Multiplexing Proteomic and Ingenuity Pathway Analysis of Attention/Working Memory in Virally Suppressed Women with HIV: A Feasibility Study
Abstract
1. Introduction
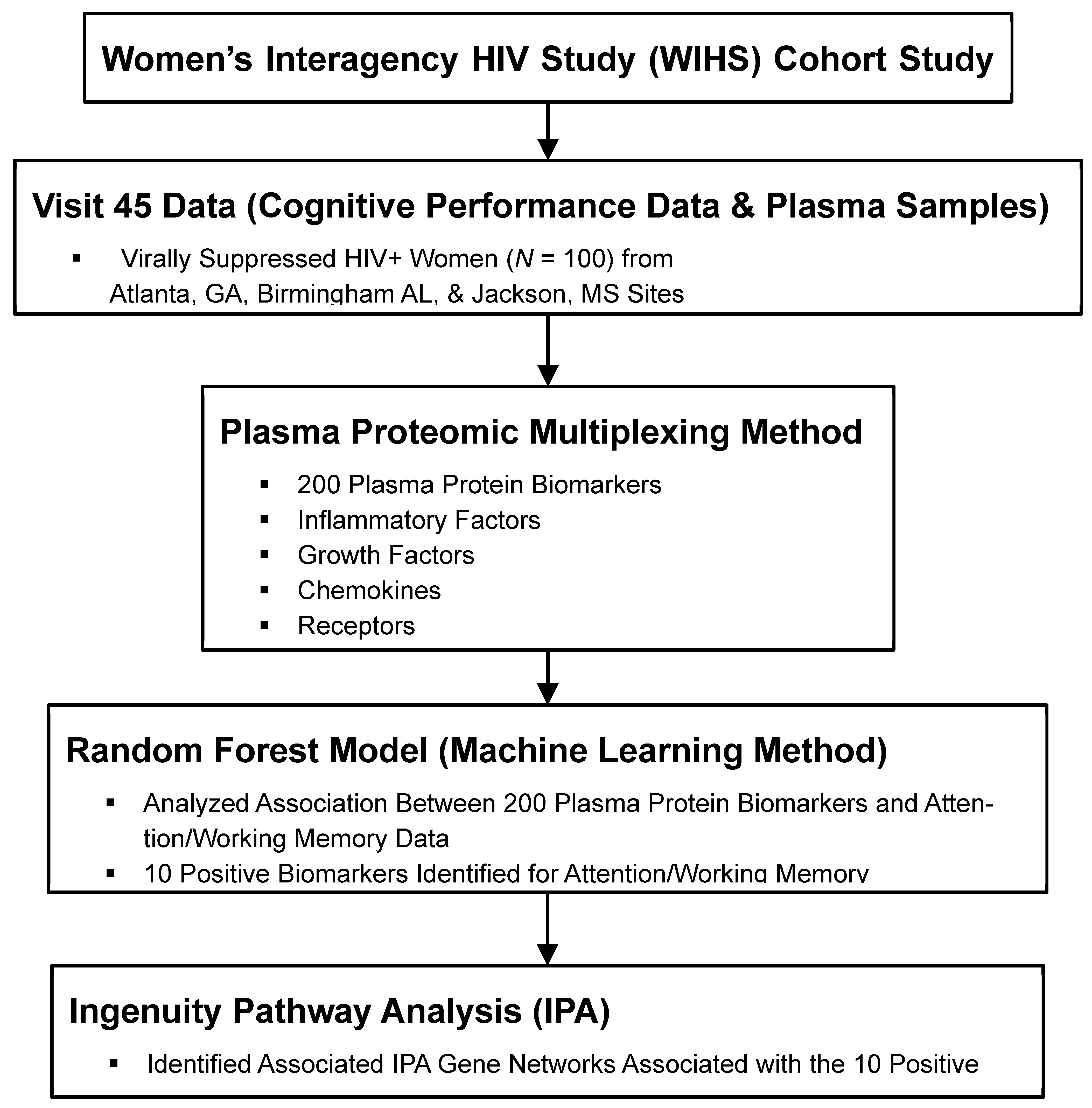
2. Methods
2.1. Participants
2.2. Measurement of Biomarkers
2.3. Attention/Working Memory
2.4. Statistical Analyses

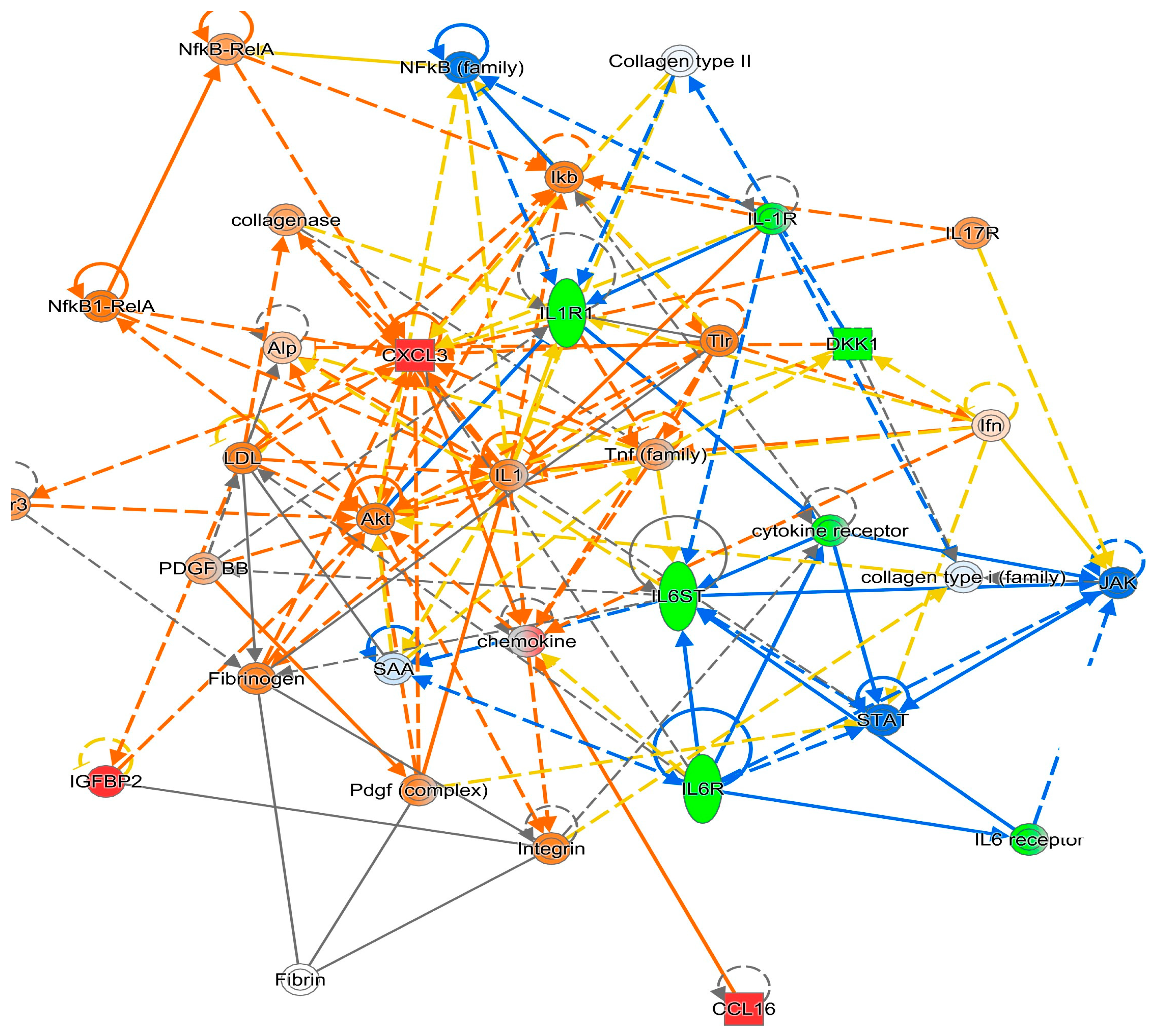
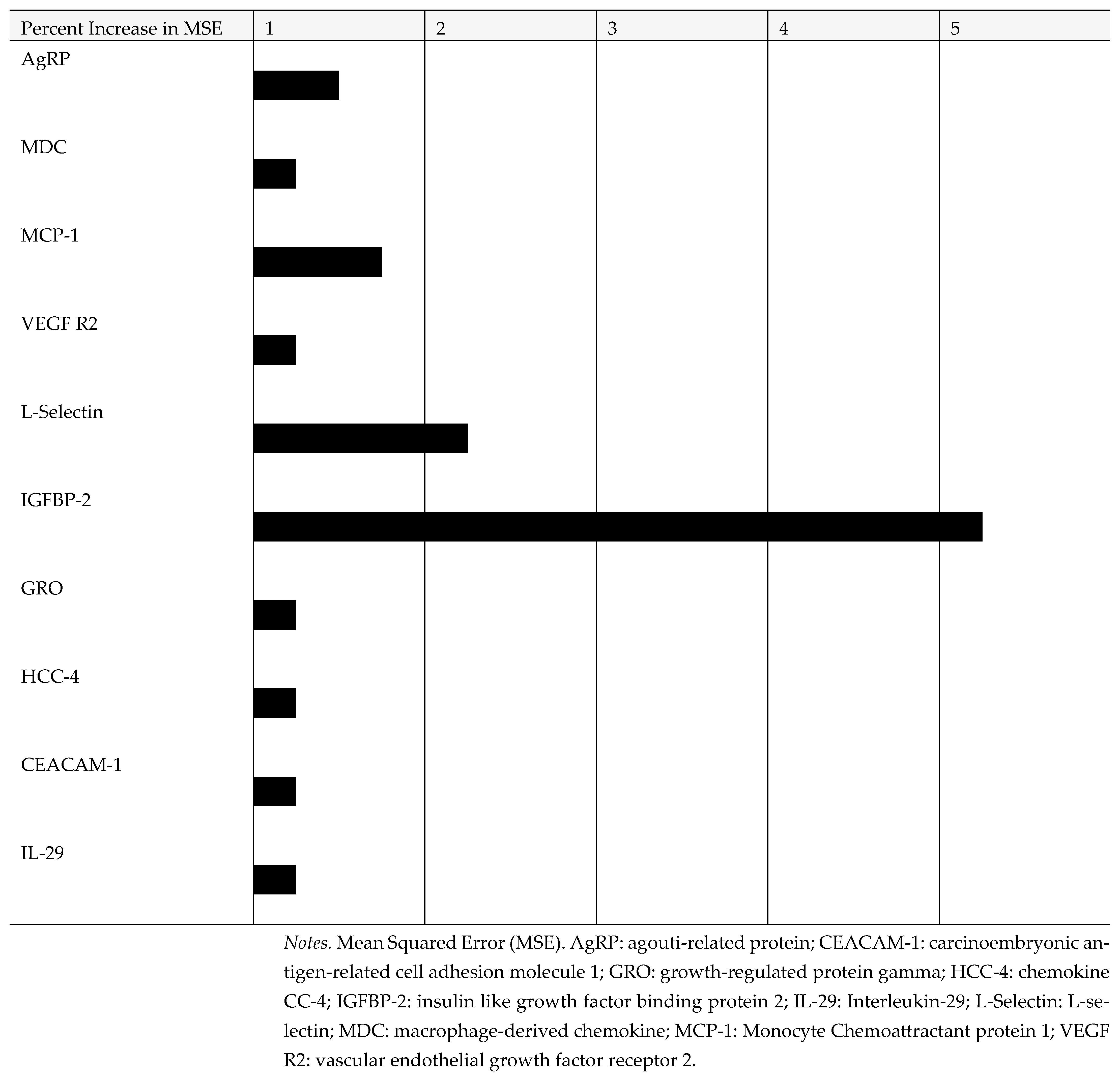
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- High, K.P.; Brennan-Ing, M.; Clifford, D.B.; Cohen, M.H.; Currier, J.; Deeks, S.G.; Deren, S.; Effros, R.B.; Gebo, K.; Goronzy, J.J.; et al. HIV and Aging: State of Knowledge and Areas of Critical Need for Research. A Report to the NIH Office of AIDS Research by the HIV and Aging Working Group. J. Acquir. Immune Defic. Syndr. 2012, 60 (Suppl. 1), S1–S18. [Google Scholar] [CrossRef]
- Veenstra, M.; León-Rivera, R.; Li, M.; Gama, L.; Clements, J.E.; Berman, J.W. Mechanisms of CNS Viral Seeding by HIV+ CD14+ CD16+ Monocytes: Establishment and Reseeding of Viral Reservoirs Contributing to HIV-Associated Neurocognitive Disorders. mBio 2017, 8, e01280-17. [Google Scholar] [CrossRef]
- Heaton, R.K.; Clifford, D.B.; Franklin, D.R., Jr.; Woods, S.P.; Ake, C.; Vaida, F.; Ellis, R.J.; Letendre, S.L.; Marcotte, T.D.; Atkinson, J.H.; et al. HIV-associated neurocognitive disorders persist in the era of potent antiretroviral therapy: CHARTER study. Neurology 2010, 75, 2087–2096. [Google Scholar] [CrossRef]
- Wei, J.; Hou, J.; Su, B.; Jiang, T.; Guo, C.; Wang, W.; Zhang, Y.; Chang, B.; Wu, H.; Zhang, T. The Prevalence of Frascati-Criteria-Based HIV-Associated Neurocognitive Disorder (HAND) in HIV-Infected Adults: A Systematic Review and Meta-Analysis. Front. Neurol. 2020, 11, 581346. [Google Scholar] [CrossRef]
- Joseph, S.B.; Gianella, S.; Burdo, T.H.; Cinque, P.; Gisslen, M.; Letendre, S.; Nath, A.; Morgello, S.; Ndhlovu, L.C.; Spudich, S. Biotypes of Central Nervous System Complications in People with Human Immunodeficiency Virus: Virology, Immunology, and Neuropathology. J. Infect. Dis. 2023, 227 (Suppl. 1), S3–S15. [Google Scholar] [CrossRef] [PubMed]
- Gabuzda, D.; Jamieson, B.D.; Collman, R.G.; Lederman, M.M.; Burdo, T.H.; Deeks, S.G.; Dittmer, D.P.; Fox, H.S.; Funderburg, N.T.; Pahwa, S.G. Pathogenesis of Aging and Age-related Comorbidities in People with HIV: Highlights from the HIV ACTION Workshop. Pathog. Immun. 2020, 5, 143–174. [Google Scholar] [CrossRef] [PubMed]
- Lee, W.J.; Liao, Y.C.; Wang, Y.F.; Lin, I.F.; Wang, S.J.; Fuh, J.L. Plasma MCP-1 and Cognitive Decline in Patients with Alzheimer’s Disease and Mild Cognitive Impairment: A Two-year Follow-up Study. Sci. Rep. 2018, 8, 1280. [Google Scholar] [CrossRef] [PubMed]
- Sanchez-Sanchez, J.L.; Giudici, K.V.; Guyonnet, S.; Delrieu, J.; Li, Y.; Bateman, R.J.; Parini, A.; Vellas, B.; de Souto Barreto, P.; MAPT/DSA Group. Plasma MCP-1 and changes on cognitive function in community-dwelling older adults. Alzheimers Res. Ther. 2022, 14, 5. [Google Scholar] [CrossRef]
- Galimberti, D.; Fenoglio, C.; Lovati, C.; Venturelli, E.; Guidi, I.; Corrà, B.; Scalabrini, D.; Clerici, F.; Mariani, C.; Bresolin, N.; et al. Serum MCP-1 levels are increased in mild cognitive impairment and mild Alzheimer’s disease. Neurobiol. Aging. 2006, 27, 1763–1768. [Google Scholar] [CrossRef]
- Jumare, J.; Akolo, C.; Ndembi, N.; Bwala, S.; Alabi, P.; Okwuasaba, K.; Adebiyi, R.; Umlauf, A.; Cherner, M.; Abimiku, A.; et al. Elevated Plasma Levels of sCD14 and MCP-1 Are Associated with HIV Associated Neurocognitive Disorders Among Antiretroviral-Naive Individuals in Nigeria. J. Acquir. Immune Defic. Syndr. 2020, 84, 196–202. [Google Scholar] [CrossRef]
- Kamat, A.; Lyons, J.L.; Misra, V.; Uno, H.; Morgello, S.; Singer, E.J.; Gabuzda, D. Monocyte activation markers in cerebrospinal fluid associated with impaired neurocognitive testing in advanced HIV infection. J. Acquir. Immune Defic. Syndr. 2012, 60, 234–243. [Google Scholar] [CrossRef]
- Anderson, A.M.; Jang, J.H.; Easley, K.A.; Fuchs, D.; Gisslen, M.; Zetterberg, H.; Blennow, K.; Ellis, R.J.; Franklin, D.; Heaton, R.K.; et al. Cognitive and Neuronal Link with Inflammation: A Longitudinal Study in People with and Without HIV Infection. J. Acquir. Immune Defic. Syndr. 2020, 85, 617–625. [Google Scholar] [CrossRef] [PubMed]
- Burdo, T.H.; Weiffenbach, A.; Woods, S.P.; Letendre, S.; Ellis, R.J.; Williams, K.C. Elevated sCD163 in plasma but not cerebrospinal fluid is a marker of neurocognitive impairment in HIV infection. AIDS 2013, 27, 1387–1395. [Google Scholar] [CrossRef] [PubMed]
- Muñoz-Nevárez, L.A.; Imp, B.M.; Eller, M.A.; Kiweewa, F.; Maswai, J.; Polyak, C.; Olwenyi, O.A.; Allen, I.E.; Rono, E.; Milanini, B.; et al. Monocyte activation, HIV, and cognitive performance in East Africa. J. Neurovirol. 2020, 26, 52–59. [Google Scholar] [CrossRef]
- Imp, B.M.; Rubin, L.H.; Tien, P.C.; Plankey, M.W.; Golub, E.T.; French, A.L.; Valcour, V.G. Monocyte Activation Is Associated with Worse Cognitive Performance in HIV-Infected Women with Virologic Suppression. J. Infect. Dis. 2017, 215, 114–121. [Google Scholar] [CrossRef] [PubMed]
- Rubin, L.H.; Neigh, G.N.; Sundermann, E.E.; Xu, Y.; Scully, E.P.; Maki, P.M. Sex differences in neurocognitive function in adults with HIV: Patterns, predictors, and mechanisms. Curr. Psychiatry Rep. 2019, 21, 94. [Google Scholar] [CrossRef]
- Farinpour, R.; Martin, E.M.; Seidenberg, M.; Pitrak, D.L.; Pursell, K.J.; Mullane, K.M.; Novak, R.M.; Harrow, M. Verbal working memory in HIV-seropositive drug users. J. Int. Neuropsychol. Soc. 2000, 6, 548–555. [Google Scholar] [CrossRef]
- Martin, E.M.; Sullivan, T.S.; Reed, R.A.; Fletcher, T.A.; Pitrak, D.L.; Weddington, W.; Harrow, M. Auditory working memory in HIV-1 infection. J. Int. Neuropsychol. Soc. 2001, 7, 20–26. [Google Scholar] [CrossRef]
- Kanmogne, G.D.; Fonsah, J.Y.; Umlauf, A.; Moul, J.; Doh, R.; Kengne, A.M.; Tang, B.; Tagny, C.T.; Nchindap, E.; Kenmogne, L.; et al. Attention/Working Memory, Learning and Memory in Adult Cameroonians: Normative Data, Effects of HIV Infection and Viral Genotype. J. Int. Neuropsychol. Soc. 2020, 26, 607–623. [Google Scholar] [CrossRef]
- Nockher, W.A.; Bergmann, L.; Scherberich, J.E. Increased soluble CD14 serum levels and altered CD14 expression of peripheral blood monocytes in HIV-infected patients. Clin. Exp. Immunol. 1994, 98, 369–374. [Google Scholar] [CrossRef]
- Rubin, L.H.; Xu, Y.; Norris, P.J.; Wang, X.; Dastgheyb, R.; Fitzgerald, K.C.; Keating, S.M.; Kaplan, R.C.; Maki, P.M.; Anastos, K.; et al. Early Inflammatory Signatures Predict Subsequent Cognition in Long-Term Virally Suppressed Women with HIV. Front. Integr. Neurosci. 2020, 14, 20. [Google Scholar] [CrossRef]
- Aparicio, J.M.; Xu, Y.; Li, Y.; Colantuoni, C.; Dastgheyb, R.; Williams, D.W.; Asahchop, E.L.; McMillian, J.M.; Power, C.; Fujiwara, E.; et al. Plasma microRNAs are associated with domain-specific cognitive function in people with HIV. AIDS 2021, 35, 1795–1804. [Google Scholar] [CrossRef]
- Cysique, L.A.; Heaton, R.K.; Kamminga, J.; Lane, T.; Gates, T.M.; Moore, D.M.; Hubner, E.; Carr, A.; Brew, B.J. HIV-associated neurocognitive disorder in Australia: A case of a high-functioning and optimally treated cohort and implications for international neuro HIV research. J. Neurovirol. 2014, 20, 258–268. [Google Scholar] [CrossRef]
- Sacktor, N.; Skolasky, R.L.; Seaberg, E.; Munro, C.; Becker, J.T.; Martin, E.; Ragin, A.; Levine, A.; Miller, E. Prevalence of HIV-associated neurocognitive disorders in the Multicenter AIDS Cohort Study. Neurology 2016, 86, 334–340. [Google Scholar] [CrossRef]
- Heaton, R.K.; Marcotte, T.D.; Mindt, M.R.; Sadek, J.; Moore, D.J.; Bentley, H.; McCutchan, J.A.; Reicks, C.; Grant, I.; HNRC Group. The impact of HIV-associated neuropsychological impairment on everyday functioning. J. Int. Neuropsychol. Soc. 2004, 10, 317–331. [Google Scholar] [CrossRef]
- Maki, P.M.; Rubin, L.H.; Valcour, V.; Martin, E.; Crystal, H.; Young, M.; Weber, K.M.; Manly, J.; Richardson, J.; Alden, C. Cognitive function in women with HIV: Findings from the Women’s Interagency HIV Study. Neurology 2015, 84, 231–240. [Google Scholar] [CrossRef] [PubMed]
- Rubin, L.H.; Sundermann, E.E.; Cook, J.A.; Martin, E.M.; Golub, E.T.; Weber, K.M.; Cohen, M.H.; Crystal, H.; Cederbaum, J.A.; Anastos, K. Investigation of menopausal stage and symptoms on cognition in human immunodeficiency virus-infected women. Menopause 2014, 21, 997–1006. [Google Scholar] [CrossRef]
- Rubin, L.H.; Pyra, M.; Cook, J.A.; Weber, K.M.; Cohen, M.H.; Martin, E.; Valcour, V.; Milam, J.; Anastos, K.; Young, M.A.; et al. Post-traumatic stress is associated with verbal learning, memory, and psychomotor speed in HIV-infected and HIV-uninfected women. J. Neurovirol. 2016, 22, 159–169. [Google Scholar] [CrossRef] [PubMed]
- Rubin, L.H.; Cook, J.A.; Weber, K.M.; Cohen, M.H.; Martin, E.; Valcour, V.; Milam, J.; Anastos, K.; Young, M.A.; Alden, C. The association of perceived stress and verbal memory is greater in HIV-infected versus HIV-uninfected women. J. Neurovirol. 2015, 21, 422–432. [Google Scholar] [CrossRef]
- Bobińska, K.; Gałecka, E.; Szemraj, J.; Gałecki, P.; Talarowska, M. Is there a link between TNF gene expression and cognitive deficits in depression? Acta Biochim. Pol. 2017, 64, 65–73. [Google Scholar] [CrossRef] [PubMed]
- Buchhave, P.; Zetterberg, H.; Blennow, K.; Minthon, L.; Janciauskiene, S.; Hansson, O. Soluble TNF receptors are associated with Aβ metabolism and conversion to dementia in subjects with mild cognitive impairment. Neurobiol. Aging 2010, 31, 1877–1884. [Google Scholar] [CrossRef] [PubMed]
- Diniz, B.S.; Teixeira, A.L.; Ojopi, E.B.; Talib, L.L.; Mendonça, V.A.; Gattaz, W.F.; Forlenza, O.V. Higher serum sTNFR1 level predicts conversion from mild cognitive impairment to Alzheimer’s disease. J. Alzheimers Dis. 2010, 22, 1305–1311. [Google Scholar] [CrossRef]
- McCoy, M.K.; Tansey, M.G. TNF signaling inhibition in the CNS: Implications for normal brain function and neurodegenerative disease. J. Neuroinflamm. 2008, 5, 45. [Google Scholar] [CrossRef]
- Kinai, E.; Komatsu, K.; Sakamoto, M.; Taniguchi, T.; Nakao, A.; Igari, H.; Takada, K.; Watanabe, A.; Takahashi-Nakazato, A.; Takano, M.; et al. Association of age and time of disease with HIV associated neurocognitive disorders: A Japanese nationwide multicenter study. J. Neurovirol. 2017, 23, 864–874. [Google Scholar] [CrossRef] [PubMed]
- Sullivan, K.J.; Blackshear, C.; Simino, J.; Tin, A.; Walker, K.A.; Sharrett, A.R. Association of Midlife Plasma Amyloid-β Levels with Cognitive Impairment in Late Life: The ARIC Neurocognitive Study. Neurology 2021, 97, e1123-31. [Google Scholar] [CrossRef] [PubMed]

| Sample Characteristics | Mean (SD) or n (%) |
|---|---|
| Age in years (Mean, SD) | 48.0 (8.9) |
| Education in years (Mean, SD) | 12.2 (2.2) |
| Race/ethnicity, n (%) Black, non-Hispanic White, non-Hispanic | 69 (90) 8 (10) |
| Current smoking status, n (%) | 29 (38) |
| Recent heavy alcohol use, n (%) | 9 (12) |
| Recent marijuana use, n (%) | 18 (23) |
| Recent Crack, cocaine, and/or heroin use, n (%) | 3 (4) |
| Nadir CD4 count in WIHS, median (IQR) | 284.5 (253) |
| Current CD4 count, median (IQR) | 743 (333) |
| Undetectable HIV RNA, <20 cp/mL, n (%) | 75 (97) |
| Adherence (≥95%) to cART, n (%) | 72 (94) |
| ART duration in years (Mean, SD) | 7.3 (2.7) |
| Prior AIDS diagnosis, n (%) | 13 (17) |
| Cognition | TNFRSF1A | TNFRSF1B | IL1R1 | IL6R | Network |
|---|---|---|---|---|---|
| Attention Working Memory | x | x | 1 | ||
| x | x | 2 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, W.; Rubin, L.H.; Xu, Y.; Wang, Y.; Dastgheyb, R.; Ptacek, T.; Wang, G.; Kempf, M.-C.; Dionne, J.A.; Konkle-Parker, D.; et al. Multiplexing Proteomic and Ingenuity Pathway Analysis of Attention/Working Memory in Virally Suppressed Women with HIV: A Feasibility Study. Diagnostics 2025, 15, 2649. https://doi.org/10.3390/diagnostics15202649
Li W, Rubin LH, Xu Y, Wang Y, Dastgheyb R, Ptacek T, Wang G, Kempf M-C, Dionne JA, Konkle-Parker D, et al. Multiplexing Proteomic and Ingenuity Pathway Analysis of Attention/Working Memory in Virally Suppressed Women with HIV: A Feasibility Study. Diagnostics. 2025; 15(20):2649. https://doi.org/10.3390/diagnostics15202649
Chicago/Turabian StyleLi, Wei, Leah H. Rubin, Yanxun Xu, Yuezhe Wang, Raha Dastgheyb, Travis Ptacek, Ge Wang, Mirjam-Colette Kempf, Jodie A. Dionne, Deborah Konkle-Parker, and et al. 2025. "Multiplexing Proteomic and Ingenuity Pathway Analysis of Attention/Working Memory in Virally Suppressed Women with HIV: A Feasibility Study" Diagnostics 15, no. 20: 2649. https://doi.org/10.3390/diagnostics15202649
APA StyleLi, W., Rubin, L. H., Xu, Y., Wang, Y., Dastgheyb, R., Ptacek, T., Wang, G., Kempf, M.-C., Dionne, J. A., Konkle-Parker, D., Li, D. Y., Sheth, A., Ofotokun, I., & Vance, D. E. (2025). Multiplexing Proteomic and Ingenuity Pathway Analysis of Attention/Working Memory in Virally Suppressed Women with HIV: A Feasibility Study. Diagnostics, 15(20), 2649. https://doi.org/10.3390/diagnostics15202649






