A ResNet-50–UNet Hybrid with Whale Optimization Algorithm for Accurate Liver Tumor Segmentation
Abstract
1. Introduction
- We propose a novel hybrid optimization-segmentation framework called LiTs-Res-Unet + WOA, which integrates the Whale Optimization Algorithm (WOA) with a UNet-based architecture to automatically and adaptively tune the hyperparameters required for liver and tumor segmentation.
- Unlike prior studies that apply WOA to optimize a single parameter, we propose a closed-loop optimization framework in which WOA tunes a set of interdependent hyperparameters (learning rate, dropout rate, batch size) dynamically during training. This introduces a meta-optimization layer that is adaptive to the changing loss landscape of the ResNet-50–U-Net model.
- The inner level updates the network weights using backpropagation, and the outer level optimizes a population-based hyperparameter search using WOA by the validation Dice score. The convergence of validation performance shows that the hyperparameter optimization (HO) by the WOA outperforms the grid search and the Bayesian optimization methods because it achieves a better exploration–exploitation trade-off.
- Extensive experiments conducted on the LiTS17 benchmark dataset showed that the proposed model demonstrated state-of-the-art performance—with achieved pixel accuracy of 99.54%, Dice coefficient of 92.38%, and Jaccard index of 86.73%, respectively, which significantly outperformed existing segmentation techniques.
- The research results indicate that LiTs-UNet-WOA exhibits superior accuracy and robustness and has the potential to be an effective solution for liver tumor detection and treatment planning in real-life clinical applications.
2. Related Work
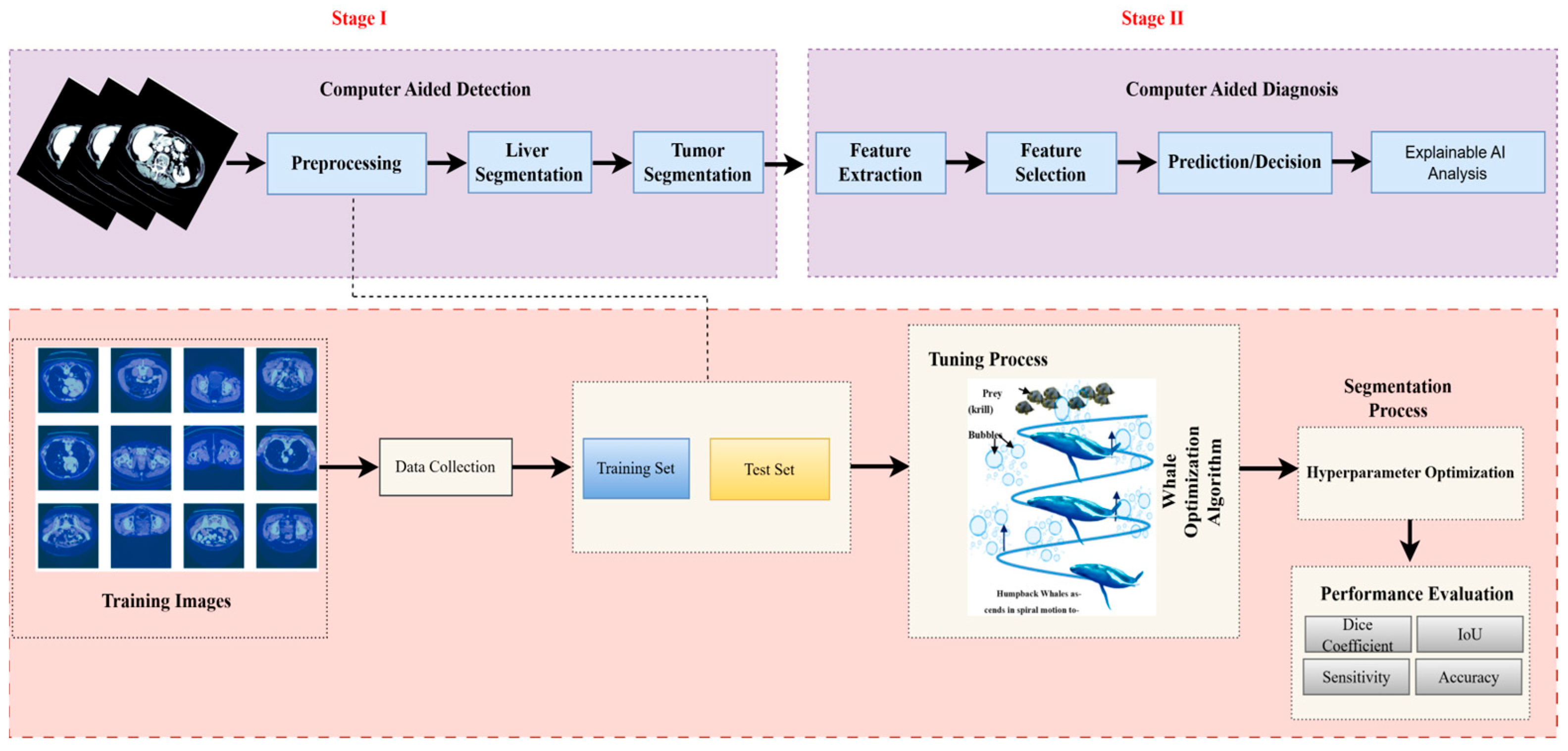
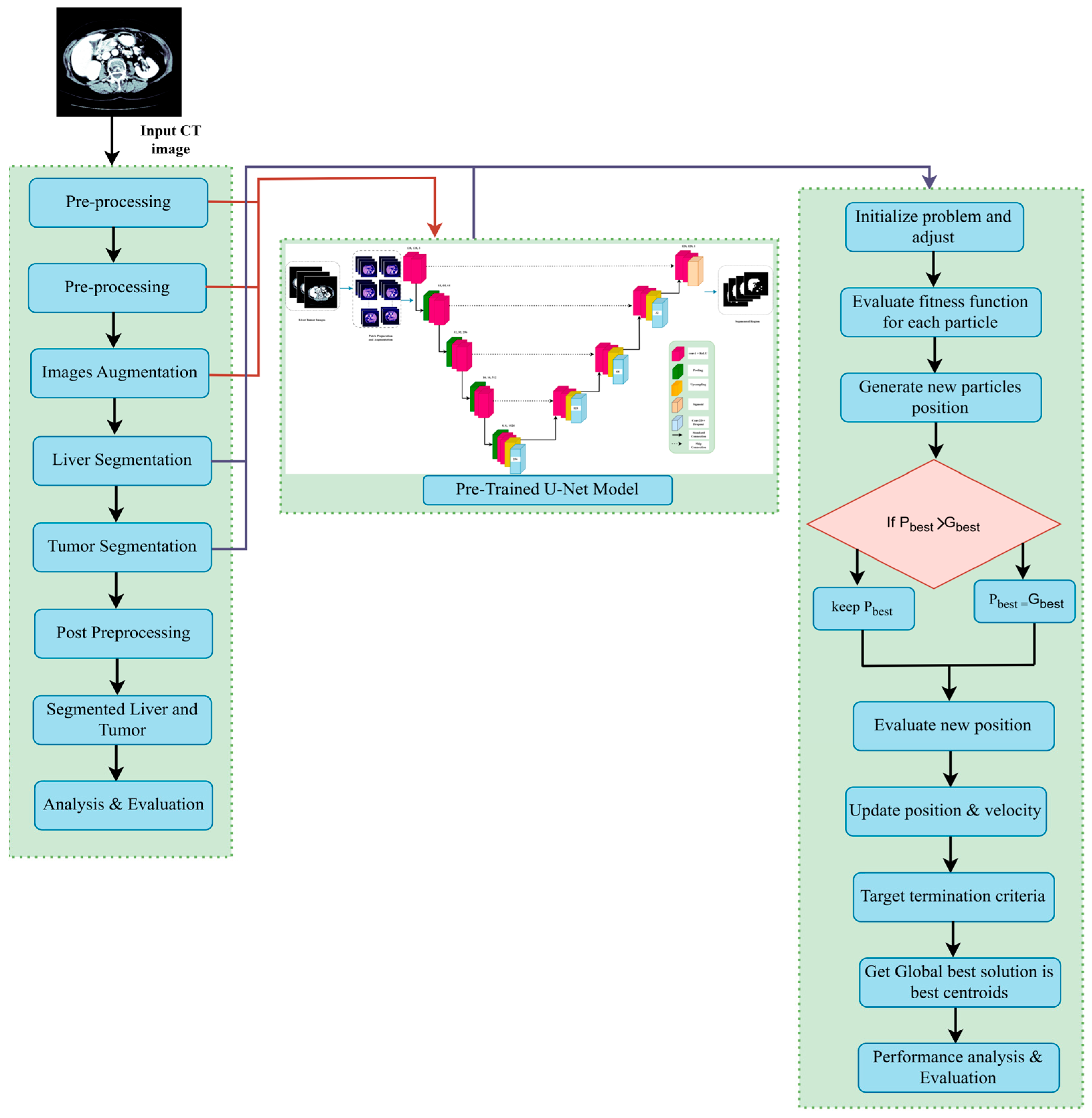
3. Methodology
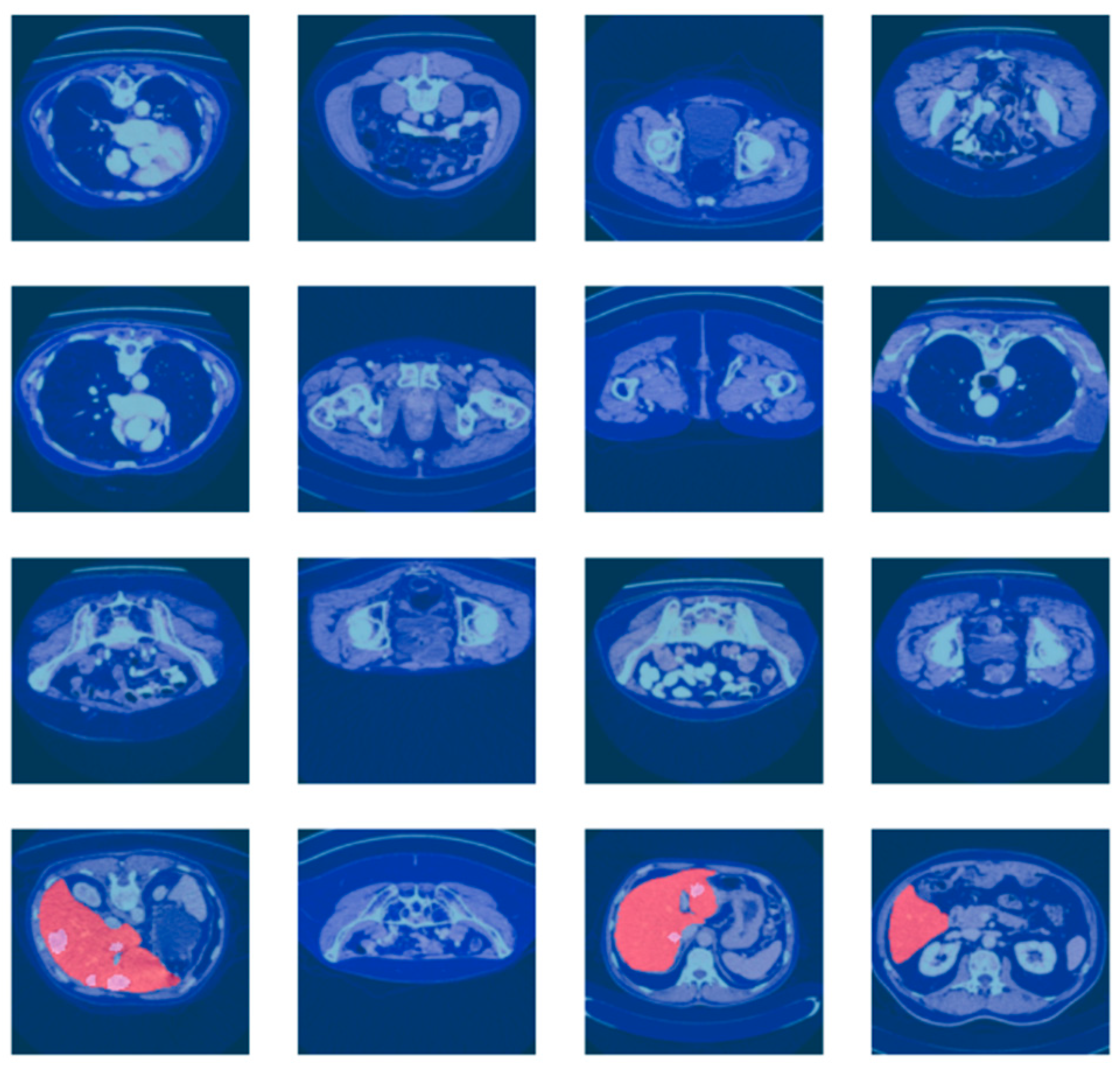
3.1. Dataset Description
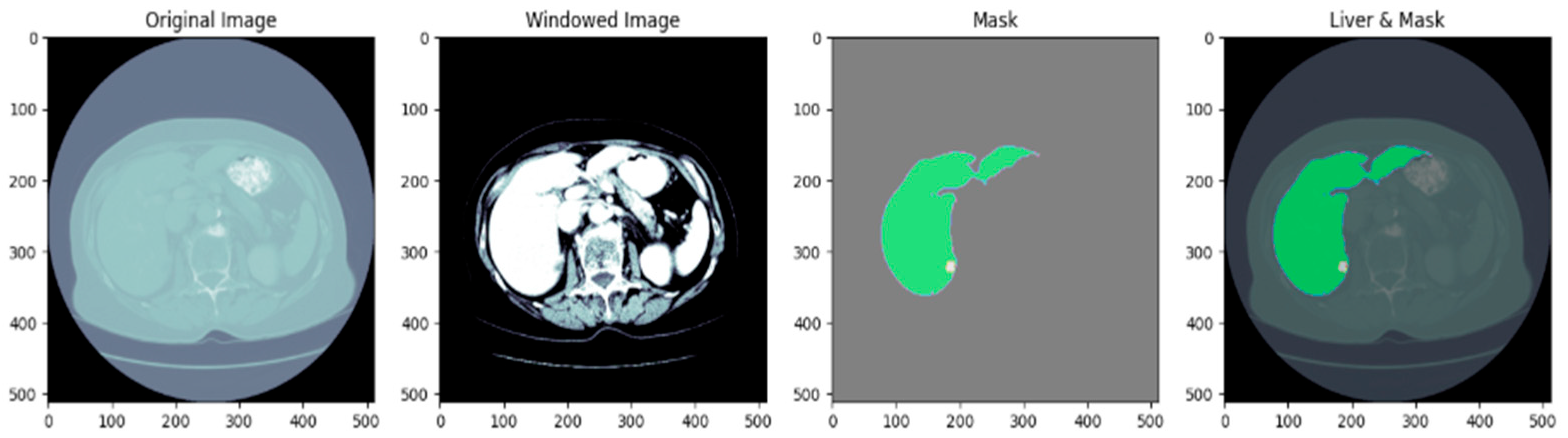
3.2. Dataset Preprocessing
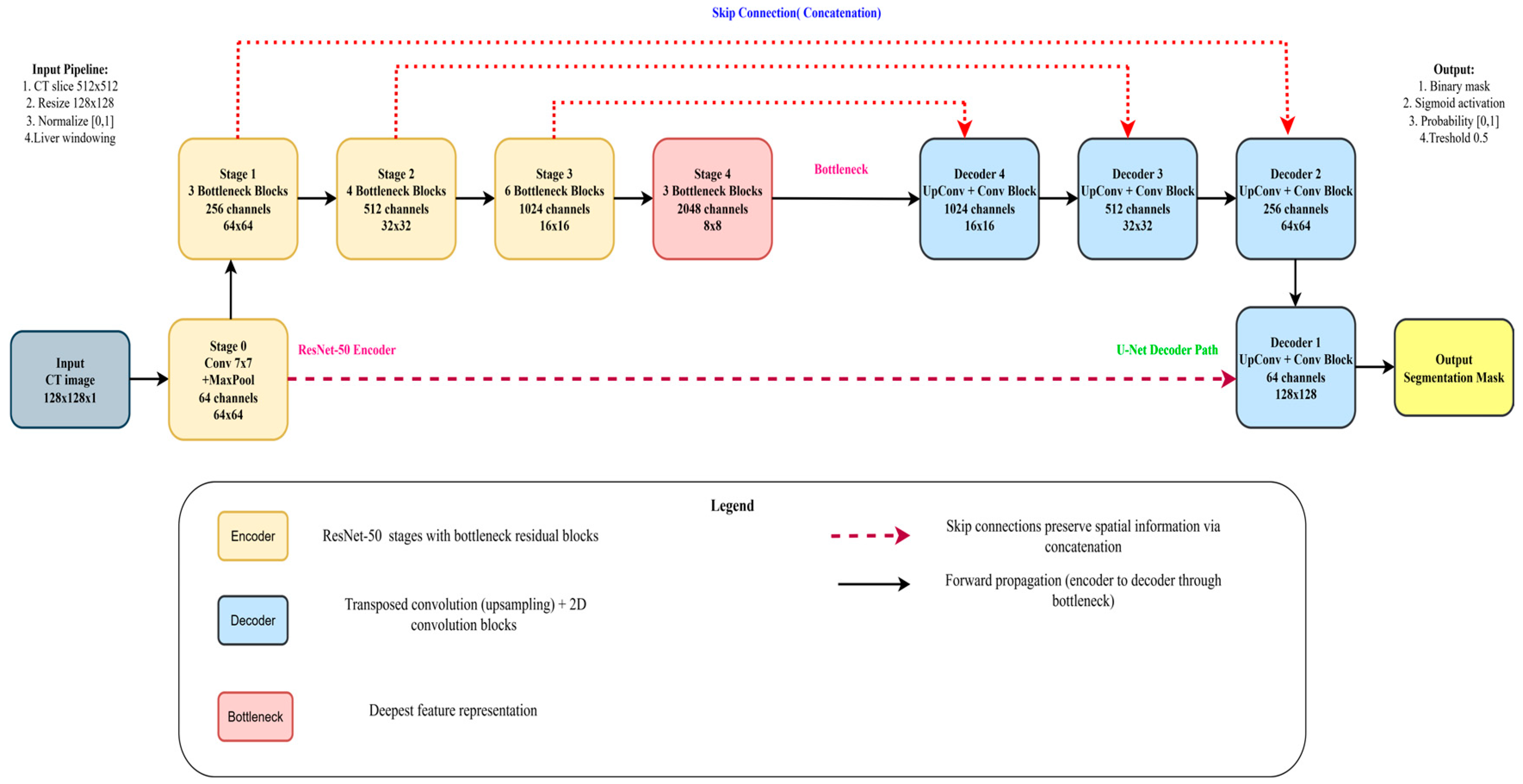
3.3. Based Model Architecture
3.3.1. Pre-Trained Encoder
3.3.2. Model Decoder Path
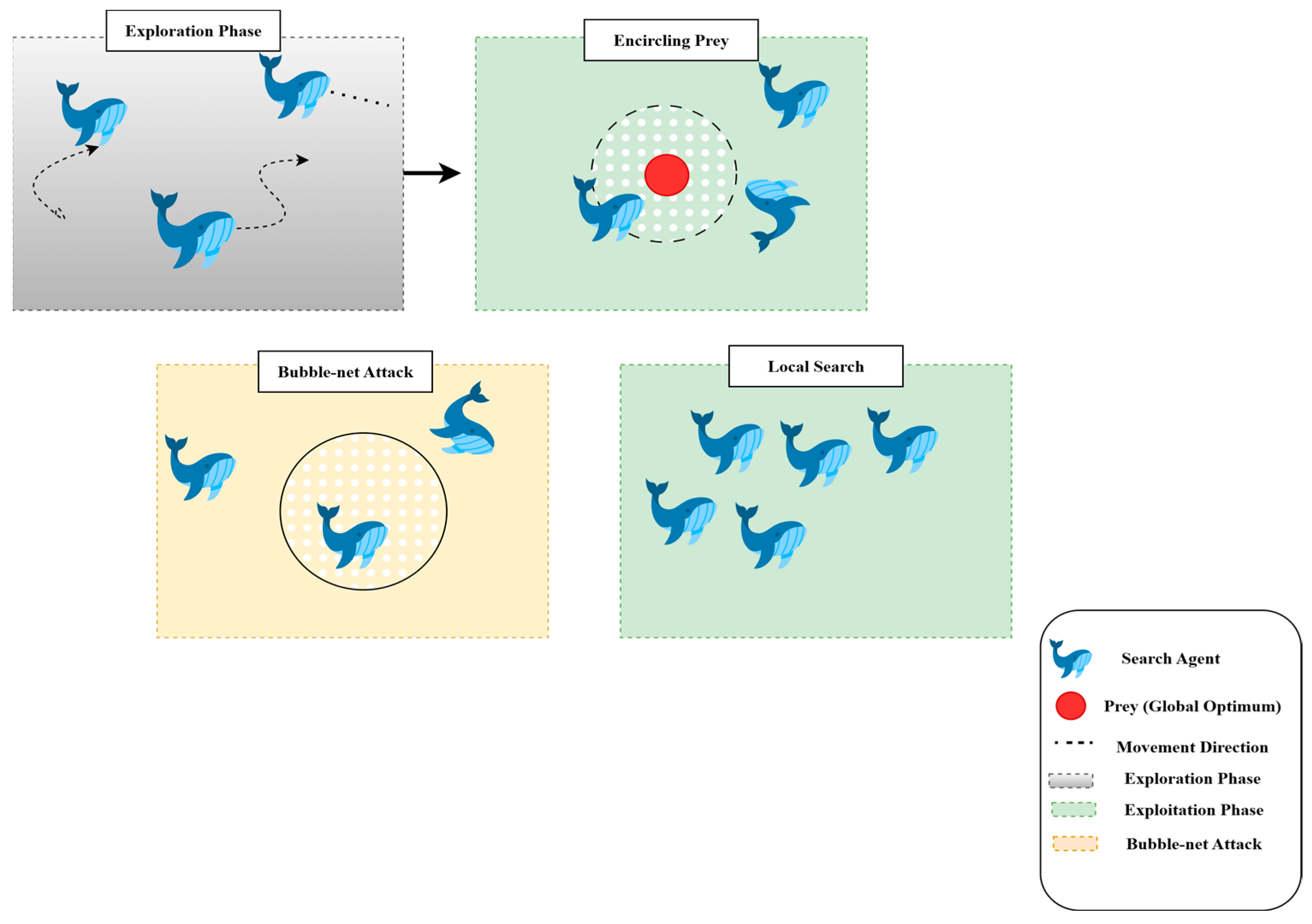
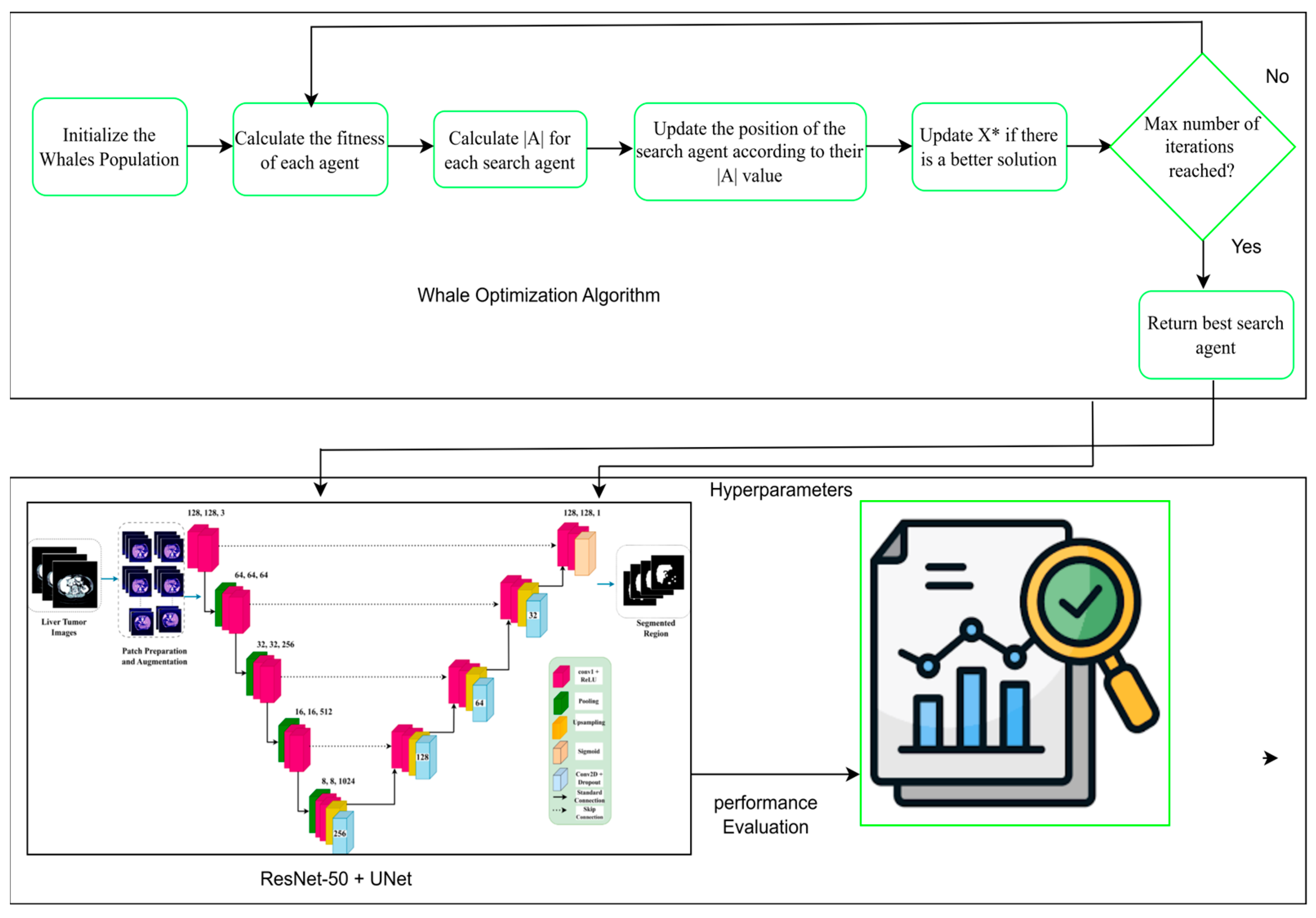
3.4. Swarm-Based Whale Optimization Algorithm (WOA)
3.4.1. Motivation from Nature
3.4.2. Exploration and Exploitation Phases
- Exploration: This process iteratively traverses the search dimensions in search of the global optimum. During this phase, whales move randomly to search for unvisited regions. The search is guided by the position of the best solution found so far, allowing the algorithm to escape local optima.
- Exploitation: This phase focuses on refining the search around the best solution detected. Whales use the shrinking (enclosing) mechanism along with the spiral path, updating their position by considering the spiral trajectory, which converges more effectively to the global optimum.
3.5. Proposed U-Net Segmentation with WOA
| Algorithm 1: Proposed WOA-driven meta-optimization framework integrated with ResNet-50–UNet for adaptive liver tumor segmentation. |
Input:
2: Set a = 2 (WOA control parameter) 3: for t = 1 to T_max do 4: for each whale i = 1 to N do 5: M_i ← Train_ResNet50_UNet (X_train, Y_train, H_i, epochs = 5) 6: Dice_i ← Evaluate_Dice (M_i, X_val, Y_val) 7: Stability_i ← 1 − std(val_loss_last_10_epochs) 8: F_i ← 0.7 × Dice_i + 0.3 × Stability_i 9: end for 10: H* ← argmax_{H_i} F_i (Identify best solution) 11: for each whale i = 1 to N do 12: Update a, A, C, l, p 13: if p < 0.5 then 14: if |A| < 1 then (Exploitation) 15: D ← |C · H* − H_i| 16: H_i ← H* − A · D 17: else 18: H_rand ← Random whale position (Exploitation) 19: D ← |C · H_rand − H_i| 20: H_i ← H_rand − A · D 21: end if 22: else 23: // Spiral update (exploitation) 24: D′ ← |H* − H_i| 25: H_i ← D′ · exp(b·l) · cos(2πl) + H* 26: end if 27: // Ensure bounds 28: H_i ← Clip(H_i, Ω) 29: end for 30: // Linear decrease of a from 2 to 0 31: a ← 2 − 2·t/T_max 32: end for 33: M* ← Train_ResNet50_UNet (X_train, Y_train, H*, epochs = 100) 34: return H*, M* |
3.5.1. Hyperparameter Tuning with Whale Optimization Algorithm
3.5.2. Mathematical Formulation of WOA-Segmentation Coupling
- Problem Formulation:
- Fitness Function Design:
- Segmentation Loss Function:
- WOA-Driven Hyperparameter Update:
- Boundary Enforcement:
- Convergence Criterion:
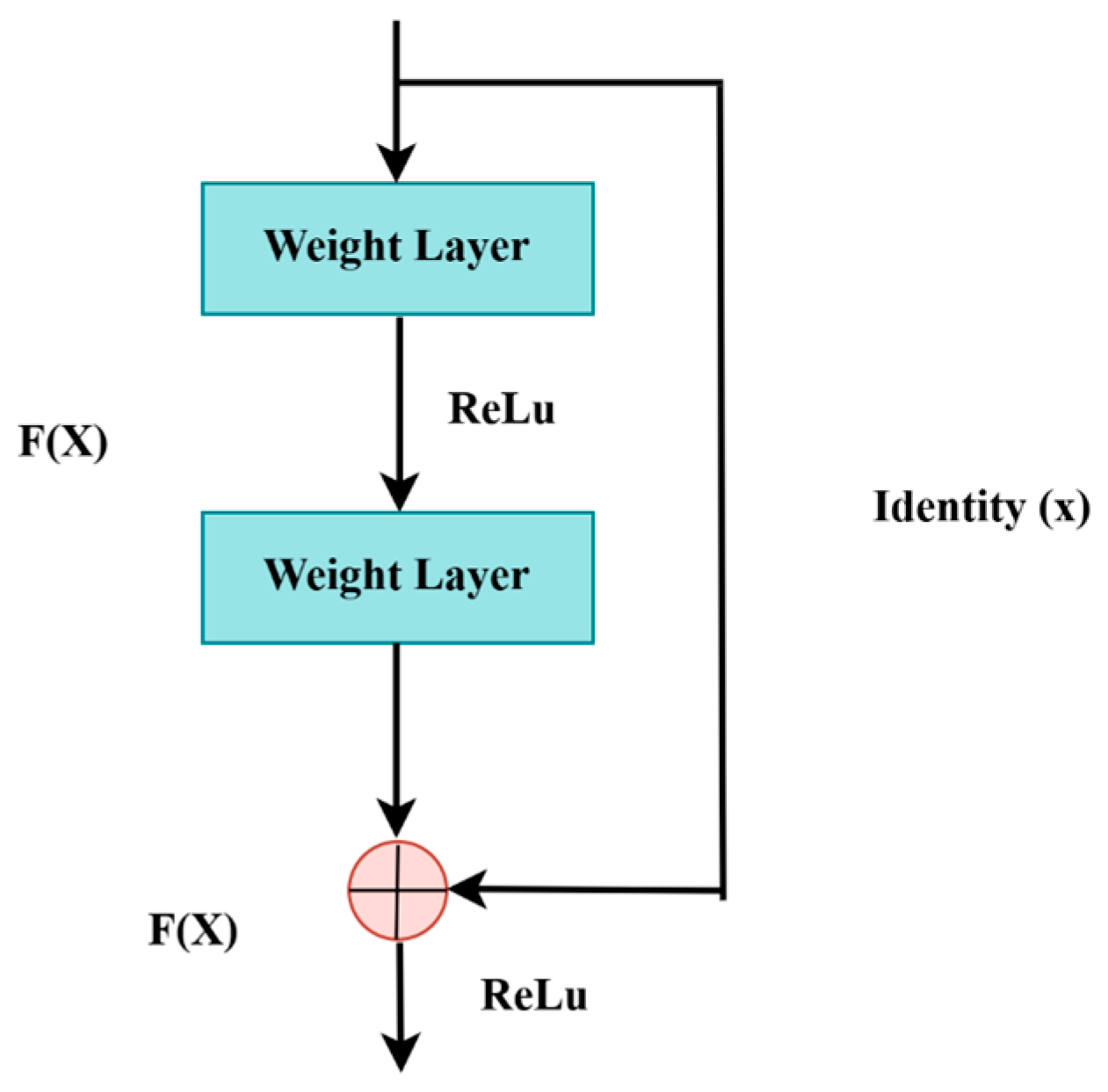
3.6. ResNet-50 + U-Net
3.6.1. Encoder
3.6.2. Decoder
3.6.3. Bottleneck
3.6.4. ResNet-50 Backbone
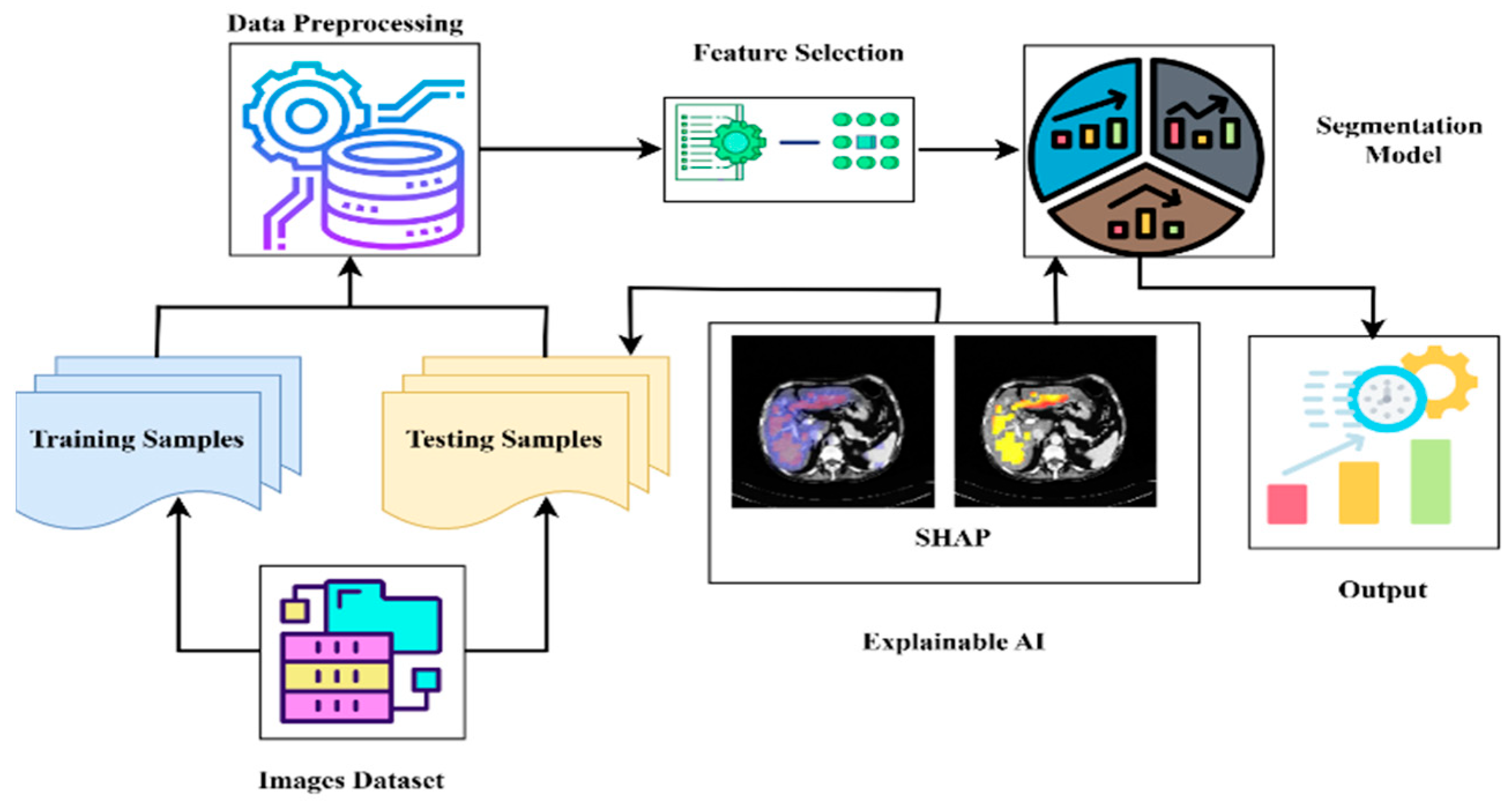
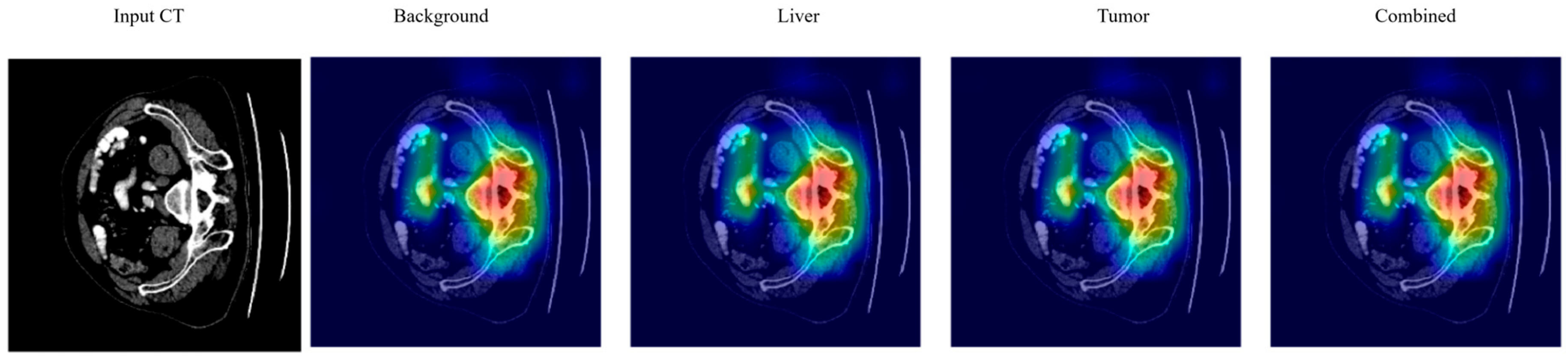
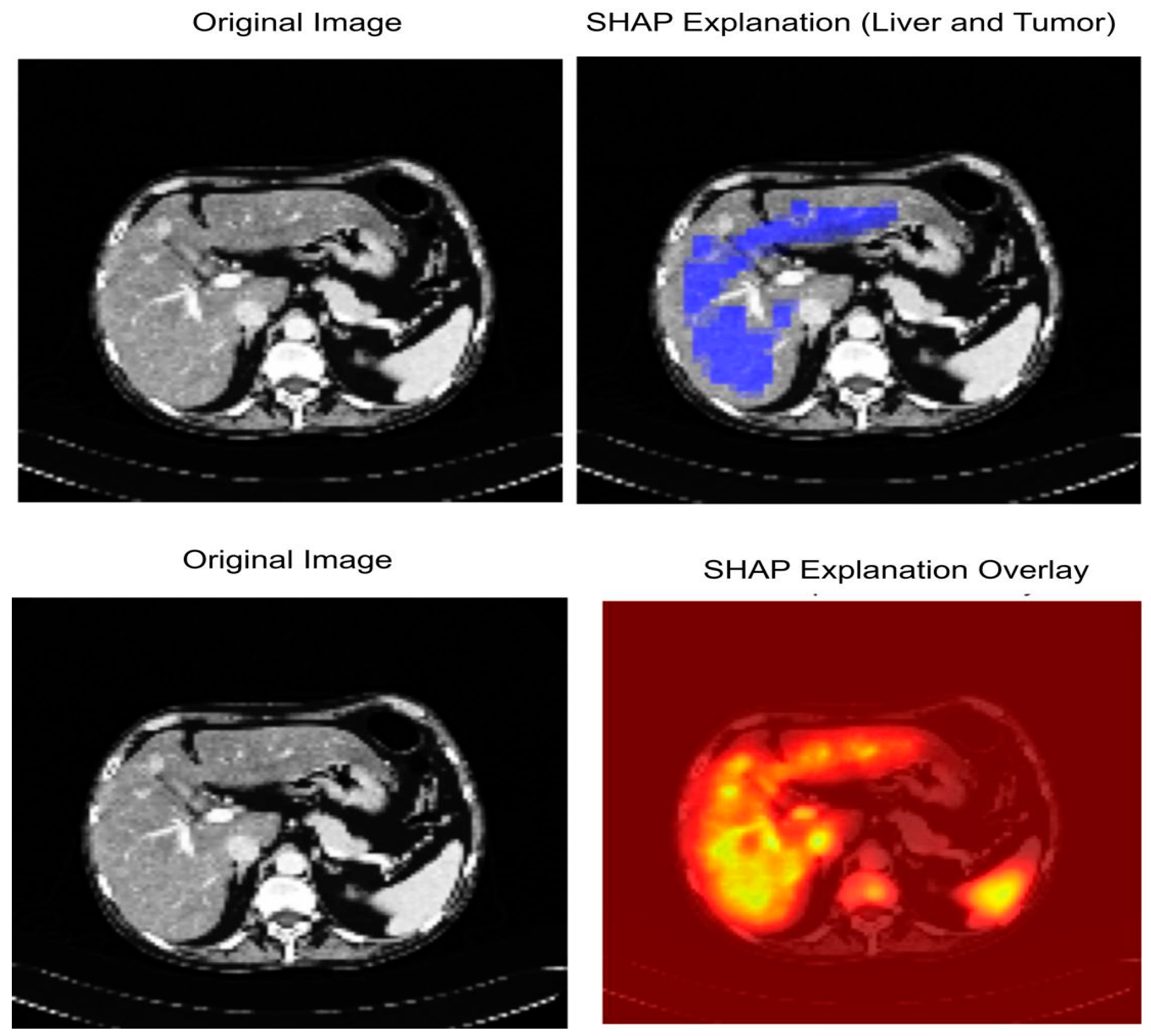
3.7. Explainable AI (XAI)
3.8. Performance Measures
4. Experimental Results and Discussions
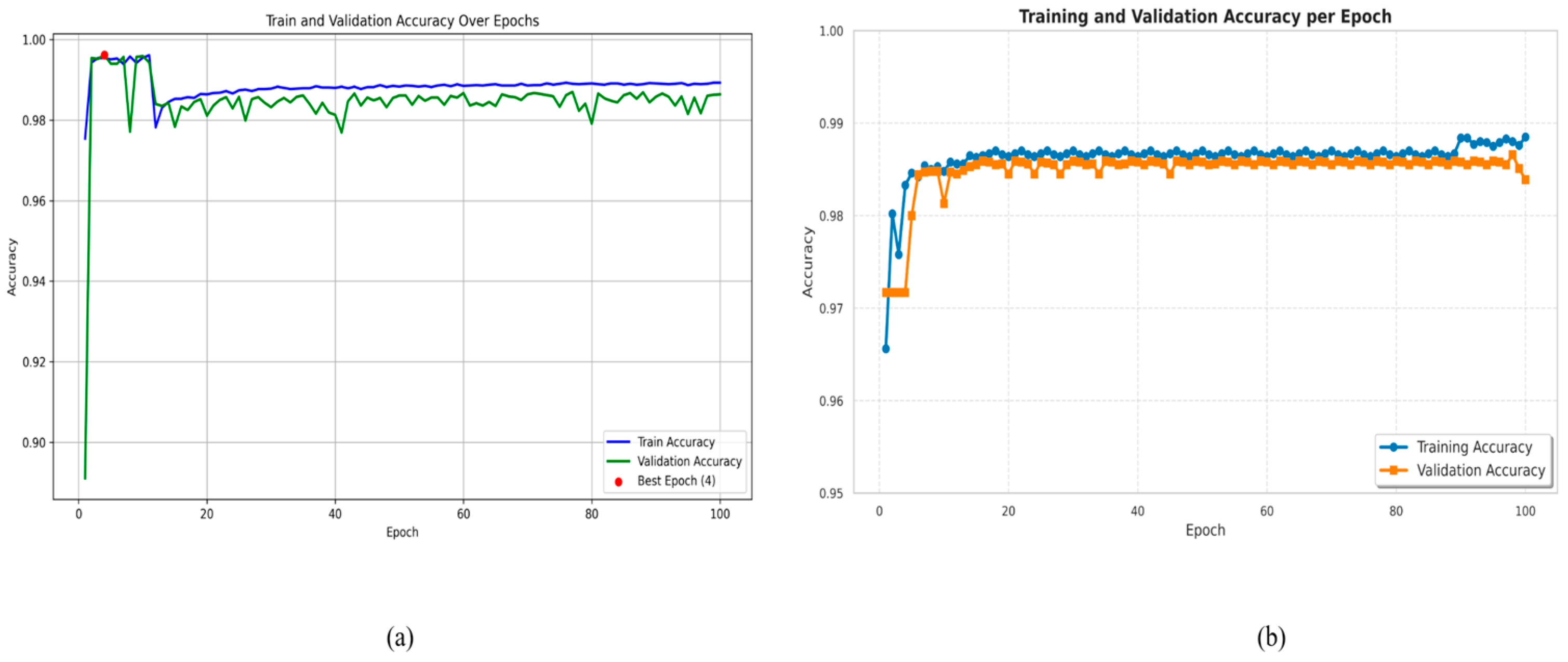
4.1. Implementation
4.2. Ablation Study
4.3. Statistical Significance Analysis
4.4. Comparison
4.5. Analysis Using Different Optimizers
4.6. Cross-Dataset Generalization Study
4.7. Explainable AI Analysis
5. Discussions
6. Limitation
7. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Almotairi, S.; Kareem, G.; Aouf, M.; Almutairi, B.; Salem, M.A.-M. Liver tumor segmentation in CT scans using modified SegNet. Sensors 2020, 20, 1516. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Xie, Y.; Zhang, P.; Chen, H.; Xia, Y.; Shen, C. Light-Weight Hybrid Convolutional Network for Liver Tumor Segmentation. IJCAI 2019, 19, 4271–4277. [Google Scholar] [CrossRef]
- Jiang, H.; Yin, Y.; Zhang, J.; Deng, W.; Li, C. Deep learning for liver cancer histopathology image analysis: A comprehensive survey. Eng. Appl. Artif. Intell. 2024, 133, 108436. [Google Scholar] [CrossRef]
- Velichko, Y.S.; Gennaro, N.; Karri, M.; Antalek, M.; Bagci, U. A comprehensive review of deep learning approaches for magnetic resonance imaging liver tumor analysis. Adv. Clin. Radiol. 2023, 5, 1–15. [Google Scholar] [CrossRef]
- Nanda, N.; Kakkar, P.; Nagpal, S. Computer-Aided Segmentation of Liver Lesions in CT Scans Using Cascaded Convolutional Neural Networks and Genetically Optimised Classifier. Arab. J. Sci. Eng. 2019, 44, 4049–4062. [Google Scholar] [CrossRef]
- Esteva, A.; Kuprel, B.; Novoa, R.A.; Ko, J.; Swetter, S.M.; Blau, H.M.; Thrun, S. Dermatologist-level classification of skin cancer with deep neural networks. Nature 2017, 542, 115–118. [Google Scholar] [CrossRef] [PubMed]
- Hu, C.; Xia, T.; Cui, Y.; Zou, Q.; Wang, Y.; Xiao, W.; Ju, S.; Li, X. Trustworthy multi-phase liver tumor segmentation via evidence-based uncertainty. Eng. Appl. Artif. Intell. 2024, 133, 108289. [Google Scholar] [CrossRef]
- Fang, L.; Jin, Y.; Huang, L.; Guo, S.; Zhao, G.; Chen, X. Iterative fusion convolutional neural networks for classification of optical coherence tomography images. J. Vis. Commun. Image Represent. 2019, 59, 327–333. [Google Scholar] [CrossRef]
- Saadh, M.J.; Bishoyi, A.K.; Ballal, S.; Singh, A.; Devi, A.; Sharma, G.C.; Valiev, A.; Naidu, K.S.; Bhakuni, P.N.; Sead, F.F. Ferroptosis and non-coding RNAs in breast cancer: Insights into CAF and TAM interactions. Discov. Oncol. 2025, 16, 1658. [Google Scholar] [CrossRef]
- Chelloug, S.A.; Mahel, A.S.B.; Alnashwan, R.; Rafiq, A.; Muthanna, M.S.A.; Aziz, A. Enhanced breast cancer diagnosis using modified InceptionNet-V3: A deep learning approach for ultrasound image classification. Front. Physiol. 2025, 16, 1558001. [Google Scholar] [CrossRef] [PubMed]
- Shen, W.; Zhou, M.; Yang, F.; Yu, D.; Dong, D.; Yang, C.; Zang, Y.; Tian, J. Multi-crop convolutional neural networks for lung nodule malignancy suspiciousness classification. Pattern Recognit. 2017, 61, 663–673. [Google Scholar] [CrossRef]
- Ghoniem, R.M.; Shaalan, K. FCSR—Fuzzy Continuous Speech Recognition Approach for Identifying Laryngeal Pathologies Using New Weighted Spectrum Features. In Advances in Intelligent Systems and Computing, Proceedings of the International Conference on Advanced Intelligent Systems and Informatics 2017, Cairo, Egypt, 9–11 September 2017; Hassanien, A.E., Shaalan, K., Gaber, T., Tolba, M.F., Eds.; Springer International Publishing: Cham, Switzerland, 2018; Volume 639, pp. 384–395. [Google Scholar] [CrossRef]
- Marini, F.; Walczak, B. Particle swarm optimization (PSO). A tutorial. Chemom. Intell. Lab. Syst. 2015, 149, 153–165. [Google Scholar] [CrossRef]
- Mirjalili, S.; Lewis, A. The whale optimization algorithm. Adv. Eng. Softw. 2016, 95, 51–67. [Google Scholar] [CrossRef]
- Bansal, J.C.; Gopal, A.; Nagar, A.K. Stability analysis of artificial bee colony optimization algorithm. Swarm Evol. Comput. 2018, 41, 9–19. [Google Scholar] [CrossRef]
- Maione, G.; Punzi, A.; Li, K. A comparative study on differential evolution with other heuristic methods for continuous optimization. In Proceedings of the 21st Mediterranean Conference on Control and Automation, Platanias, Greece, 25–28 June 2013; IEEE: New York, NY, USA, 2013; pp. 1356–1361. Available online: https://ieeexplore.ieee.org/abstract/document/6608896/ (accessed on 16 June 2025).
- Geem, Z.W.; Kim, J.H.; Loganathan, G.V. A new heuristic optimization algorithm: Harmony search. Simulation 2001, 76, 60–68. [Google Scholar] [CrossRef]
- Rashedi, E.; Nezamabadi-Pour, H.; Saryazdi, S. GSA: A gravitational search algorithm. Inf. Sci. 2009, 179, 2232–2248. [Google Scholar] [CrossRef]
- Emary, E.; Zawbaa, H.M.; Grosan, C.; Hassenian, A.E. Feature Subset Selection Approach by Gray-Wolf Optimization. In Afro-European Conference for Industrial Advancement, Advances in Intelligent Systems and Computing; Abraham, A., Krömer, P., Snasel, V., Eds.; Springer International Publishing: Cham, Switzerland, 2015; Volume 334, pp. 1–13. [Google Scholar] [CrossRef]
- Mostafa, A.; Houssein, E.H.; Houseni, M.; Hassanien, A.E.; Hefny, H. Evaluating Swarm Optimization Algorithms for Segmentation of Liver Images. In Advances in Soft Computing and Machine Learning in Image Processing, Studies in Computational Intelligence; Hassanien, A.E., Oliva, D.A., Eds.; Springer International Publishing: Cham, Switzerland, 2018; Volume 730, pp. 41–62. [Google Scholar] [CrossRef]
- Decerle, J.; Grunder, O.; El Hassani, A.H.; Barakat, O. A hybrid memetic-ant colony optimization algorithm for the home health care problem with time window, synchronization and working time balancing. Swarm Evol. Comput. 2019, 46, 171–183. [Google Scholar] [CrossRef]
- Garro, B.A.; Rodríguez, K.; Vázquez, R.A. Classification of DNA microarrays using artificial neural networks and ABC algorithm. Appl. Soft Comput. 2016, 38, 548–560. [Google Scholar] [CrossRef]
- Qi, A.-L.; Zhang, G.-M.; Dong, M.; Ma, H.-W.; Harvey, D.M. An artificial bee colony optimization based matching pursuit approach for ultrasonic echo estimation. Ultrasonics 2018, 88, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Thiagarajan, B.; Bremananth, R. Brain image segmentation using conditional random field based on modified artificial bee colony optimization algorithm. Recent Adv. Comput. Sci. 2014, 344, 93–106. [Google Scholar]
- Mostafa, A.; Fouad, A.; Elfattah, M.A.; Hassanien, A.E.; Hefny, H.; Zhu, S.Y.; Schaefer, G. CT liver segmentation using artificial bee colony optimisation. Procedia Comput. Sci. 2015, 60, 1622–1630. [Google Scholar] [CrossRef]
- Liu, X.; Wang, X.; Shi, N.; Li, C. Image segmentation algorithm based on improved ant colony algorithm. Int. J. Signal Process. Image Process. Pattern Recognit. 2014, 7, 433–442. [Google Scholar] [CrossRef]
- Emon, M.H.; Mondal, P.K.; Mozumder, M.A.; Kim, H.C.; Lapina, M.; Babenko, M.; Muthanna, M.S. An Integrated Architecture for Colorectal Polyp Segmentation: The µ-Net Framework with Explainable AI. Diagnostics 2025, 15, 2890. [Google Scholar] [CrossRef]
- Horng, M.-H. Multilevel thresholding selection based on the artificial bee colony algorithm for image segmentation. Expert Syst. Appl. 2011, 38, 13785–13791. [Google Scholar] [CrossRef]
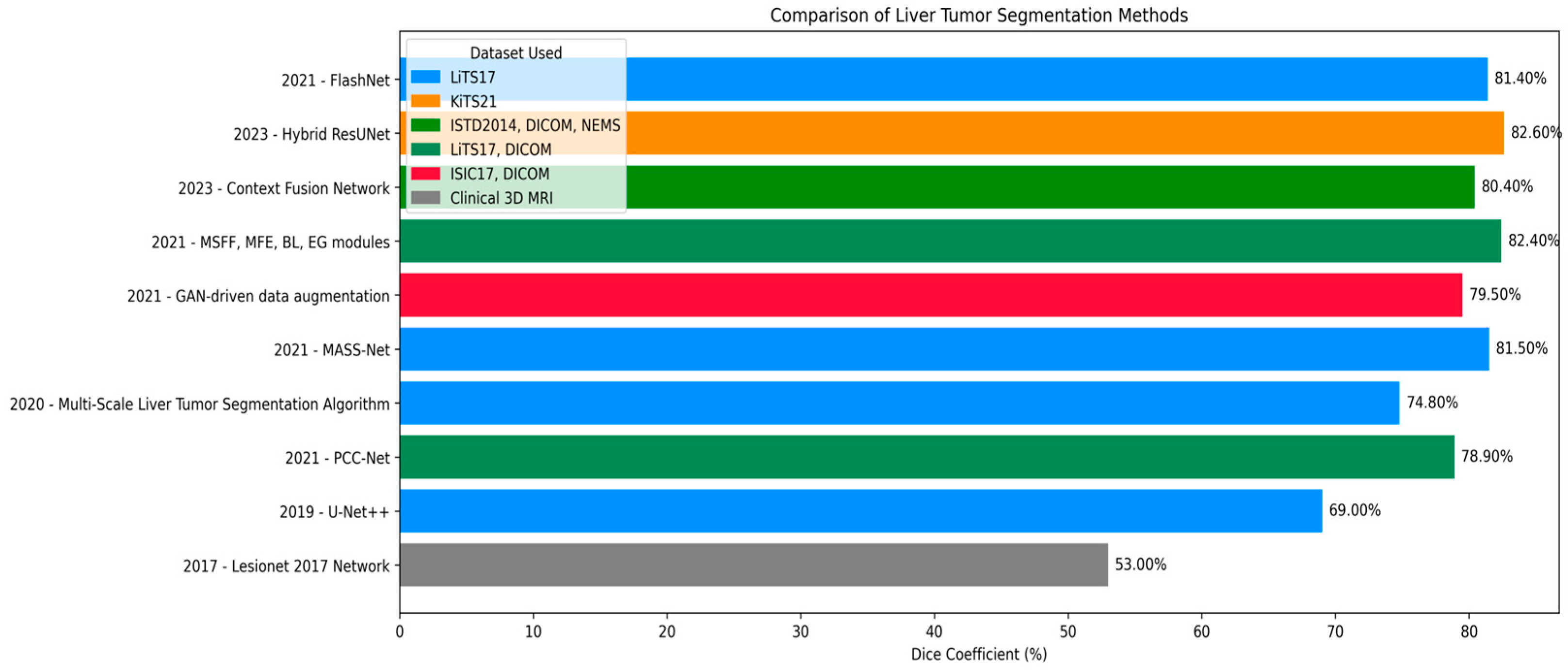
- Chen, L.; Luo, S. Multi-Scale Liver Tumor Segmentation Algorithm by Fusing Convolution and Transformer. J. Comput. Eng. Appl. 2024, 60, 4. Available online: https://search.ebscohost.com/login.aspx?direct=true&profile=ehost&scope=site&authtype=crawler&jrnl=10028331&AN=175598615&h=HRR4o5hklnz6GycZGS81hvavLVl1CdqCk8KUsgjapy9BP4k7ihKB0qeY7qHwjlSSYKtl58xhRF27xcR9%2BG5iWA%3D%3D&crl=c (accessed on 23 June 2025).
- You, Y.; Bai, Z.; Zhang, Y.; Li, Z. Contour-induced parallel graph reasoning for liver tumor segmentation. Biomed. Signal Process. Control. 2024, 92, 106111. [Google Scholar] [CrossRef]
- He, J.; Luo, Z.; Lian, S.; Su, S.; Li, S. Towards accurate abdominal tumor segmentation: A 2D model with Position-Aware and Key Slice Feature Sharing. Comput. Biol. Med. 2024, 179, 108743. [Google Scholar] [CrossRef]
- Mondal, P.K.; Mozumder, M.A.; Kim, H.C. Deep Learning Framework for Automated Detection of Diabetic Retinopathy in Retinal Fundus Images. Korea Inst. Inf. Commun. Eng. 2025, 29, 511–514. [Google Scholar]
- Shui, Y.; Wang, Z.; Liu, B.; Wang, W.; Fu, S.; Li, Y. A three-path network with multi-scale selective feature fusion, edge-inspiring and edge-guiding for liver tumor segmentation. Comput. Biol. Med. 2024, 168, 107841. [Google Scholar] [CrossRef]
- Wang, Z.; Zhu, J.; Fu, S.; Ye, Y. Context fusion network with multi-scale-aware skip connection and twin-split attention for liver tumor segmentation. Med. Biol. Eng. Comput. 2023, 61, 3167–3180. [Google Scholar] [CrossRef] [PubMed]
- Muhammad, S.; Zhang, J. Segmentation of Liver Tumors by Monai and PyTorch in CT Images with Deep Learning Techniques. Appl. Sci. 2024, 14, 5144. [Google Scholar] [CrossRef]
- Fallahpoor, M.; Nguyen, D.; Montahaei, E.; Hosseini, A.; Nikbakhtian, S.; Naseri, M.; Salahshour, F.; Farzanefar, S.; Abbasi, M. Segmentation of liver and liver lesions using deep learning. Phys. Eng. Sci. Med. 2024, 47, 611–619. [Google Scholar] [CrossRef]
- Wang, J.; Peng, Y.; Jing, S.; Han, L.; Li, T.; Luo, J. A deep-learning approach for segmentation of liver tumors in magnetic resonance imaging using UNet++. BMC Cancer 2023, 23, 1060. [Google Scholar] [CrossRef]
- Zhen, S.-H.; Cheng, M.; Tao, Y.-B.; Wang, Y.-F.; Juengpanich, S.; Jiang, Z.-Y.; Jiang, Y.-K.; Yan, Y.-Y.; Lu, W.; Lue, J.-M.; et al. Deep learning for accurate diagnosis of liver tumor based on magnetic resonance imaging and clinical data. Front. Oncol. 2020, 10, 680. [Google Scholar] [CrossRef]
- Zahra, S.B.; Athar, A.; Khan, M.A.; Abbas, S.; Ahmad, G. Automated diagnosis of liver disorder using multilayer neu-ro-fuzzy. Int. J. Adv. Appl. Sci. 2019, 6, 23–32. [Google Scholar]
- McGrath, H.; Li, P.; Dorent, R.; Bradford, R.; Saeed, S.; Bisdas, S.; Ourselin, S.; Shapey, J.; Vercauteren, T. Manual segmentation versus semi-automated segmentation for quantifying vestibular schwannoma volume on MRI. Int. J. Comput. Assist. Radiol. Surg. 2020, 15, 1445–1455. [Google Scholar] [CrossRef] [PubMed]
- Mojtahed, A.; Núñez, L.; Connell, J.; Fichera, A.; Nicholls, R.; Barone, A.; Marieiro, M.; Puddu, A.; Arya, Z.; Ferreira, C.; et al. Repeatability and reproducibility of deep-learning-based liver volume and Couinaud segment volume measurement tool. Abdom. Imaging 2021, 47, 143–151. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Xu, Y.; Song, J.; Liu, X.; Liu, S.; Yang, N.; Wang, L.; Liu, Y.; Zhao, Y.; Zhou, W.; et al. Tumor Cell-Targeting and Tumor Microenvironment–Responsive Nanoplatforms for the Multimodal Imaging-Guided Photodynamic/Photothermal/Chemodynamic Treatment of Cervical Cancer. Int. J. Nanomed. 2024, ume 19, 5837–5858. [Google Scholar] [CrossRef]
- Venkadesh, K.V.; Aleef, T.A.; Scholten, E.T.; Saghir, Z.; Silva, M.; Sverzellati, N.; Pastorino, U.; van Ginneken, B.; Prokop, M.; Jacobs, C. Prior CT Improves Deep Learning for Malignancy Risk Estimation of Screening-detected Pulmonary Nodules. Radiology 2023, 308, e223308. [Google Scholar] [CrossRef]
- Alzahrani, A.A.; Ahmed, A.; Raza, A. Content-Based Medical Image Retrieval Method Using Multiple Pre-Trained Convolutional Neural Networks Feature Extraction Models. 2024. Available online: https://www.researchgate.net/profile/Ali-Ahmed-46/publication/382005755_Content-based_medical_image_retrieval_method_using_multiple_pre-trained_convolutional_neural_networks_feature_extraction_models/links/668c4975b15ba55907497936/Content-based-medical-image-retrieval-method-using-multiple-pre-trained-convolutional-neural-networks-feature-extraction-models.pdf (accessed on 16 June 2025).
- Singh, R.; Gupta, S.; Almogren, A.; Rehman, A.U.; Bharany, S.; Altameem, A.; Choi, J. FasNet: A hybrid deep learning model with attention mechanisms and uncertainty estimation for liver tumor segmentation on LiTS17. Sci. Rep. 2025, 15, 1–19. [Google Scholar] [CrossRef]
- Biswas, A.; Maity, S.P.; Banik, R.; Bhattacharya, P.; Debbarma, J. GAN-Driven liver tumor segmentation: Enhancing accuracy in biomedical imaging. SN Comput. Sci. 2024, 5, 652. [Google Scholar] [CrossRef]
- Liver Tumor Segmentation—Part 2. Available online: https://www.kaggle.com/datasets/andrewmvd/liver-tumor-segmentation-part-2 (accessed on 20 October 2025).
- Medicine. Available online: https://journals.lww.com/md-journal/fulltext/2025/05300/mammogram_mastery__breast_cancer_image.30.aspx?context=latestarticles (accessed on 16 June 2025).
- Deep Learning Based Liver Cancer Detection Using Watershed Transform and Gaussian Mixture Model Techniques—Sci-enceDirect. Available online: https://www.sciencedirect.com/science/article/pii/S1389041718310143 (accessed on 16 June 2025).
- Raza, M.; Farwa, U.E.; Mozumder, M.A.; Mon-il, J.; Kim, H.C. ETRC-net: Efficient transformer for grading renal cell carcinoma in histopathological images. Comput. Electr. Eng. 2025, 128, 110747. [Google Scholar] [CrossRef]
- Ghoniem, R.M.; Algarni, A.D.; Shaalan, K. Multi-Modal Emotion Aware System Based on Fusion of Speech and Brain Information. Information 2019, 10, 239. [Google Scholar] [CrossRef]
- Sumon, R.I.; Mozumdar, M.A.; Akter, S.; Uddin, S.M.; Al-Onaizan, M.H.; Alkanhel, R.I.; Muthanna, M.S. Comparative Study of Cell Nuclei Segmentation Based on Computational and Handcrafted Features Using Machine Learning Algorithms. Diagnostics 2025, 15, 1271. [Google Scholar] [CrossRef] [PubMed]
- Soler, L.; Hostettler, A.; Agnus, V.; Charnoz, A.; Fasquel, J.; Moreau, J.; Osswald, A.; Bouhadjar, M.; Marescaux, J. 3D Image Reconstruction for Comparison of Algorithm Database: A Patient-Specific Anatomical and Medical Image Database. IRCAD, Strasbourg, France, Tech. Rep. 2010. Available online: https://www.ircad.fr/research/data-sets/liver-segmentation-3d-ircadb-01 (accessed on 16 June 2025).
- Kavur, A.E.; Gezer, N.S.; Barış, M.; Aslan, S.; Conze, P.H.; Groza, V.; Selver, M.A. CHAOS challenge–combined (CT-MR) healthy abdominal organ segmentation. Med. Image Anal. 2021, 69, 101950. [Google Scholar] [CrossRef] [PubMed]
| Category | Parameter | Search Range |
|---|---|---|
| Model | Encoder depth | 5 levels |
| Initial filters | 64 | |
| Kernel size | 3 × 3 | |
| WOA Hyperparameters | Population size | 20 whales |
| Max iterations | 100 | |
| Spiral constant (b) | 1.0 | |
| Control parameter (a) | Linear decrease [2 → 0] | |
| Trainable Hyperparameters | Learning rate (η) | [10−4, 10−2] |
| Dropout rate (ρ) | [0.0, 0.5] | |
| Batch size (β) | [4, 32] |
| Model Variant | Dice Coefficient (%) | Jaccard Index (%) | Total Params (M) | Trainable Params (M) | Pixel Accuracy (%) | Training Time (h) |
|---|---|---|---|---|---|---|
| Baseline U-Net | 78.45 ± 1.23 | 64.67 ± 1.45 | 7.8 | 7.8 | 97.23 ± 0.34 | 8.2 ± 0.4 |
| U-Net + WOA | 82.13 ± 0.98 | 69.85 ± 1.12 | 7.8 | 7.8 | 98.11 ± 0.28 | 7.1 ± 0.3 |
| ResNet-50 U-Net | 87.66 ± 1.05 | 78.24 ± 1.34 | 31.3 | 29.1 | 98.87 ± 0.31 | 12.5 ± 0.6 |
| LiTs-Res-UNet + WOA | 92.38 ± 0.76 | 86.73 ± 0.89 | 33.8 | 31.2 | 99.54 ± 0.18 | 10.8 ± 0.5 |
| Comparison | Dice Δ | p-Value | 95% CI | Effect Size (Cohen’s d) |
|---|---|---|---|---|
| U-Net + WOA vs. Baseline | +3.68% | 0.003 | [1.82, 5.54] | 2.14 (large) |
| ResNet-50 U-Net vs. Baseline | +9.21% | <0.001 | [7.45, 10.97] | 4.87 (very large) |
| Proposed vs. Baseline | +13.93% | <0.001 | [12.34, 15.52] | 7.23 (very large) |
| Proposed vs. ResNet-50 U-Net | +4.72% | 0.001 | [2.98, 6.46] | 3.45 (very large) |
| Ref | Technique | Dice Coefficient (%) | Jaccard Index (IoU) (%) |
|---|---|---|---|
| [30] | Multi-Scale Liver Tumor Segmentation Algorithm | 74.3 | – |
| [31] | PGC-Net | 73.63 | – |
| [34] | PAKS-Net | 76.9 | – |
| [33] | MSFF, MFF, EI, EG modules | 85.55 | 81.11 |
| [46] | GAN-driven data augmentation strategy | 60.5 | – |
| [35] | Context Fusion Network with TSA & MSA skip connections | 85.97 | 81.56 |
| [45] | Hybrid “FasNet” Model with Attention and Monte Carlo Dropout | 87.66 | 84.87 |
| Our proposed | LiTs-Res-Unet + WOA | 92.38 | 86.73 |
| Optimizer | Loss | Accuracy | Dice Coefficient | Jaccard Index |
|---|---|---|---|---|
| AdaGrad | 0.0435 | 0.9531 | 0.8066 | 0.7787 |
| SGD | 0.0299 | 0.9344 | 0.7913 | 0.7743 |
| Adam | 0.0251 | 0.9954 | 0.8766 | 0.8487 |
| RMSProp | 0.0435 | 0.9231 | 0.7766 | 0.7587 |
| AdaDelta | 0.3119 | 0.9540 | 0.8043 | 0.7772 |
| Metaheuristic Optimizer (WOA) | 0.0121 | 0.9954 | 0.9238 | 0.8673 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mondol, P.K.; Islam Mozumder, M.A.; Cheol Kim, H.; Hassan Ali Al-Onaizan, M.; Hassan, D.S.M.; Al-Bahri, M.; Muthanna, M.S.A. A ResNet-50–UNet Hybrid with Whale Optimization Algorithm for Accurate Liver Tumor Segmentation. Diagnostics 2025, 15, 2975. https://doi.org/10.3390/diagnostics15232975
Mondol PK, Islam Mozumder MA, Cheol Kim H, Hassan Ali Al-Onaizan M, Hassan DSM, Al-Bahri M, Muthanna MSA. A ResNet-50–UNet Hybrid with Whale Optimization Algorithm for Accurate Liver Tumor Segmentation. Diagnostics. 2025; 15(23):2975. https://doi.org/10.3390/diagnostics15232975
Chicago/Turabian StyleMondol, Proloy Kumar, Md Ariful Islam Mozumder, Hee Cheol Kim, Mohammad Hassan Ali Al-Onaizan, Dina S. M. Hassan, Mahmood Al-Bahri, and Mohammed Saleh Ali Muthanna. 2025. "A ResNet-50–UNet Hybrid with Whale Optimization Algorithm for Accurate Liver Tumor Segmentation" Diagnostics 15, no. 23: 2975. https://doi.org/10.3390/diagnostics15232975
APA StyleMondol, P. K., Islam Mozumder, M. A., Cheol Kim, H., Hassan Ali Al-Onaizan, M., Hassan, D. S. M., Al-Bahri, M., & Muthanna, M. S. A. (2025). A ResNet-50–UNet Hybrid with Whale Optimization Algorithm for Accurate Liver Tumor Segmentation. Diagnostics, 15(23), 2975. https://doi.org/10.3390/diagnostics15232975









