.; Wang, W. Two-dimensional Shear-Wave Elastography Performance in the Noninvasive Evaluation of Liver Fibrosis in Patients with Chronic Hepatitis B: Comparison with Serum Fibrosis Indexes. Radiology 2017, 283, 873–882. [Google Scholar] [CrossRef] [PubMed]
- Zheng, J.; Guo, H.; Zeng, J.; Huang, Z.; Zheng, B.; Ren, J.; Xu, E.; Li, K.; Zheng, R. Two-dimensional shear-wave elastography and conventional US: The optimal evaluation of liver fibrosis and cirrhosis. Radiology 2015, 275, 290–300. [Google Scholar] [CrossRef]
- Gillies, R.J.; Kinahan, P.E.; Hricak, H. Radiomics: Images Are More than Pictures, They Are Data. Radiology 2016, 278, 563–577. [Google Scholar] [CrossRef]
- Mayerhoefer, M.E.; Materka, A.; Langs, G.; Häggström, I.; Szczypiński, P.; Gibbs, P.; Cook, G. Introduction to Radiomics. J. Nucl. Med. 2020, 61, 488–495. [Google Scholar] [CrossRef]
- Li, J.; Qureshi, M.; Gupta, A.; Anderson, S.W.; Soto, J.; Li, B. Quantification of Degree of Liver Fibrosis Using Fibrosis Area Fraction Based on Statistical Chi-Square Analysis of Heterogeneity of Liver Tissue Texture on Routine Ultrasound Images. Acad. Radiol. 2019, 26, 1001–1007. [Google Scholar] [CrossRef]
- Acharya, U.R.; Raghavendra, U.; Koh, J.E.W.; Meiburger, K.M.; Ciaccio, E.J.; Hagiwara, Y.; Molinari, F.; Leong, W.L.; Vijayananthan, A.; Yaakup, N.A.; et al. Automated detection and classification of liver fibrosis stages using contourlet transform and nonlinear features. Comput. Methods Programs Biomed. 2018, 166, 91–98. [Google Scholar] [CrossRef]
- Li, W.; Huang, Y.; Zhuang, B.W.; Liu, G.J.; Hu, H.T.; Li, X.; Liang, J.Y.; Wang, Z.; Huang, X.W.; Zhang, C.Q.; et al. Multiparametric ultrasomics of significant liver fibrosis: A machine learning-based analysis. Eur. Radiol. 2019, 29, 1496–1506. [Google Scholar] [CrossRef]
- Wang, K.; Lu, X.; Zhou, H.; Gao, Y.; Zheng, J.; Tong, M.; Wu, C.; Liu, C.; Huang, L.; Jiang, T.; et al. Deep learning Radiomics of shear wave elastography significantly improved diagnostic performance for assessing liver fibrosis in chronic hepatitis B: A prospective multicentre study. Gut 2019, 68, 729–741. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Tan, Z.; Li, C.; Mou, L.; Shi, Y.L.; Zhu, X.X.; Luo, Y. A Deep Learning Model Based on High-Frequency Ultrasound Images for Classification of Different Stages of Liver Fibrosis. Liver Int. 2025, 45, e70148. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Wen, H.; Zhu, Z.; Li, Q.; Liu, L.; Li, T.; Xu, W.; Hou, C.; Huang, B.; Li, Z.; et al. Diagnosis of significant liver fibrosis in patients with chronic hepatitis B using a deep learning-based data integration network. Hepatol. Int. 2022, 16, 526–536. [Google Scholar] [CrossRef] [PubMed]
- Xue, L.Y.; Jiang, Z.Y.; Fu, T.T.; Wang, Q.M.; Zhu, Y.L.; Dai, M.; Wang, W.P.; Yu, J.H.; Ding, H. Transfer learning radiomics based on multimodal ultrasound imaging for staging liver fibrosis. Eur. Radiol. 2020, 30, 2973–2983. [Google Scholar] [CrossRef]
- Yu, Y.; Wang, J.; Ng, C.W.; Ma, Y.; Mo, S.; Fong, E.L.S.; Xing, J.; Song, Z.; Xie, Y.; Si, K.; et al. Deep learning enables automated scoring of liver fibrosis stages. Sci. Rep. 2018, 8, 16016. [Google Scholar] [CrossRef]
- Garg, S.; Singh, P. Transfer Learning Based Lightweight Ensemble Model for Imbalanced Breast Cancer Classification. IEEE/ACM Trans. Comput. Biol. Bioinform. 2023, 20, 1529–1539. [Google Scholar] [CrossRef]
- Song, Q.; Zheng, Y.J.; Sheng, W.G.; Yang, J. Tridirectional Transfer Learning for Predicting Gastric Cancer Morbidity. IEEE Trans. Neural Netw. Learn. Syst. 2021, 32, 561–574. [Google Scholar] [CrossRef]
- Banerjee, I.; Crawley, A.; Bhethanabotla, M.; Daldrup-Link, H.E.; Rubin, D.L. Transfer learning on fused multiparametric MR images for classifying histopathological subtypes of rhabdomyosarcoma. Comput. Med. Imaging Graph. 2018, 65, 167–175. [Google Scholar] [CrossRef]
- Scheuer, P.J. The nomenclature of chronic hepatitis: Time for a change. J. Hepatol. 1995, 22, 112–114. [Google Scholar] [CrossRef]
- Wai, C.T.; Greenson, J.K.; Fontana, R.J.; Kalbfleisch, J.D.; Marrero, J.A.; Conjeevaram, H.S.; Lok, A.S. A simple noninvasive index can predict both significant fibrosis and cirrhosis in patients with chronic hepatitis C. Hepatology 2003, 38, 518–526. [Google Scholar] [CrossRef]
- Sterling, R.K.; Lissen, E.; Clumeck, N.; Sola, R.; Correa, M.C.; Montaner, J.; M, S.S.; Torriani, F.J.; Dieterich, D.T.; Thomas, D.L.; et al. Development of a simple noninvasive index to predict significant fibrosis in patients with HIV/HCV coinfection. Hepatology 2006, 43, 1317–1325. [Google Scholar] [CrossRef]
- Zeng, J.; Liu, G.J.; Huang, Z.P.; Zheng, J.; Wu, T.; Zheng, R.Q.; Lu, M.D. Diagnostic accuracy of two-dimensional shear wave elastography for the non-invasive staging of hepatic fibrosis in chronic hepatitis B: A cohort study with internal validation. Eur. Radiol. 2014, 24, 2572–2581. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; He, Y.; Wan, L.; Li, C.; Li, Z.; Li, Z.; Xu, H.; Tu, C. Deep learning models in classifying primary bone tumors and bone infections based on radiographs. NPJ Precis. Oncol. 2025, 9, 72. [Google Scholar] [CrossRef] [PubMed]
- Sun, K.; He, M.; Xu, Y.; Wu, Q.; He, Z.; Li, W.; Liu, H.; Pi, X. Multi-label classification of fundus images with graph convolutional network and LightGBM. Comput. Biol. Med. 2022, 149, 105909. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Luo, J.; Yang, Y.; Wang, W.; Deng, J.; Yu, L. Automatic lung segmentation in chest X-ray images using improved U-Net. Sci. Rep. 2022, 12, 8649. [Google Scholar] [CrossRef]
- Lin, Z.; Yang, W.; Zhang, W.; Jiang, C.; Chu, J.; Yang, J.; Yuan, X. Recognizing pathology of renal tumor from macroscopic cross-section image by deep learning. Biomed. Eng. 2023, 22, 3. [Google Scholar] [CrossRef]
- Wang, C.; Zheng, L.; Li, Y.; Xia, S.; Lv, J.; Hu, X.; Zhan, W.; Yan, F.; Li, R.; Ren, X. Noninvasive Assessment of Liver Fibrosis and Inflammation in Chronic Hepatitis B: A Dual-task Convolutional Neural Network (DtCNN) Model Based on Ultrasound Shear Wave Elastography. J. Clin. Transl. Hepatol. 2022, 10, 1077–1085. [Google Scholar] [CrossRef]
- Imajo, K.; Honda, Y.; Kobayashi, T.; Nagai, K.; Ozaki, A.; Iwaki, M.; Kessoku, T.; Ogawa, Y.; Takahashi, H.; Saigusa, Y.; et al. Direct Comparison of US and MR Elastography for Staging Liver Fibrosis in Patients With Nonalcoholic Fatty Liver Disease. Clin. Gastroenterol. Hepatol. 2022, 20, 908–917.e911. [Google Scholar] [CrossRef]
- Indre, M.G.; Leucuta, D.C.; Lupsor-Platon, M.; Turco, L.; Ferri, S.; Hashim, A.; Orasan, O.H.; Procopet, B.; Stefanescu, H.; Morelli, M.C.; et al. Diagnostic accuracy of 2D-SWE ultrasound for liver fibrosis assessment in MASLD: A multilevel random effects model meta-analysis. Hepatology 2024, 82, 454–469. [Google Scholar] [CrossRef]
- Hu, W.; Li, H.; Wang, C.; Gou, S.; Fu, L. Characterization of collagen fibers by means of texture analysis of second harmonic generation images using orientation-dependent gray level co-occurrence matrix method. J. Biomed. Opt. 2012, 17, 026007. [Google Scholar] [CrossRef]
- Utino, F.L.; Garcia, M.; Velho, P.; França, A.; Stelini, R.F.; Pelegati, V.B.; Cesar, C.L.; de Souza, E.M.; Cintra, M.L.; Damiani, G.V. Second-harmonic generation imaging analysis can help distinguish sarcoidosis from tuberculoid leprosy. J. Biomed. Opt. 2018, 23, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Cicchi, R.; Kapsokalyvas, D.; De Giorgi, V.; Maio, V.; Van Wiechen, A.; Massi, D.; Lotti, T.; Pavone, F.S. Scoring of collagen organization in healthy and diseased human dermis by multiphoton microscopy. J. Biophotonics 2010, 3, 34–43. [Google Scholar] [CrossRef] [PubMed]
- Chunmei, X.; Mei, H.; Yan, Z.; Haiying, W. Diagnostic Method of Liver Cirrhosis Based on MR Image Texture Feature Extraction and Classification Algorithm. J. Med. Syst. 2019, 44, 11. [Google Scholar] [CrossRef]
- Zhang, X.; Gao, X.; Liu, B.J.; Ma, K.; Yan, W.; Liling, L.; Yuhong, H.; Fujita, H. Effective staging of fibrosis by the selected texture features of liver: Which one is better, CT or MR imaging? Comput. Med. Imaging Graph. 2015, 46, 227–236. [Google Scholar] [CrossRef] [PubMed]
- Gracia-Sancho, J.; Marrone, G.; Fernández-Iglesias, A. Hepatic microcirculation and mechanisms of portal hypertension. Nat. Rev. Gastroenterol. Hepatol. 2019, 16, 221–234. [Google Scholar] [CrossRef]
- Guixé-Muntet, S.; Quesada-Vázquez, S.; Gracia-Sancho, J. Pathophysiology and therapeutic options for cirrhotic portal hypertension. Lancet Gastroenterol. Hepatol. 2024, 9, 646–663. [Google Scholar] [CrossRef]
- Ginès, P.; Krag, A.; Abraldes, J.G.; Solà, E.; Fabrellas, N.; Kamath, P.S. Liver cirrhosis. Lancet 2021, 398, 1359–1376. [Google Scholar] [CrossRef]
- Muratori, L.; Lohse, A.W.; Lenzi, M. Diagnosis and management of autoimmune hepatitis. Bmj 2023, 380, e070201. [Google Scholar] [CrossRef]
- EASL Clinical Practice Guidelines: Autoimmune hepatitis. J. Hepatol. 2015, 63, 971–1004. [CrossRef]
- Youk, J.H.; Kwak, J.Y.; Lee, E.; Son, E.J.; Kim, J.A. Grayscale Ultrasound Radiomic Features and Shear-Wave Elastography Radiomic Features in Benign and Malignant Breast Masses. Ultraschall Med. 2020, 41, 390–396. [Google Scholar] [CrossRef]
- Alghamdi, D.; Kernohan, N.; Li, C.; Nabi, G. Comparative Assessment of Different Ultrasound Technologies in the Detection of Prostate Cancer: A Systematic Review and Meta-Analysis. Cancers 2023, 15, 4105. [Google Scholar] [CrossRef] [PubMed]
- Özer, H.; Yılmaz, S.; Bozkurt, B.; Tezcan, D.; Yazol, M.; Hakbilen, S.; Topaloğlu, Ö.F.; Durmaz, M.S. Assessment of lacrimal gland involvement in primary Sjögren’s syndrome using gray-scale ultrasonography and shear wave elastography. Eur. Radiol. 2023, 33, 9368–9377. [Google Scholar] [CrossRef] [PubMed]
- Caviglia, J.M.; Yan, J.; Jang, M.K.; Gwak, G.Y.; Affo, S.; Yu, L.; Olinga, P.; Friedman, R.A.; Chen, X.; Schwabe, R.F. MicroRNA-21 and Dicer are dispensable for hepatic stellate cell activation and the development of liver fibrosis. Hepatology 2018, 67, 2414–2429. [Google Scholar] [CrossRef] [PubMed]
- Kandil, R.; Merkel, O.M. Recent Progress of Polymeric Nanogels for Gene Delivery. Curr. Opin. Colloid. Interface Sci. 2019, 39, 11–23. [Google Scholar] [CrossRef]
- Zhang, Y.; Hamada, M. DeepM6ASeq: Prediction and characterization of m6A-containing sequences using deep learning. BMC Bioinform. 2018, 19, 524. [Google Scholar] [CrossRef]
- Hosseiniyan Khatibi, S.M.; Najjarian, F.; Homaei Rad, H.; Ardalan, M.; Teshnehlab, M.; Zununi Vahed, S.; Pirmoradi, S. Key therapeutic targets implicated at the early stage of hepatocellular carcinoma identified through machine-learning approaches. Sci. Rep. 2023, 13, 3840. [Google Scholar] [CrossRef]
- Zhong, Y.; Peng, Y.; Lin, Y.; Chen, D.; Zhang, H.; Zheng, W.; Chen, Y.; Wu, C. MODILM: Towards better complex diseases classification using a novel multi-omics data integration learning model. BMC Med. Inform. Decis. Mak. 2023, 23, 82. [Google Scholar] [CrossRef]

| Variable | All Patients | Training Cohort | Validation Cohort | p-Value |
|---|---|---|---|---|
| Number of patients | 198 (100.0%) | 136 (68.7%) | 62 (31.3%) | - |
| Gender (male) | 117(59.1%) | 82(60.3%) | 35(56.5%) | 0.610 |
| Age (years) | 51.1 ± 15.6 | 50.3 ± 16.3 | 52.9 ± 14.5 | 0.624 |
| SD (mm) | 145.2 ± 36.7 | 140.6 ± 33.8 | 156.5 ± 42.3 | 0.210 |
| ST (mm) | 49.8 ± 14.0 | 47.8 ± 12.7 | 54.7 ± 16.2 | 0.154 |
| SVD (mm) | 9.6 ± 3.9 | 9.5 ± 3.8 | 9.9 ± 4.4 | 0.770 |
| PVD (mm) | 12.8 ± 2.8 | 13.0 ± 3.0 | 12.4 ± 2.4 | 0.535 |
| PVV (cm/s) | 25.3 ± 10.4 | 25.2 ± 10.8 | 25.6 ± 9.8 | 0.925 |
| AST (U/L) | 43.6 ± 33.2 | 40.9 ± 36.1 | 49.8 ± 25.3 | 0.425 |
| ALT (U/L) | 34.5 ± 31.3 | 35.3 ± 35.9 | 32.7 ± 17.7 | 0.808 |
| GGT (U/L) | 83.6 ±112.1 | 64.7 ± 67.7 | 127.1 ± 173.0 | 0.094 |
| ALB (g/L) | 38.4 ± 5.8 | 39.1 ± 6.2 | 36.8 ± 4.7 | 0.254 |
| TBIL (μmol/L) | 35.4 ± 33.1 | 34.9 ± 36.1 | 36.6 ± 26.3 | 0.878 |
| DBIL (μmol/L) | 11.4 ± 16.8 | 12.6 ± 19.7 | 8.7 ± 5.8 | 0.493 |
| IBIL (μmol/L) | 24.1 ± 19.5 | 22.4 ± 18.5 | 28.1 ± 21.8 | 0.387 |
| PLT (109/L) | 126.9 ±72.5 | 134.0 ± 76.1 | 110.6 ± 63.4 | 0.338 |
| Etiology | 0.295 | |||
| HBV | 101 (51.0%) | 69 (50.7%) | 32 (51.6%) | |
| AILD | 97 (49.0%) | 67 (49.3%) | 30 (48.4%) | |
| Fibrosis stages | 0.997 | |||
| S2 | 58 (29.3%) | 40 (29.4%) | 18 (29.0%) | |
| S3 | 51 (25.8%) | 35 (25.7%) | 16 (25.8%) | |
| S4 | 89 (44.9%) | 61 (44.9%) | 28 (45.2%) |
| AUC | Sensitivity (%) | Specificity (%) | PPV (%) | NPV (%) | LR+ | LR− | ||
|---|---|---|---|---|---|---|---|---|
| liver grayscale images | ||||||||
| S4 | ||||||||
| TL | T | 0.60 (0.55–0.66) | 0.16 | 0.92 | 0.49 | 0.71 | 2.10 | 0.91 |
| V | 0.55 (0.43–0.67) | 0.13 | 0.93 | 0.33 | 0.80 | 1.83 | 0.94 | |
| ≥S3 | ||||||||
| TL | T | 0.54 (0.48–0.60) | 0.34 | 0.75 | 0.42 | 0.69 | 1.37 | 0.88 |
| V | 0.51 (0.39–0.63) | 0.33 | 0.72 | 0.30 | 0.75 | 1.19 | 0.93 | |
| ≥S2 | ||||||||
| TL | T | 0.59 (0.54–0.65) | 0.69 | 0.42 | 0.39 | 0.72 | 1.20 | 0.73 |
| V | 0.46 (0.35–0.58) | 0.62 | 0.37 | 0.51 | 0.48 | 0.99 | 1.02 | |
| spleen grayscale images | ||||||||
| S4 | ||||||||
| TL | T | 0.79 (0.72–0.84) | 0.76 | 0.68 | 0.58 | 0.83 | 2.35 | 0.36 |
| V | 0.55 (0.38–0.71) | 0.47 | 0.64 | 0.26 | 0.82 | 1.31 | 0.83 | |
| ≥S3 | ||||||||
| TL | T | 0.63 (0.56–0.70) | 0.24 | 0.84 | 0.43 | 0.69 | 1.54 | 0.90 |
| V | 0.42 (0.26–0.57) | 0.20 | 0.92 | 0.50 | 0.75 | 2.56 | 0.87 | |
| ≥S2 | ||||||||
| TL | T | 0.75 (0.68–0.81) | 0.55 | 0.76 | 0.49 | 0.80 | 2.31 | 0.59 |
| V | 0.61 (0.48–0.75) | 0.69 | 0.69 | 0.69 | 0.68 | 2.21 | 0.45 | |
| liver 2D-SWE images | ||||||||
| S4 | ||||||||
| TL | T | 0.99 (0.99–1.00) | 0.83 | 1.00 | 1.00 | 0.91 | - | 0.17 |
| V | 0.92 (0.81–1.00) | 0.84 | 0.89 | 0.93 | 0.76 | 7.59 | 0.18 | |
| ≥S3 | ||||||||
| TL | T | 0.99 (0.99–1.00) | 1.00 | 0.94 | 0.85 | 1.00 | 16.63 | 0.00 |
| V | 0.94 (0.84–1.00) | 0.78 | 0.93 | 0.70 | 0.95 | 10.63 | 0.24 | |
| ≥S2 | ||||||||
| TL | T | 0.99 (0.99–1.00) | 1.00 | 0.97 | 0.96 | 1.00 | 37.33 | 0.00 |
| V | 0.94 (0.79–1.00) | 0.89 | 0.93 | 0.73 | 0.97 | 12.15 | 0.12 | |
| spleen 2D-SWE images | ||||||||
| S4 | ||||||||
| TL | T | 0.95 (0.92–0.98) | 0.84 | 0.89 | 0.78 | 0.92 | 7.79 | 0.18 |
| V | 0.84 (0.70–0.95) | 0.74 | 0.73 | 0.81 | 0.65 | 2.78 | 0.36 | |
| ≥S3 | ||||||||
| TL | T | 0.99 (0.97–1.00) | 0.88 | 0.99 | 0.98 | 0.93 | 75.00 | 0.12 |
| V | 0.92 (0.78–1.00) | 0.78 | 0.97 | 0.88 | 0.93 | 22.56 | 0.23 | |
| ≥S2 | ||||||||
| TL | T | 0.97 (0.94–0.99) | 0.90 | 0.94 | 0.86 | 0.96 | 14.17 | 0.10 |
| V | 0.76 (0.57–0.91) | 0.33 | 0.78 | 0.22 | 0.86 | 1.52 | 0.85 | |
| CTL(grayscale) | ||||||||
| S4 | ||||||||
| T | 0.99 (0.99–1.00) | 0.98 | 0.98 | 0.95 | 0.99 | 47.47 | 0.02 | |
| V | 0.62 (0.45–0.80) | 0.38 | 0.74 | 0.31 | 0.80 | 1.50 | 0.83 | |
| ≥S3 | ||||||||
| T | 0.98 (0.96–0.99) | 0.96 | 0.87 | 0.82 | 0.97 | 7.45 | 0.04 | |
| V | 0.57 (0.39–0.75) | 0.47 | 0.68 | 0.35 | 0.78 | 1.47 | 0.78 | |
| ≥S2 | ||||||||
| T | 0.98 (0.97–0.99) | 0.75 | 0.99 | 0.96 | 0.89 | 49.88 | 0.25 | |
| V | 0.48 (0.33–0.63) | 0.39 | 0.68 | 0.55 | 0.53 | 1.22 | 0.89 | |
| CTL(2D-SWE) | ||||||||
| S4 | ||||||||
| T | 0.99 (0.98–1.00) | 0.93 | 1.00 | 1.00 | 0.97 | - | 0.07 | |
| V | 0.99 (0.96–1.00) | 0.84 | 1.00 | 1.00 | 0.78 | - | 0.16 | |
| ≥S3 | ||||||||
| T | 1.00 (1.00–1.00) | 1.00 | 0.97 | 0.96 | 1.00 | 37.67 | 0.00 | |
| V | 0.98 (0.95–1.00) | 1.00 | 0.90 | 0.69 | 1.00 | 10.25 | 0.00 | |
| ≥S2 | ||||||||
| T | 0.99 (0.99–1.00) | 1.00 | 0.99 | 0.98 | 1.00 | 122.00 | 0.00 | |
| V | 1.00 (1.00–1.00) | 1.00 | 0.98 | 0.90 | 1.00 | 41.00 | 0.00 | |
| APRI | ||||||||
| S4 | ||||||||
| T | 0.63 (0.48–0.78) | 0.38 | 0.79 | 0.78 | 0.39 | 1.80 | 0.79 | |
| V | 0.62 (0.39–0.85) | 0.50 | 0.89 | 0.89 | 0.50 | 4.50 | 0.56 | |
| ≥S3 | ||||||||
| T | 0.58 (0.39–0.77) | 0.17 | 0.91 | 0.33 | 0.80 | 1.83 | 0.92 | |
| V | 0.58 (0.31–0.85) | 0.33 | 0.89 | 0.50 | 0.81 | 3.17 | 0.75 | |
| ≥S2 | ||||||||
| T | 0.65 (0.42–0.89) | 0.71 | 0.45 | 0.16 | 0.92 | 1.30 | 0.64 | |
| V | 0.70 (0.35–1.00) | 0.67 | 0.55 | 0.17 | 0.92 | 1.47 | 0.61 | |
| FIB-4 | ||||||||
| S4 | ||||||||
| T | 0.67 (0.52–0.81) | 0.92 | 0.42 | 0.76 | 0.73 | 1.59 | 0.19 | |
| V | 0.67 (0.42–0.89) | 0.88 | 0.22 | 0.67 | 0.50 | 1.12 | 0.56 | |
| ≥S3 | ||||||||
| T | 0.49 (0.31–0.67) | 0.42 | 0.66 | 0.25 | 0.81 | 1.22 | 0.89 | |
| V | 0.37 (0.16–0.59) | 0.17 | 0.58 | 0.11 | 0.69 | 0.40 | 1.44 | |
| ≥S2 | ||||||||
| T | 0.69 (0.44–0.90) | 0.86 | 0.37 | 0.17 | 0.95 | 1.40 | 0.37 | |
| V | 0.59 (0.25–1.00) | 1.00 | 0.32 | 0.17 | 1.00 | 1.47 | 0.00 | |
| AUC | Sensitivity (%) | Specificity (%) | PPV (%) | NPV (%) | LR+ | LR− | ||
|---|---|---|---|---|---|---|---|---|
| Liver | ||||||||
| RF | T | 1.00(1.00–1.00) | 1.00 | 1.00 | 1.00 | 1.00 | - | 0.00 |
| V | 0.78(0.72–0.85) | 0.71 | 0.70 | 0.70 | 0.70 | 2.33 | 0.42 | |
| DT | T | 0.99(0.99–1.00) | 0.98 | 0.99 | 0.99 | 0.98 | 389.00 | 0.02 |
| V | 0.56(0.46–0.66) | 0.54 | 0.57 | 0.55 | 0.55 | 1.23 | 0.82 | |
| LR | T | 0.66(0.63–0.70) | 0.62 | 0.65 | 0.64 | 0.63 | 1.74 | 0.59 |
| V | 0.69(0.61–0.76) | 0.61 | 0.68 | 0.65 | 0.63 | 1.88 | 0.58 | |
| SVM | T | 0.80(0.77–0.83) | 0.76 | 0.73 | 0.74 | 0.75 | 2.78 | 0.33 |
| V | 0.76(0.79–0.83) | 0.68 | 0.68 | 0.68 | 0.68 | 2.09 | 0.48 | |
| GBM | T | 1.00(1.00–1.00) | 1.00 | 1.00 | 1.00 | 1.00 | - | 0.00 |
| V | 0.76(0.70–0.83) | 0.72 | 0.70 | 0.70 | 0.71 | 2.37 | 0.41 | |
| XGB | T | 1.00(1.00–1.00) | 1.00 | 1.00 | 1.00 | 1.00 | - | 0.00 |
| V | 0.76(0.70–0.83) | 0.68 | 0.70 | 0.69 | 0.68 | 2.23 | 0.46 | |
| TL | T | 0.76(0.72–0.80) | 0.52 | 0.82 | 0.73 | 0.64 | 2.82 | 0.59 |
| V | 0.73(0.64–0.81) | 0.48 | 0.81 | 0.62 | 0.71 | 2.52 | 0.64 | |
| Spleen | ||||||||
| RF | T | 1.00(1.00–1.00) | 1.00 | 1.00 | 1.00 | 1.00 | - | 0.00 |
| V | 0.78(0.70–0.86) | 0.65 | 0.73 | 0.71 | 0.68 | 2.44 | 0.48 | |
| DT | T | 1.00(0.99–1.00) | 0.99 | 1.00 | 1.00 | 0.99 | - | 0.01 |
| V | 0.61(0.47–0.74) | 0.57 | 0.63 | 0.61 | 0.59 | 1.55 | 0.68 | |
| LR | T | 0.80(0.76–0.84) | 0.73 | 0.69 | 0.70 | 0.72 | 2.35 | 0.40 |
| V | 0.68(0.59–0.78) | 0.65 | 0.55 | 0.59 | 0.61 | 1.44 | 0.64 | |
| SVM | T | 0.99(0.99–1.00) | 0.95 | 0.98 | 0.98 | 0.95 | 45.40 | 0.06 |
| V | 0.78(0.70–0.87) | 0.62 | 0.82 | 0.77 | 0.68 | 3.36 | 0.47 | |
| GBM | T | 1.00(1.00–1.00) | 1.00 | 1.00 | 1.00 | 1.00 | - | 0.00 |
| V | 0.81(0.73–0.88) | 0.70 | 0.78 | 0.76 | 0.72 | 3.23 | 0.38 | |
| XGB | T | 1.00(1.00–1.00) | 1.00 | 1.00 | 1.00 | 1.00 | - | 0.00 |
| V | 0.74(0.66–0.83) | 0.63 | 0.73 | 0.70 | 0.67 | 2.38 | 0.50 | |
| TL | T | 0.72(0.66–0.77) | 0.45 | 0.82 | 0.71 | 0.61 | 2.56 | 0.66 |
| V | 0.57(0.44–0.68) | 0.36 | 0.83 | 0.58 | 0.67 | 2.15 | 0.77 | |
| Combined- TL | T | 0.96(0.94–0.98) | 0.78 | 0.97 | 0.94 | 0.87 | 22.64 | 0.23 |
| V | 0.79(0.73–0.86) | 0.61 | 0.82 | 0.68 | 0.76 | 3.35 | 0.48 | |
| AUC | Sensitivity (%) | Specificity (%) | PPV (%) | NPV (%) | LR+ | LR− | ||
|---|---|---|---|---|---|---|---|---|
| Liver | ||||||||
| RF | T | 1.00(1.00–1.00) | 1.00 | 1.00 | 1.00 | 1.00 | - | 0.00 |
| V | 0.78(0.65–0.91) | 0.62 | 0.81 | 0.76 | 0.68 | 3.20 | 0.48 | |
| DT | T | 0.99(0.99–1.00) | 0.98 | 0.99 | 0.99 | 0.98 | 389.00 | 0.02 |
| V | 0.77(0.58–0.96) | 0.62 | 0.81 | 0.76 | 0.68 | 3.20 | 0.48 | |
| LR | T | 0.78(0.71–0.84) | 0.62 | 0.65 | 0.64 | 0.63 | 1.74 | 0.59 |
| V | 0.72(0.58–0.86) | 0.62 | 0.73 | 0.70 | 0.66 | 2.29 | 0.53 | |
| SVM | T | 0.99(0.98–1.00) | 0.76 | 0.73 | 0.74 | 0.75 | 2.78 | 0.33 |
| V | 0.84(0.73–0.95) | 0.73 | 0.77 | 0.76 | 0.74 | 3.17 | 0.35 | |
| GBM | T | 1.00(1.00–1.00) | 1.00 | 1.00 | 1.00 | 1.00 | - | 0.00 |
| V | 0.82(0.70–0.93) | 0.65 | 0.81 | 0.77 | 0.70 | 3.40 | 0.43 | |
| XGB | T | 1.00(1.00–1.00) | 1.00 | 1.00 | 1.00 | 1.00 | - | 0.00 |
| V | 0.79(0.67–0.92) | 0.65 | 0.73 | 0.71 | 0.68 | 2.43 | 0.47 | |
| TL | T | 0.99(0.99–1.00) | 0.99 | 0.99 | 0.99 | 0.99 | 94.75 | 0.01 |
| V | 0.97(0.93–1.00) | 0.87 | 0.92 | 0.91 | 0.89 | 11.30 | 0.14 | |
| Spleen | ||||||||
| RF | T | 1.00(1.00–1.00) | 1.00 | 1.00 | 1.00 | 1.00 | - | 0.00 |
| V | 0.94(0.88–1.00) | 0.71 | 0.95 | 0.94 | 0.77 | 15.00 | 0.30 | |
| DT | T | 1.00(1.00–1.00) | 1.00 | 1.00 | 1.00 | 1.00 | - | 0.00 |
| V | 0.83(0.62–1.00) | 0.76 | 0.90 | 0.89 | 0.79 | 8.00 | 0.26 | |
| LR | T | 0.93(0.89–0.96) | 0.80 | 0.87 | 0.86 | 0.81 | 6.09 | 0.23 |
| V | 0.81(0.68–0.94) | 0.71 | 0.76 | 0.75 | 0.73 | 3.00 | 0.38 | |
| SVM | T | 1.00(1.00–1.00) | 0.99 | 1.00 | 1.00 | 0.99 | - | 0.01 |
| V | 0.84(0.70–0.98) | 0.67 | 0.95 | 0.93 | 0.74 | 14.00 | 0.35 | |
| GBM | T | 1.00(1.00–1.00) | 1.00 | 1.00 | 1.00 | 1.00 | - | 0.00 |
| V | 0.93(0.86–0.99) | 0.71 | 0.86 | 0.83 | 0.75 | 5.00 | 0.33 | |
| XGB | T | 1.00(1.00–1.00) | 1.00 | 1.00 | 1.00 | 1.00 | - | 0.00 |
| V | 0.88(0.77–0.98) | 0.76 | 0.90 | 0.89 | 0.79 | 8.00 | 0.26 | |
| TL | T | 0.98(0.96–0.99) | 0.94 | 0.91 | 0.91 | 0.94 | 10.64 | 0.07 |
| V | 0.92(0.82–0.99) | 0.82 | 0.86 | 0.82 | 0.86 | 5.76 | 0.21 | |
| Combined- TL | T | 1.00(1.00–1.00) | 0.98 | 1.00 | 1.00 | 0.98 | - | 0.03 |
| V | 0.94(0.85–1.00) | 0.78 | 0.96 | 0.95 | 0.83 | 20.35 | 0.23 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, K.; Chen, F.; Tian, A.; Deng, L.; Mao, X. A Novel Deep Learning Framework for Liver Fibrosis Staging and Etiology Diagnosis Using Integrated Liver–Spleen Elastography. Diagnostics 2025, 15, 2986. https://doi.org/10.3390/diagnostics15232986
Yang K, Chen F, Tian A, Deng L, Mao X. A Novel Deep Learning Framework for Liver Fibrosis Staging and Etiology Diagnosis Using Integrated Liver–Spleen Elastography. Diagnostics. 2025; 15(23):2986. https://doi.org/10.3390/diagnostics15232986
Chicago/Turabian StyleYang, Kai, Fei Chen, Aiping Tian, Long Deng, and Xiaorong Mao. 2025. "A Novel Deep Learning Framework for Liver Fibrosis Staging and Etiology Diagnosis Using Integrated Liver–Spleen Elastography" Diagnostics 15, no. 23: 2986. https://doi.org/10.3390/diagnostics15232986
APA StyleYang, K., Chen, F., Tian, A., Deng, L., & Mao, X. (2025). A Novel Deep Learning Framework for Liver Fibrosis Staging and Etiology Diagnosis Using Integrated Liver–Spleen Elastography. Diagnostics, 15(23), 2986. https://doi.org/10.3390/diagnostics15232986




