Emerging Technologies for the Diagnosis of Urinary Tract Infections: Advances in Molecular Detection and Resistance Profiling
Abstract
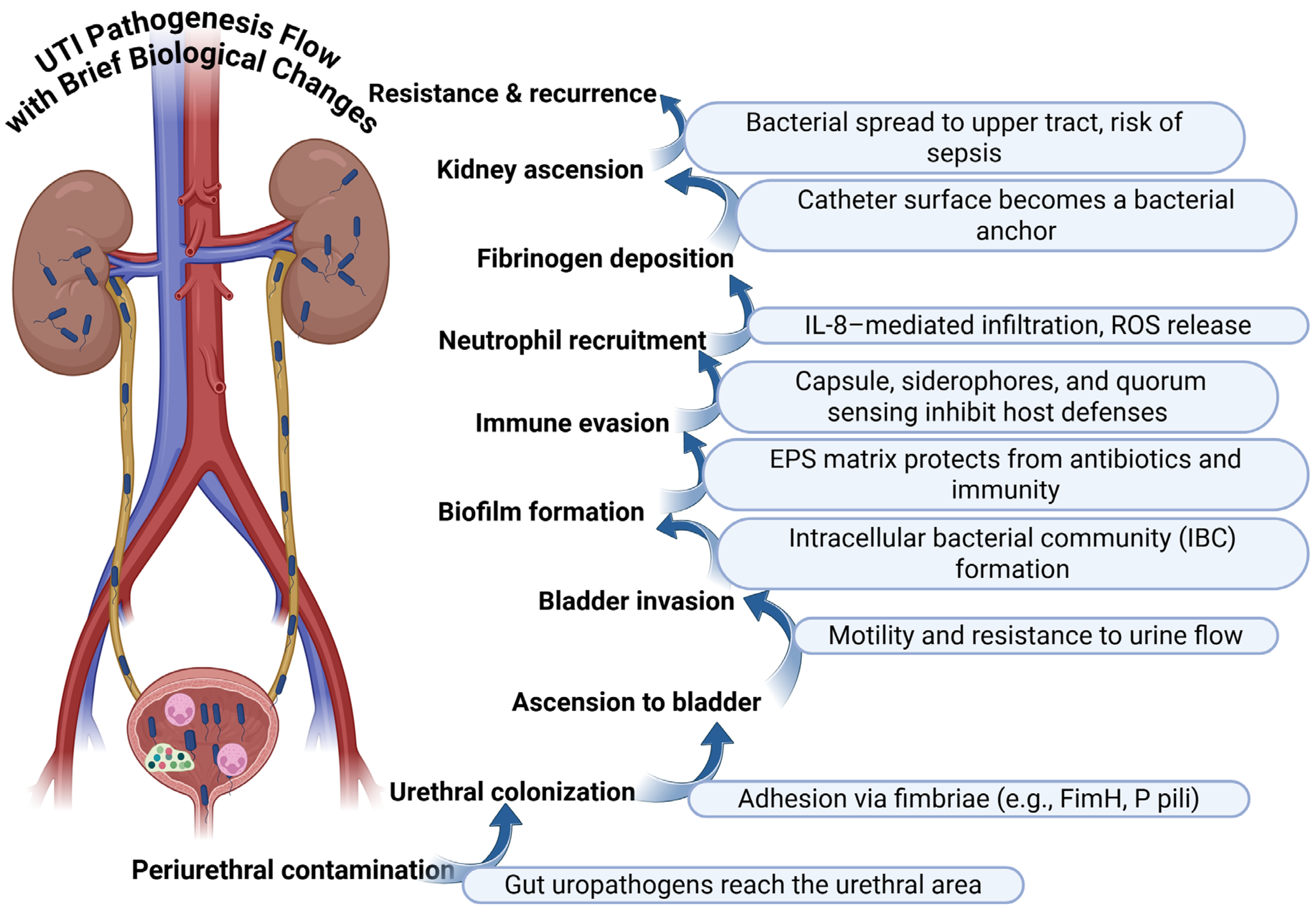
1. Introduction
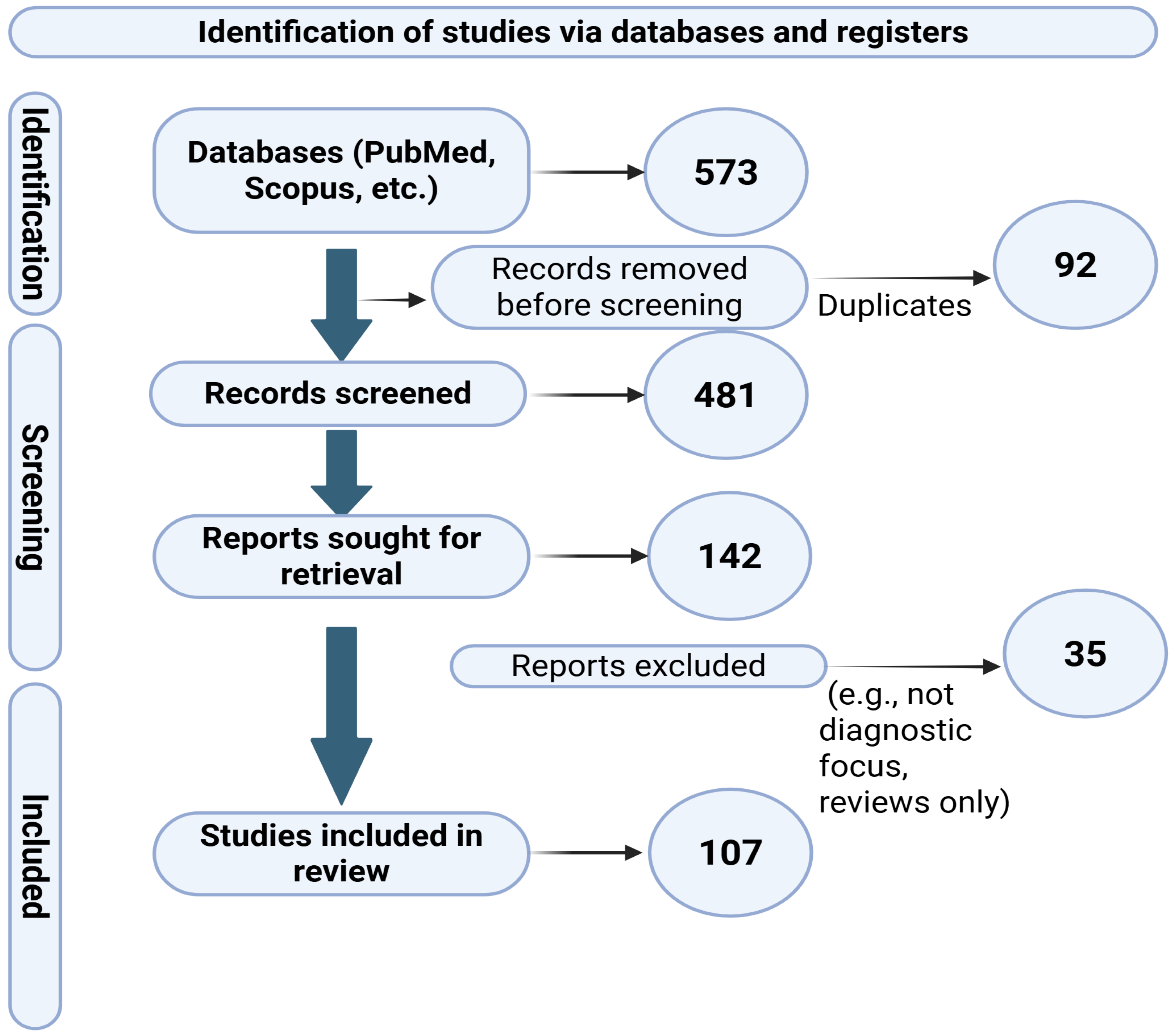
2. Materials and Methods
3. Urinary Diagnostics in Transition: Standard Culture, Enhanced Methods, and the Urobiome
4. Urinary Diagnostics: Improved Bacteriuria Screens, Multiplex Nucleic Acids, Sequencing, and Phenotypic Methods
5. Advances in Molecular Diagnostics for Urinary Tract Infections: Amplification, Hybridization, and Sequencing Approaches
6. Future Perspectives
7. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- He, Y.; Zhao, J.; Wang, L.; Han, C.; Yan, R.; Zhu, P.; Qian, T.; Yu, S.; Zhu, X.; He, W. Epidemiological trends and predictions of urinary tract infections in the Global Burden of Disease Study 2021. Sci. Rep. 2025, 15, 4702. [Google Scholar] [CrossRef]
- Zeng, Z.; Zhan, J.; Zhang, K.; Chen, H.; Cheng, S. Global, regional, and national burden of urinary tract infections from 1990 to 2019: An analysis of the Global Burden of Disease Study 2019. World J. Urol. 2022, 40, 755–763. [Google Scholar] [CrossRef]
- Salvatore, S.; Salvatore, S.; Cattoni, E.; Siesto, G.; Serati, M.; Sorice, P.; Torella, M. Urinary tract infections in women. Eur. J. Obstet. Gynecol. Reprod. Biol. 2011, 156, 131–136. [Google Scholar] [CrossRef]
- Baimakhanova, B.; Sadanov, A.; Trenozhnikova, L.; Balgimbaeva, A.; Baimakhanova, G.; Orasymbet, S.; Tleubekova, D.; Amangeldi, A.; Turlybaeva, Z.; Nurgaliyeva, Z.; et al. Understanding the Burden and Management of Urinary Tract Infections in Women. Diseases 2025, 13, 59. [Google Scholar] [CrossRef]
- Anton, C.-I.; Ștefan, I.; Zamfir, M.; Ghiațău, C.F.; Sima, C.S.; Osman, C.L.; Ștefan, T.A.; Streinu-Cercel, A. Etiology and Risk Factors of Recurrent Urinary Tract Infections in Women in a Multidisciplinary Hospital in Romania. Microorganisms 2025, 13, 626. [Google Scholar] [CrossRef] [PubMed]
- Miller, E.A.; Beasley, D.E.; Dunn, R.R.; Archie, E.A. Lactobacilli dominance and vaginal pH: Why is the human vaginal microbiome unique? Front. Microbiol. 2016, 7, 1936. [Google Scholar] [CrossRef] [PubMed]
- Neugent, M.L.; Kumar, A.; Hulyalkar, N.V.; Lutz, K.C.; Nguyen, V.H.; Fuentes, J.L.; Zhang, C.; Nguyen, A.; Sharon, B.M.; Kuprasertkul, A.; et al. Recurrent urinary tract infection and estrogen shape the taxonomic ecology and function of the postmenopausal urogenital microbiome. Cell Rep. Med. 2022, 3, 100753. [Google Scholar] [CrossRef] [PubMed]
- Maharjan, G.; Khadka, P.; Siddhi Shilpakar, G.; Chapagain, G.; Dhungana, G.R. Catheter-associated urinary tract infection and obstinate biofilm producers. Can. J. Infect. Dis. Med. Microbiol. 2018, 2018, 7624857. [Google Scholar] [CrossRef]
- Zorc, J.J.; Kiddoo, D.A.; Shaw, K.N. Diagnosis and management of pediatric urinary tract infections. Clin. Microbiol. Rev. 2005, 18, 417–422. [Google Scholar] [CrossRef]
- Dutta, C.; Pasha, K.; Paul, S.; Abbas, M.S.; Nassar, S.T.; Tasha, T.; Desai, A.; Bajgain, A.; Ali, A.; Mohammed, L. Urinary tract infection induced delirium in elderly patients: A systematic review. Cureus 2022, 14, e32321. [Google Scholar] [CrossRef]
- Mayne, S.; Bowden, A.; Sundvall, P.D.; Gunnarsson, R. The scientific evidence for a potential link between confusion and urinary tract infection in the elderly is still confusing—A systematic literature review. BMC Geriatr. 2019, 19, 32. [Google Scholar] [CrossRef]
- Schmiemann, G.; Kniehl, E.; Gebhardt, K.; Matejczyk, M.M.; Hummers-Pradier, E. The diagnosis of urinary tract infection: A systematic review. Dtsch. Arztebl. Int. 2010, 107, 361–367. [Google Scholar] [CrossRef]
- Mancuso, G.; Midiri, A.; Gerace, E.; Marra, M.; Zummo, S.; Biondo, C. Urinary Tract Infections: The Current Scenario and Future Prospects. Pathogens 2023, 12, 623. [Google Scholar] [CrossRef]
- Resistance Patterns Varied Among Different Age Categories: A Retrospective Study From a Tertiary General Hospital During a 12-Year Period. Front. Microbiol. 2021, 12, 813145.
- Johnson, J.R. Definitions of complicated urinary tract infection and pyelonephritis. Clin. Infect. Dis. 2017, 64, 390. [Google Scholar] [CrossRef] [PubMed]
- Bader, M.S.; Loeb, M.; Brooks, A.A. An update on the management of urinary tract infections in the era of antimicrobial resistance. Postgrad. Med. 2017, 129, 242–258. [Google Scholar] [CrossRef] [PubMed]
- Nicolle, L.E.; Gupta, K.; Bradley, S.F.; Colgan, R.; DeMuri, G.P.; Drekonja, D.; Eckert, L.O.; Geerlings, S.E.; Köves, B.; Hooton, T.M.; et al. Clinical practice guideline for the management of asymptomatic bacteriuria: 2019 update by the Infectious Diseases Society of America. Clin. Infect. Dis. 2019, 68, e83–e110. [Google Scholar] [CrossRef]
- Gołębiewska, J.E.; Krawczyk, B.; Wysocka, M.; Dudziak, A.; Dębska-Ślizień, A. Asymptomatic Bacteriuria in Kidney Transplant Recipients—A Narrative Review. Medicina 2023, 59, 198. [Google Scholar] [CrossRef]
- Colella, M.; Topi, S.; Palmirotta, R.; D’Agostino, D.; Charitos, I.A.; Lovero, R.; Santacroce, L. An Overview of the Microbiota of the Human Urinary Tract in Health and Disease: Current Issues and Perspectives. Life 2023, 13, 1486. [Google Scholar] [CrossRef]
- Aragón, I.M.; Herrera-Imbroda, B.; Queipo-Ortuño, M.I.; Castillo, E.; Del Moral, J.S.; Gómez-Millán, J.; Yucel, G.; Lara, M.F. The Urinary Tract Microbiome in Health and Disease. Eur. Urol. Focus 2018, 4, 128–138. [Google Scholar] [CrossRef]
- Flores-Mireles, A.L.; Walker, J.N.; Caparon, M.; Hultgren, S.J. Urinary tract infections: Epidemiology, mechanisms of infection and treatment options. Nat. Rev. Microbiol. 2015, 13, 269–284. [Google Scholar] [CrossRef]
- Mody, L.; Juthani-Mehta, M. Urinary tract infections in older women: A clinical review. JAMA 2014, 26, 844–854. [Google Scholar] [CrossRef]
- McLellan, L.K.; Hunstad, D.A. Urinary Tract Infection: Pathogenesis and Outlook. Trends Mol. Med. 2016, 22, 946–957. [Google Scholar] [CrossRef] [PubMed]
- Mestrovic, T.; Matijasic, M.; Peric, M.; Cipcic Paljetak, H.; Baresic, A.; Verbanac, D. The Role of Gut, Vaginal, and Urinary Microbiome in Urinary Tract Infections: From Bench to Bedside. Diagnostics 2020, 11, 7. [Google Scholar] [CrossRef] [PubMed]
- Mason, C.Y.; Sobti, A.; Goodman, A.L. Staphylococcus aureus bacteriuria: Implications and management. JAC Antimicrob. Resist. 2023, 5, dlac123. [Google Scholar] [CrossRef]
- Dias, V. Candida species in the urinary tract: Is it a fungal infection or not? Future Microbiol. 2020, 15, 81–83. [Google Scholar] [CrossRef]
- Naber, K.G.; Tiran-Saucedo, J.; Wagenlehner, F.M.E. Psychosocial burden of recurrent uncomplicated urinary tract infections. GMS Infect. Dis. 2022, 10, Doc01. [Google Scholar]
- Lewis, D.A.; Brown, R.; Williams, J.; White, P.; Jacobson, S.K.; Marchesi, J.R.; Drake, M.J. The human urinary microbiome; bacterial DNA in voided urine of asymptomatic adults. Front. Cell. Infect. Microbiol. 2013, 3, 41. [Google Scholar] [CrossRef]
- Barraud, O.; Ravry, C.; François, B.; Daix, T.; Ploy, M.-C.; Vignon, P. Shotgun metagenomics for microbiome and resistome detection in septic patients with urinary tract infection. Int. J. Antimicrob. Agents 2019, 54, 803–808. [Google Scholar] [CrossRef]
- Serapide, F.; Pallone, R.; Quirino, A.; Marascio, N.; Barreca, G.S.; Davoli, C.; Lionello, R.; Matera, G.; Russo, A. Impact of Multiplex PCR on Diagnosis of Bacterial and Fungal Infections and Choice of Appropriate Antimicrobial Therapy. Diagnostics 2025, 15, 1044. [Google Scholar] [CrossRef]
- Page, M.J.; McKenzie, J.E.; Bossuyt, P.M.; Boutron, I.; Hoffmann, T.C.; Mulrow, C.D.; Shamseer, L.; Tetzlaff, J.M.; Akl, E.A.; Brennan, S.E.; et al. The PRISMA 2020 statement: An updated guideline for reporting systematic reviews. BMJ 2021, 372, n71. [Google Scholar] [CrossRef]
- Dabkowski, J.; Dodds, P.; Hughes, K.; Bush, M. A persistent, symptomatic urinary tract infection with multiple “negative” urine cultures. Conn. Med. 2013, 77, 27–29. [Google Scholar]
- Litster, A.; Moss, S.; Platell, J.; Trott, D.J. Occult bacterial lower urinary tract infections in cats—Urinalysis and culture findings. Vet. Microbiol. 2009, 136, 130–134. [Google Scholar] [CrossRef] [PubMed]
- Anger, J.; Lee, U.; Ackerman, A.L.; Chou, R.; Chughtai, B.; Clemens, J.Q.; Hickling, D.; Kapoor, A.; Kenton, K.S.; Kaufman, M.R.; et al. Recurrent uncomplicated urinary tract infections in women: AUA/CUA/SUFU guideline. J. Urol. 2019, 202, 282–289. [Google Scholar] [CrossRef] [PubMed]
- Claeys, K.C.; Blanco, N.; Morgan, D.J.; Leekha, S.; Sullivan, K.V. Advances and challenges in the diagnosis and treatment of urinary tract infections: The need for diagnostic stewardship. Curr. Infect. Dis. Rep. 2019, 21, 11. [Google Scholar] [CrossRef] [PubMed]
- Pearce, M.M.; Hilt, E.E.; Rosenfeld, A.B.; Zilliox, M.J.; Thomas-White, K.; Fok, C.; Kliethermes, S.; Schreckenberger, P.C.; Brubaker, L.; Gai, X.; et al. The female urinary microbiome: A comparison of women with and without urgency urinary incontinence. mBio 2014, 5, e01283-14. [Google Scholar] [CrossRef]
- Wolfe, A.J.; Toh, E.; Shibata, N.; Rong, R.; Kenton, K.; Fitzgerald, M.; Mueller, E.R.; Schreckenberger, P.; Dong, Q.; Nelson, D.E.; et al. Evidence of uncultivated bacteria in the adult female bladder. J. Clin. Microbiol. 2012, 50, 1376–1383. [Google Scholar] [CrossRef]
- Hilt, E.E.; McKinley, K.; Pearce, M.M.; Rosenfeld, A.B.; Zilliox, M.J.; Mueller, E.R.; Brubaker, L.; Gai, X.; Wolfe, A.J.; Schreckenberger, P.C. Urine is not sterile: Use of enhanced urine culture techniques to detect resident bacterial flora in the adult female bladder. J. Clin. Microbiol. 2014, 52, 871–876. [Google Scholar] [CrossRef]
- Coorevits, L.; Heytens, S.; Boelens, J.; Claeys, G. The resident microflora of voided midstream urine of healthy controls: Standard versus expanded urine culture protocols. Eur. J. Clin. Microbiol. Infect. Dis. 2017, 36, 635–639. [Google Scholar] [CrossRef]
- Boshell, B.R.; Sanford, J.P. A screening method for the evaluation of urinary tract infections in female patients without catheterization. Ann. Intern. Med. 1958, 48, 1040–1045. [Google Scholar] [CrossRef]
- Miller, J.M.; Binnicker, M.J.; Campbell, S.; Carroll, K.C.; Chapin, K.C.; Gonzalez, M.D.; Harrington, A.; Jerris, R.C.; Kehl, S.C.; Leal, S.M., Jr.; et al. Guide to utilization of the microbiology laboratory for diagnosis of infectious diseases: 2024 update by the Infectious Diseases Society of America (IDSA) and the American Society for Microbiology (ASM). Clin. Infect. Dis. 2024, 67, e1–e94. [Google Scholar] [CrossRef] [PubMed]
- Lum, K.T.; Meers, P.D. Boric acid converts urine into an effective bacteriostatic transport medium. J. Infect. 1989, 18, 51–58. [Google Scholar] [CrossRef] [PubMed]
- Yarbrough, M.L.; Potter, R.F. Urine cultures. In Clinical Microbiology Procedures Handbook; ASM Press: Washington, DC, USA, 2023; pp. 3.11.1.1–3.11.1.15. [Google Scholar]
- Porter, I.A.; Brodie, J. Boric acid preservation of urine samples. BMJ 1969, 2, 353–355. [Google Scholar] [CrossRef] [PubMed]
- McCarter, Y.S.; Burd, E.M.; Hall, G.S.; Zervos, M. Cumitech 2C: Laboratory Diagnosis of Urinary Tract Infections; ASM Press: Washington, DC, USA, 2009. [Google Scholar]
- Sanford, J.P.; Favour, C.B.; Mao, F.H.; Harrison, J.H. Evaluation of the positive urine culture: An approach to the differentiation of significant bacteria from contaminants. Am. J. Med. 1956, 20, 88–93. [Google Scholar] [CrossRef]
- Gaston, J.R.; Johnson, A.O.; Bair, K.L.; White, A.N.; Armbruster, C.E. Polymicrobial interactions in the urinary tract: Is the enemy of my enemy my friend? Infect. Immun. 2021, 89, e00652-20. [Google Scholar] [CrossRef]
- Mazzulli, T. Diagnosis and management of simple and complicated urinary tract infections (UTIs). Can. J. Urol. 2012, 19 (Suppl. S1), 42–48. [Google Scholar]
- Clemens, J.Q.; Erickson, D.R.; Varela, N.P.; Lai, H.H. Diagnosis and treatment of interstitial cystitis/bladder pain syndrome. J. Urol. 2022, 208, 34–42. [Google Scholar] [CrossRef]
- Hooton, T.M.; Roberts, P.L.; Cox, M.E.; Stapleton, A.E. Voided midstream urine culture and acute cystitis in premenopausal women. N. Engl. J. Med. 2013, 369, 1883–1891. [Google Scholar] [CrossRef]
- Price, T.K.; Dune, T.; Hilt, E.E.; Thomas-White, K.J.; Kliethermes, S.; Brincat, C.; Brubaker, L.; Wolfe, A.J.; Mueller, E.R.; Schreckenberger, P.C. The clinical urine culture: Enhanced techniques improve detection of clinically relevant microorganisms. J. Clin. Microbiol. 2016, 54, 1216–1222. [Google Scholar] [CrossRef]
- Lainhart, W.; Gonzalez, M.D. Aerococcus urinae, Alloscardovia omnicolens, and Actinotignum schaalii: The AAA minor league team of urinary tract infection pathogens. Clin. Microbiol. Newsl. 2018, 40, 77–82. [Google Scholar] [CrossRef]
- Costales, J.; Alsyouf, M.; Napolitan, P.; Wang, S.; Hu, B. Corynebacterium urealyticum: Rare urinary tract infection with serious complications. Can. J. Urol. 2019, 26, 9680–9682. [Google Scholar] [PubMed]
- Reasoner, S.A.; Flores, V.; Van Horn, G.; Morales, G.; Peard, L.M.; Abelson, B.; Manuel, C.; Lee, J.; Baker, B.; Williams, T.; et al. Survey of the infant male urobiome and genomic analysis of Actinotignum spp. NPJ Biofilms Microbiomes 2023, 9, 91. [Google Scholar] [CrossRef]
- Deen, N.S.; Ahmed, A.; Tasnim, N.T.; Khan, N. Clinical relevance of expanded quantitative urine culture in health and disease. Front. Cell. Infect. Microbiol. 2023, 13, 1210161. [Google Scholar] [CrossRef] [PubMed]
- Vaughan, M.H.; Zemtsov, G.E.; Dahl, E.M.; Karstens, L.; Ma, L.; Siddiqui, N.Y. Concordance of urinary microbiota detected by 16S ribosomal RNA amplicon sequencing vs expanded quantitative urine culture. Am. J. Obstet. Gynecol. 2022, 227, 773–775. [Google Scholar] [CrossRef]
- Marshall, C.W.; Kurs-Lasky, M.; McElheny, C.L.; Bridwell, S.; Liu, H.; Shaikh, N. Performance of conventional urine culture compared to 16S rRNA gene amplicon sequencing in children with suspected urinary tract infection. Microbiol. Spectr. 2021, 9, e0186121. [Google Scholar] [CrossRef]
- Montgomery, S.; Roman, K.; Ngyuen, L.; Cardenas, A.M.; Knox, J.; Tomaras, A.P.; Graf, E.H. Prospective evaluation of light scatter technology paired with matrix-assisted laser desorption ionization–time of flight mass spectrometry for rapid diagnosis of urinary tract infections. J. Clin. Microbiol. 2017, 55, 1802–1811. [Google Scholar] [CrossRef]
- Davaro, E.; Tomaras, A.P.; Chamberland, R.R.; Isbell, T.S. Evaluation of a novel light scattering methodology for the detection of pathogenic bacteria in urine. J. Appl. Lab. Med. 2020, 5, 370–376. [Google Scholar] [CrossRef]
- Ilki, A.; Bekdemir, P.; Ulger, N.; Soyletir, G. Rapid reporting of urine culture results: Impact of the Uro-Quick screening system. New Microbiol. 2010, 33, 147–153. [Google Scholar]
- Athamna, A.; Zbriger, A.; Avadov, S.; Shapira, M.; Tal, Y.; Freimann, S. Rapid identification of uropathogens by combining Alfred 60 system with matrix-assisted laser desorption/ionization–time-of-flight mass spectrometry technology. Eur. J. Clin. Microbiol. Infect. Dis. 2020, 39, 1855–1863. [Google Scholar] [CrossRef]
- Ganguly, A.; Basu, U.; Zimmern, P.; De Nisco, N.; Prasad, S. LBA01-10 uSense: A label-free, rapid, non-culture based biosensing platform for urinary tract infection diagnosis and management. J. Urol. 2023, 209, e1180. [Google Scholar] [CrossRef]
- Teppa, R.J.; Roberts, J.M. The Uriscreen test to detect significant asymptomatic bacteriuria during pregnancy. J. Soc. Gynecol. Investig. 2005, 12, 50–53. [Google Scholar] [CrossRef]
- Boon, H.A.; De Burghgraeve, T.; Verbakel, J.Y.; Van den Bruel, A. Point-of-care tests for pediatric urinary tract infections in general practice: A diagnostic accuracy study. Fam. Pract. 2022, 39, 616–622. [Google Scholar] [CrossRef] [PubMed]
- Iseri, E.; Nilsson, S.; van Belkum, A.; van der Wijngaart, W.; Özenci, V. Performance of an innovative culture-based digital dipstick for detection of bacteriuria. Microbiol. Spectr. 2024, 12, e0361323. [Google Scholar] [CrossRef] [PubMed]
- Dhada, K.S.; Abdallah, M.A.; Chamberland, R.R.; Buchan, B.W.; Faron, M.; Manuel, C.; Maynard, R.; Dietrich, J.; Cole, S.D.; Wallace, M.A.; et al. Characterization of Pattern Bioscience single-cell microbiology platform for bacteriuria screening. In ASM Microbe 2024 Abstracts; American Society for Microbiology: Atlanta, GA, USA, 2024. [Google Scholar]
- Advani, S.D.; Turner, N.A.; Schmader, K.E.; Wrenn, R.H.; Moehring, R.W.; Polage, C.R.; Vaughn, V.M.; Anderson, D.J. Optimizing reflex urine cultures: Using a population-specific approach to diagnostic stewardship. Infect. Control Hosp. Epidemiol. 2023, 44, 206–209. [Google Scholar] [CrossRef]
- Robledo, X.G.; Arcila, K.V.O.; Riascos, S.H.M.; García-Perdomo, H.A. Accuracy of molecular diagnostic techniques in patients with a confirmed urine culture: A systematic review and meta-analysis. Can. Urol. Assoc. J. 2022, 16, E484–E489. [Google Scholar] [CrossRef]
- Ashraf, M.S.; Gaur, S.; Bushen, O.Y.; Chopra, T.; Chung, P.; Clifford, K.; Hames, E.; Hertogh, C.M.P.M.; Krishna, A.; Mahajan, D.; et al. Diagnosis, treatment, and prevention of urinary tract infections in post-acute and long-term care settings: A consensus statement. J. Am. Med. Dir. Assoc. 2020, 21, 12–24. [Google Scholar] [CrossRef]
- Zering, J.; Stohs, E.J. Urine polymerase chain reaction tests: Stewardship helper or hindrance? Antimicrob. Steward. Healthc. Epidemiol. 2024, 4, e77. [Google Scholar] [CrossRef]
- Palat, S.-I.T.; Biehle, L.; Adler, L. Rapid molecular testing for UTIs: A diagnostic stewardship perspective. J. Am. Med. Dir. Assoc. 2024, 25, 105031. [Google Scholar] [CrossRef]
- Szlachta-McGinn, A.; Douglass, K.M.; Chung, U.Y.R.; Jackson, N.J.; Nickel, J.C.; Ackerman, A.L. Molecular diagnostic methods versus conventional urine culture for diagnosis and treatment of urinary tract infection: A systematic review and meta-analysis. Eur. Urol. Open Sci. 2022, 44, 113–124. [Google Scholar] [CrossRef]
- Moreland, R.B.; Choi, B.I.; Geaman, W.; Gonzalez, C.; Hochstedler-Kramer, B.R.; John, J.; Kaindl, J.; Kesav, N.; Lamichhane, J.; Lucio, L.; et al. Beyond the usual suspects: Emerging uropathogens in the microbiome age. Front. Urol. 2023, 3, 1212590. [Google Scholar] [CrossRef]
- Brubaker, L.; Gourdine, J.-P.F.; Siddiqui, N.Y.; Holland, A.; Halverson, T.; Limeria, R.; Pride, D.; Ackerman, L.; Forster, C.S.; Jacobs, K.M.; et al. Forming consensus to advance urobiome research. mSystems 2021, 6, e0137120. [Google Scholar] [CrossRef]
- Arora-Williams, K.; Mangifesta, M.; Stinnett, R.; Broadbent, K.; Schlaberg, R. Target capture versus shotgun metagenomics for uropathogen detection: Analyzing pee in HD. In ASM Microbe 2023 Abstracts; American Society for Microbiology: Houston, TX, USA, 2023. [Google Scholar]
- McDonald, M.; Kameh, D.; Johnson, M.E.; Johansen, T.E.B.; Albala, D.; Mouraviev, V. A head-to-head comparative phase II study of standard urine culture and sensitivity versus DNA next-generation sequencing testing for urinary tract infections. Rev. Urol. 2017, 19, 213–220. [Google Scholar]
- Yoo, J.J.; Shin, H.B.; Moon, J.E.; Lee, S.H.; Jeong, H.; Yang, H.J.; Kim, W.B.; Lee, K.W.; Kim, J.H.; Kim, Y.H. Korean urobiome platform (KUROM) study for acute uncomplicated sporadic versus recurrent cystitis in women: Clinical significance. Investig. Clin. Urol. 2024, 65, 378–390. [Google Scholar] [CrossRef]
- Choi, H.W.; Lee, K.W.; Kim, Y.H. Microbiome in urological diseases: Axis crosstalk and bladder disorders. Investig. Clin. Urol. 2023, 64, 126–139. [Google Scholar] [CrossRef]
- Baltekin, Ö.; Boucharin, A.; Tano, E.; Andersson, D.I.; Elf, J. Antibiotic susceptibility testing in less than 30 min using direct single-cell imaging. Proc. Natl. Acad. Sci. USA 2017, 114, 9170–9175. [Google Scholar] [CrossRef]
- Dubey, R.; Khan, U.; Chaudhari, R.; KV, M. Method for Testing of Antibiotic Susceptibility in Microorganisms. U.S. Patent WO2020109986A1, 26 November 2019. [Google Scholar]
- Alonso-Tarrés, C.; Benjumea Moreno, C.; Navarro, F.; Habison, A.C.; Gonzàlez-Bertran, E.; Blanco, F.; Borràs, J.; Garrigó, M.; Saker, J. Bacteriuria and phenotypic antimicrobial susceptibility testing in 45 min by point-of-care Sysmex PA-100 system: First clinical evaluation. Eur. J. Clin. Microbiol. Infect. Dis. 2024, 43, 1533–1543. [Google Scholar] [CrossRef] [PubMed]
- Tomlinson, E.; Jones, H.E.; James, R.; Cooper, C.; Stokes, C.; Begum, S.; Watson, J.; Hay, A.D.; Ward, M.; Thom, H.; et al. Clinical effectiveness of point-of-care tests for diagnosing urinary tract infection: A systematic review. Clin. Microbiol. Infect. 2024, 30, 197–205. [Google Scholar] [CrossRef]
- Spencer, D.C.; Paton, T.F.; Mulroney, K.T.; Inglis, T.J.J.; Sutton, J.M.; Morgan, H. A fast impedance-based antimicrobial susceptibility test. Nat. Commun. 2020, 11, 5328. [Google Scholar] [CrossRef]
- Bock, L. O04 A novel, fast, accurate, cost-effective antimicrobial susceptibility test for urine samples. JAC-Antimicrob. Resist. 2023, 5, dlac133.004. [Google Scholar] [CrossRef]
- Tarlton, N.J.; Petrovic, D.-F.; Frazee, B.W.; Borges, C.A.; Pham, E.M.; Milton, A.K.; Jackson, N.; deBoer, T.R.; Murthy, N.; Riley, L.W. A dual enzyme-based biochemical test rapidly detects third-generation cephalosporin-resistant CTX-M–producing uropathogens in clinical urine samples. Microb. Drug Resist. 2021, 27, 450–461. [Google Scholar] [CrossRef]
- Banerjee, R.; Humphries, R. Rapid antimicrobial susceptibility testing methods for blood cultures and their clinical impact. Front. Med. 2021, 8, 635831. [Google Scholar] [CrossRef]
- Brown, N.M.; Goodman, A.L.; Horner, C.; Jenkins, A.; Brown, E.M. Treatment of methicillin-resistant Staphylococcus aureus (MRSA): Updated guidelines from the UK. JAC-Antimicrob. Resist. 2021, 3, dlaa114. [Google Scholar] [CrossRef] [PubMed]
- Heintz, B.H.; Halilovic, J.; Christensen, C.L. Vancomycin-resistant enterococcal urinary tract infections. Pharmacotherapy 2010, 30, 1136–1149. [Google Scholar] [CrossRef] [PubMed]
- De Oca, J.E.M.; Veve, M.P.; Zervos, M.J.; Kenney, R.M. Aminopenicillins vs non-aminopenicillins for treatment of enterococcal lower urinary tract infections. Int. J. Antimicrob. Agents 2023, 61, 106800. [Google Scholar] [CrossRef]
- Tamma, P.D.; Aitken, S.L.; Bonomo, R.A.; Mathers, A.J.; van Duin, D.; Clancy, C.J. Infectious Diseases Society of America 2022 guidance on the treatment of extended-spectrum β-lactamase–producing Enterobacterales (ESBL-E), carbapenem-resistant Enterobacterales (CRE), and Pseudomonas aeruginosa with difficult-to-treat resistance (DTR-P. aeruginosa). Clin. Infect. Dis. 2022, 75, 187–212. [Google Scholar] [CrossRef]
- Rahman, M.M.; Hossain, M.M.K.; Rubaya, R.; Halder, J.; Karim, M.E.; Bhuiya, A.A.; Khatun, A.; Alam, J. Association of antibiotic resistance traits in uropathogenic Escherichia coli (UPEC) isolates. Can. J. Infect. Dis. Med. Microbiol. 2022, 2022, 4251486. [Google Scholar] [CrossRef]
- Falsey, A.R.; Branche, A.R.; Croft, D.P.; Formica, M.A.; Peasley, M.R.; Walsh, E.E. Real-life assessment of BioFire FilmArray pneumonia panel in adults hospitalized with respiratory illness. J. Infect. Dis. 2024, 229, 214–222. [Google Scholar] [CrossRef]
- Wojno, K.J.; Baunoch, D.; Luke, N.; Opel, M.; Korman, H.; Kelly, C.; Jafri, S.M.A.; Keating, P.; Hazelton, D.; Hindu, S.; et al. Multiplex PCR based urinary tract infection (UTI) analysis compared to traditional urine culture in identifying significant pathogens in symptomatic patients. Urology 2020, 136, 119–126. [Google Scholar] [CrossRef]
- Heytens, S.; De Sutter, A.; Coorevits, L.; Cools, P.; Boelens, J.; Van Simaey, L.; Christiaens, T.; Vaneechoutte, M.; Claeys, G. Women with symptoms of a urinary tract infection but a negative urine culture: PCR-based quantification of Escherichia coli suggests infection in most cases. Clin. Microbiol. Infect. 2017, 23, 647–652. [Google Scholar] [CrossRef]
- Garrett, E.M.; Bobenchik, A.M. Recent advances in direct blood culture phenotypic antimicrobial susceptibility testing. Clin. Microbiol. Newsl. 2022, 44, 209–216. [Google Scholar] [CrossRef]
- Patel, R.; Polage, C.R.; Dien Bard, J.; May, L.; Lee, F.M.; Fabre, V.; Hayden, M.K.; Doernberg, S.D.B.; Haake, D.A.; Trautner, B.W.; et al. Envisioning future urinary tract infection diagnostics. Clin. Infect. Dis. 2022, 74, 1284–1292. [Google Scholar] [CrossRef]
- Yu, Y.; Zielinski, M.D.; Rolfe, M.A.; Kuntz, M.M.; Nelson, H.; Nelson, K.E.; Pieper, R. Similar neutrophil-driven inflammatory and antibacterial responses in elderly patients with symptomatic and asymptomatic bacteriuria. Infect. Immun. 2015, 83, 4142–4153. [Google Scholar] [CrossRef]
- Flores, E.; Martínez-Racaj, L.; Blasco, Á.; Diaz, E.; Esteban, P.; López-Garrigós, M.; Salinas, M. A step forward in the diagnosis of urinary tract infections: From machine learning to clinical practice. Comput. Struct. Biotechnol. J. 2024, 24, 533–541. [Google Scholar] [CrossRef]
- Taylor, R.A.; Moore, C.L.; Cheung, K.H.; Brandt, C. Predicting urinary tract infections in the emergency department with machine learning. PLoS ONE 2018, 13, e0194085. [Google Scholar] [CrossRef]
| Preanalytical Limitation | Typical Indicator at Accession | Risk to Interpretation | Reflex Action When Suspected | Companion Data to Consider | Expected Effect of Mitigation |
|---|---|---|---|---|---|
| Delay to plating at room temperature beyond about 30 min to 2 h | Undocumented transport time or reported delay | Overgrowth of periurethral flora and shift in relative abundance | Repeat collection with reinforced instructions and process immediately or hold at 4 °C; if repeat is not feasible, interpret with caution | Pyuria on microscopy, leukocyte esterase or nitrite on dipstick, symptom severity | Reduces mixed growth and false positives; improves quantitative fidelity [41,42,43,44] |
| Prolonged cold storage or extended exposure to boric acid preservative | Time in cold storage exceeds local policy or approaches 48 h | Loss of viability for some fastidious organisms and underdetection | Repeat collection and process promptly; consider enhanced quantitative urine culture with larger inoculum and extended incubation | Pyuria and clinical trajectory | Restores recovery of slow-growing or fastidious taxa [38,41,42,43,44,45] |
| Uncertain midstream clean catch quality | Abundant squamous epithelial cells or visible contamination | Mixed growth that mimics contamination and obscures true pathogen | Repeat collection with coached clean catch; for children, consider catheter or suprapubic aspiration when clinically justified | Pyuria, symptom pattern, fever | Improves purity and clarity of the culture readout [40,42,46,47] |
| Infant bag collection | Bag method documented | High contamination rate and unreliable quantitation | Prefer catheter or suprapubic aspiration for diagnostic purposes; repeat if feasible | Pyuria and clinical assessment | Lowers contamination and supports reliable interpretation [40] |
| Menstruation or heavy vaginal discharge at collection | History at accession or visible blood or discharge | Increased mixed growth and misclassification of contamination | Repeat after menstruation when feasible; reinforced clean catch; consider larger inoculum with extended conditions if urgent | Pyuria, nitrite, symptom context | Lowers false positives due to contamination [42,46] |
| Long catheter dwell or sampling from drainage bag | Sample taken from bag or unknown dwell time | Biofilm-derived organisms and misleading counts | Resample from catheter port after appropriate disinfection or replace catheter prior to collection | Pyuria, systemic signs, device history | Improves specificity for catheter-associated infection [42,46,47] |
| Recent antibiotic exposure within the previous 24 to 48 h | Medication history positive for recent antibiotics | Suppressed growth despite ongoing symptoms and false negatives | Repeat collection after antibiotic washout when safe; increase inoculated volume and apply enhanced culture conditions | Pyuria, C-reactive protein if available, symptom trajectory | Increases yield when viable organisms are present at low level [35,45] |
| Low inoculated volume in a patient with high clinical probability | One microliter protocol documented with borderline counts | Counts near the detection limit and missed low-level infection | Increase inoculated volume to ten microliters or more, and consider enhanced quantitative urine culture | Pyuria, nitrite, prior results | Lowers false negatives near the threshold of detection [35,42,46,47] |
| Mixed growth with three or more morphotypes | Report notes multiple distinct colony types | Assumed contamination may mask true polymicrobial infection in selected hosts | Repeat collection and increase inoculum; if clinical probability is high, apply enrichment media and atmospheres with longer incubation | Pyuria, host risk factors, recent procedures | Distinguishes contamination from clinically meaningful polymicrobial infection [35,47] |
| Borderline quantitative counts near 102 to 103 CFU per mL with symptoms | Quantitation close to the laboratory limit of detection | Ambiguity between contamination and early infection | Repeat collection, increase inoculum, and use enhanced culture conditions; interpret alongside inflammation and symptoms | Pyuria, leukocyte esterase, nitrite, pain pattern | Clarifies low-level results and reduces both false negatives and false positives [34,35,42,46] |
| Clinical Scenario | Collection Method | Preferred Inoculum for Plating | Threshold for High Likelihood of Clinical Significance | High-Risk Preanalytical Pitfalls to Watch | Diagnostic Flags at Accession | Reflex Actions if Borderline or Conflicting | Companion Clinical Data to Weigh | Expected Effect of Mitigation | Notes and References |
|---|---|---|---|---|---|---|---|---|---|
| Adult outpatient without catheter | Midstream clean catch | ≥10 µL when clinical probability is high | ≥105 CFU per mL for a single uropathogen | Delay to plating at room temperature, uncertain cleansing, menstruation or heavy discharge, very small inoculum | Undocumented transport time, abundant squamous cells, visible blood or discharge | Repeat coached clean catch, process immediately or hold at 4 °C, increase inoculum, consider enhanced culture if symptoms persist | Pyuria, nitrite, symptom severity and trajectory | Reduces mixed growth and false positives, improves detection near the limit | Storage/preservative practices [36,37,38,39]; thresholds and mixed growth interpretation [34,42]; enhanced culture utility [51,52,53,54] |
| Pregnancy | Midstream clean catch | ≥10 µL | ≥105 CFU per mL; lower tolerance for mixed growth | Same as adult outpatient; added emphasis on prompt processing | History of delay or suboptimal collection | Repeat promptly, increase inoculum, consider enhanced culture when symptoms persist | Pyuria and clinical assessment | Minimizes contamination and supports timely decisions | Thresholds/collection [34,42]; preanalytical control [36,37,38,39] |
| Pediatric outpatient | Clean catch if feasible; catheter or suprapubic for diagnosis | ≥10 µL for clean catch; standard loop for catheter or suprapubic | Clean catch ≥105 CFU per mL; catheter ≥104 CFU per mL | Bag collection, delay to plating, small inoculum | Bag method documented, abundant squamous cells | Prefer catheter or suprapubic aspiration for diagnosis; repeat if contamination suspected | Pyuria, fever, systemic signs | Lowers contamination and improves quantitative fidelity | Pediatric sampling and thresholds [40,42,46] |
| Infant diagnostic sampling | Catheter or suprapubic aspiration | Laboratory standard | Catheter ≥104 CFU per mL; any growth from suprapubic aspirate is significant | Any use of bag collection; transport delays | Bag collection recorded; long transport time | Replace bag with catheter or suprapubic aspiration; process immediately or hold at 4 °C | Pyuria and clinical assessment | Improves specificity and reduces false positives | Collection method impact [40,42,46] |
| Indwelling catheter in place | From catheter port, not drainage bag | Laboratory standard; increase to ≥10 µL if borderline | ≥104 CFU per mL for a single uropathogen | Sampling from drainage bag; long catheter dwell time | Sample source recorded as bag; unknown dwell time | Resample from disinfected port or after catheter change; consider enhanced culture | Pyuria, device history, systemic signs | Better discrimination of catheter-associated infection | Catheter sampling and biofilm issues [40,42,46] |
| After catheter replacement or suprapubic aspiration | Sterile port after replacement or suprapubic aspirate | Laboratory standard | Catheter ≥104 CFU per mL; suprapubic any significant growth | Prolonged cold storage beyond policy; preservative exposure near 48 h | Time in cold storage approaching 48 h | Process promptly; if delayed, repeat collection; consider enhanced culture | Pyuria, pain, fever | Restores recovery of fastidious organisms | Storage/preservative effects [41,42,43,44]; enhanced culture [44,45,46] |
| Neurogenic bladder or urinary diversion | As appropriate for device and anatomy | ≥10 µL if borderline | Often interpret ≥104 CFU per mL with greater weight on symptoms and inflammation | Inadequate cleansing; device colonization; transport delays | Mixed flora with multiple morphotypes | Repeat collection; increase inoculum; apply enhanced media and atmospheres with longer incubation | Pyuria, residual volume, device history | Clarifies mixed growth and detects low-level infection | Host context and polymicrobial risk [46,47,51,52,53] |
| Immunosuppressed or transplant | Method tailored to clinical status | ≥10 µL if borderline | ≥104 CFU per mL may be meaningful with symptoms or inflammation | Recent antibiotics; prolonged storage; small inoculum | Recent antimicrobial exposure recorded | Repeat after brief washout when safe; increase inoculum; apply enhanced culture | Pyuria, systemic markers, symptom course | Increases yield when viable organisms are present at low level | Low-count symptomatic infection [44,45,46,47]; antibiotic impact [42,50] |
| Recent antibiotics within 24–48 h | Any method with exposure documented | ≥10 µL | Numeric thresholds are less reliable due to suppressed growth | Antibiotic exposure; transport delay | Medication history positive for recent antibiotics | Repeat after washout when safe; increase inoculum; use enhanced culture | Pyuria, C-reactive protein if available, symptom trajectory | Restores detection near the limit and reduces false negatives | Antibiotic suppression and reflex strategy [46,47,51,52,53] |
| Recurrent symptoms with prior negative culture | Same or upgraded collection method | ≥10 µL | Treat any quantitative result with caution; pattern over time matters | Any of the above depending on setting | Prior negative culture with high clinical probability | Repeat with larger inoculum and enhanced culture; consider molecular profiling if available; integrate symptoms and inflammation | Pyuria, prior results, analgesic or antibiotic use | Clarifies discordant results and guides targeted work-up | Reflex rules and enhanced detection [46,47,51,52,53] |
| Mixed growth reported | Three or more distinct morphotypes or mixed flora | ≥10 µL on repeat | Often contamination in low-risk settings; can be polymicrobial disease in selected hosts | Uncertain clean catch quality; menstruation; device colonization | Abundant squamous cells; mixed colony types | Repeat coached clean catch; increase inoculum; in high-risk hosts apply additional media, alternative atmospheres, and longer incubation | Pyuria, host risk factors, procedure history | Distinguishes contamination from clinically meaningful polymicrobial infection | Mixed growth interpretation and enhanced culture [46,47,51,52,53] |
| Method | Detection Principle | Typical Time to Result | Strengths | Limitations | Best-Fit Clinical Use | Example Platforms and Notes | References |
|---|---|---|---|---|---|---|---|
| Bacteriuria screening assays | Surrogate markers of infection or non-specific bacterial detection by light scattering, enzyme activity, or electrochemical signals | Minutes to ~3 h | Very high negative predictive value; reduces unnecessary cultures; can fast-track downstream testing | Positive predictive value for infection can be low; does not identify the organism | Triage of routine specimens; early rule-out; selection of samples for direct mass-spectrometry identification | BacterioScan 216Dx (~3 h; sensitivity ~96.5%, specificity ~72%; cleared by the United States Food and Drug Administration); Alifax Uro-Quick (~3 h; available outside the United States); Usense Jimini (~5 min; host inflammatory and bacterial targets); Uriscreen (catalase activity; modest performance in pregnancy and pediatrics); UTRIPlex (three urinary inflammatory markers; high specificity, low sensitivity in pediatrics); UTI-lizer (microreactor “digital dipstick,” ~8 h; very high negative predictive value, widely variable positive predictive value depending on organism); Pattern single-cell platform (negative predictive value ~98% for bacteriuria and promising Gram classification) | [58,59,60,61,62,63,64,65,66,67] |
| Rapid molecular identification (multiplex nucleic acid amplification) | Direct amplification of microbial nucleic acids from urine; multiple targets per panel | Hours | Rapid; high analytical sensitivity; can detect difficult-to-culture organisms | May detect nucleic acids from nonviable organisms after recent antibiotics; target lists are finite; interpretation requires clinical correlation when inflammation is absent or signals are low | High clinical probability with negative or borderline culture; recent antibiotics; situations where early organism-directed therapy could change outcomes | Commercial multiplex panels; reports should state target coverage and unresolved gaps | [68,69,70,71,72] |
| Next-generation sequencing | Community profiling by sequencing (for example, 16S ribosomal RNA gene sequencing or shotgun sequencing) | ~1–3 days (laboratory dependent) | Detects difficult-to-grow and non-culturable organisms; useful in complicated or recurrent urinary infections | May detect commensals in people with and without symptoms; interpretation is complex; does not provide phenotypic susceptibility | Complex or recurrent presentations; discordant culture findings; targeted discovery rather than routine frontline testing | Requires standardized workflows and outcome-linked reporting; integrate with symptoms, inflammation, and collection method | [71,72,73,74,75,76,77,78] |
| Rapid phenotypic antimicrobial susceptibility testing | Measures bacterial growth or metabolic activity in the presence of antibiotics directly in urine | Hours | Rapid evaluation of antimicrobial susceptibility; potential to move from empirical to targeted treatment on the same day | Requires sufficient bacterial burden; scope of antibiotic panels varies; usually needs accompanying culture for organism identification and linkage of result | High-probability cases where early escalation or de-escalation could change outcomes | Pattern Biosciences single-cell droplet imaging under development; additional optical and electrochemical platforms are emerging | [79,80,81,82,83,84,85] |
| Test and Platform (Manufacturer) | Regulatory Status | Detection Principle | Clinical Role This Test Answers | Time to Result | Specimen Handling and Throughput | Stated Limit of Detection (Colony-Forming Units per Milliliter) | Suitable for Near-Patient Use | Can Feed Same-Day Matrix-Assisted Laser Desorption Ionization Time-of-Flight Mass Spectrometry? | Reported Performance Highlights | Notes and Caveats | References |
|---|---|---|---|---|---|---|---|---|---|---|---|
| UTI-lizer Digital Dipstick (Utilizer, Quotient Diagnostics Ltd., Camberley, UK) | Research use only | Miniaturized culturing in sealed microreactors with species-linked color change | Point-of-care mini-culture that estimates bacterial load and triages need for full culture | About 8 h | Single specimen | About 253 colony-forming units per milliliter | Yes | No | Negative predictive value above 97 percent; positive predictive value varies widely by organism from 0 to 90 percent | Requires short incubation; visual readout demands training and quality control | [65] |
| Urine screen (Pattern Biosciences, Austin, TX, USA) | Research use only | Single-cell compartmentalization in droplets with fluorescence imaging and metabolic profiling | Rapid bacteriuria detection and organism class prediction; platform under development for direct susceptibility testing | About 2 h | Single specimen | Not applicable | Yes | Potentially, depending on biomass and workflow | Negative predictive value near 98 percent; promising accuracy for Gram classification | Development pathway includes direct organism identification and rapid phenotypic susceptibility testing | [66] |
| 216Dx UTI System (BacterioScan, Inc., St. Louis, MI, USA) | Cleared as an in vitro diagnostic device by the United States Food and Drug Administration | Laser light scattering to quantify bacterial biomass | Rapid rule-out or rule-in of bacteriuria and triage to culture or downstream testing | About 3 h | Urine; up to about 20 specimens per day | About 50,000 colony-forming units per milliliter | No | Yes, when biomass is sufficient | Sensitivity about 96.5 percent; specificity about 72 percent versus culture | Does not identify organism; positive screens still need organism identification and susceptibility testing | [58,59,60,61] |
| Rapid Urine Culture Test (Alifax S.r.l., Padua, Italy) | Conformité Européenne in vitro diagnostic marking | Light scattering with dedicated analyzers | Same as above; also used to gate direct mass-spectrometry identification | Less than 4 h | Urine; about 60 to 420 specimens depending on analyzer | About 30,000 colony-forming units per milliliter | No | Yes | Widely adopted outside the United States; performance depends on instrument and workflow | Requires analyzer platform (for example, Alfred or HB&L) | [60,61] |
| Uriscreen (Savyon Diagnostics Ltd., Ashdod, Israel) | Conformité Européenne in vitro diagnostic marking | Detection of catalase activity as a proxy for bacteria in somatic cells | Very rapid triage where culture capacity is limited | About 2 min | Single specimen | Not applicable | Yes | No | Pregnancy study: sensitivity 60.7 percent, specificity 89.3 percent; pediatric ambulatory study: sensitivity 67 percent, specificity 69 percent | Modest accuracy relative to culture; best used as a fast rule-out when prevalence is low | [63,64] |
| UTRIPlex (Global Access Diagnostics, Thurleigh, Bedfordshire, UK) | Research use only | Lateral-flow detection of three urinary inflammatory markers, including matrix metalloproteinase-8 and human neutrophil elastase | Host-response screen to prioritize culture in children | About 6 min | Single specimen | Not applicable | Yes | No | In pediatrics: sensitivity 21 percent and specificity 94 percent | High specificity but low sensitivity; useful only as a rule-in when positive | [64] |
| Automated urinalysis analyzers (flow cytometry or digital microscopy, (e.g., Sysmex Corporation, Kobe, Japan; Beckman Coulter, Brea, CA, USA; Siemens Healthineers, Erlangen, Germany) | Varies by model; cleared as in vitro diagnostic devices in many regions | Automated counting of bacteria and leukocytes and imaging of urinary sediment | High-throughput screening and triage with programmable thresholds; supports early rule-out of bacteriuria | About 2 min | Urine; high throughput with continuous loading depending on analyzer | Instrument-specific; typically reported as counts per microliter rather than colony-forming units per milliliter | No | No | High negative predictive value when thresholds are optimized; enables laboratory-scale triage before culture | Requires benchtop analyzer, calibration, and local validation of thresholds; does not identify organism | [67] |
| Test and Platform (Manufacturer) | Regulatory Status | Method (Spelled Out) | Targets Reported (Identification and Resistance) | Approximate Time to Result | Specimen and Throughput | Analytical Sensitivity (Limit of Detection) | Platform Requirements and Near-Patient Suitability | Distinctive Notes |
|---|---|---|---|---|---|---|---|---|
| Nucleic acid amplification assays (NAATs) | ||||||||
| CLONeT E. coli Assay for urinary infection | Research use only | Multiplex polymerase chain reaction with lateral-flow readout | Organism identification: E. coli; resistance genes: none reported | Less than 45 min | Urine; one specimen per run | About 100 to 1000 CFU/mL | Multiple real-time thermocyclers; suitable for near-patient use | Narrow organism coverage with very rapid time to result |
| Usense module (Module Innovations Pvt. Ltd., Pune, India) | Research use only | Signal amplification by nanoparticles (size-dependent) | Organism identification: not disclosed; resistance genes: none reported | About 15 min | Urine; one specimen per run | No data | Designed for near-patient use | Very rapid screen; target list not disclosed in provided source |
| Lodestar urinary infection test (Llusern Scientific Ltd., Cardiff, UK) | United Kingdom Conformity Assessment | Loop-mediated amplification | Organism identification: six targets; resistance genes: none reported | About 30 min | Urine; one specimen per run | About 10,000 CFU/mL | Llusen Lodestar DX analyzer; suitable for near-patient use | Short turnaround with dedicated analyzer |
| Randox urinary infection panel (Randox Laboratories Ltd., Crumlin, Northern Ireland, UK) | Research use only | Multiplex real-time polymerase chain reaction | Organism identification: twenty-three targets, including one Candida species; resistance genes: eight targets | Less than 4 h | Urine; one specimen per run | About 10,000 CFU/mL | Vivalytic platform; designed for near-patient use | Broad panel for organism identification and resistance markers |
| PathoKey MP urinary infection identification and antimicrobial resistance polymerase chain reaction test (Vela Diagnostics Pte. Ltd., Singapore (headquarters); also with operations in Hamburg, Germany) | Research use only | Multiplex real-time polymerase chain reaction | Organism identification: thirteen targets, including one Candida species; resistance genes: fourteen targets | At least 4 h | About forty samples per run | About 1000 CFU/mL | Multiple real-time thermocyclers; not designed for near-patient use | Combined organism and resistance panel |
| OpenArray urinary tract microbiota (Thermo Fisher Scientific, Waltham, MA, USA) | Research use only | Multiplex polymerase chain reaction on OpenArray plates | Organism identification: sixteen bacterial targets and one Candida species; resistance genes: none reported | About 5 h | Forty-eight to one hundred ninety-two specimens per run | No data | QuantStudio 12K Flex with OpenArray block; not designed for near-patient use | High-throughput research-focused panel |
| Multiple primer sets targeting urinary infection pathogens (BioGX, Inc., Birmingham, AL, USA) | Research use only | Multiplex real-time polymerase chain reaction | Organism identification: forty-two bacterial and eight fungal targets; resistance genes: eleven targets | About 2 h | Twenty-four to ninety-six specimens per run; urine neat or preserved in boric acid | No data | Platforms include BD Max, ABI, and Bio-Rad; not designed for near-patient use | Large multi-organism panel for laboratory analyzers |
| QIAstat-Dx complicated urinary infection plus antimicrobial resistance panel (Qiagen N.V., Hilden, Germany) | Research use only | Multiplex real-time polymerase chain reaction | Organism identification: no data; resistance genes: no data in source | Day-scale throughput about 160 specimens | System-integrated cartridge platform | No data | QIAstat-Dx instrument; not designed for near-patient use | Integrated cartridge analyzer; details not provided in source |
| Unyvero urinary infection panel (Curetis GmbH (a subsidiary of OpGen, Inc.), Bodelshausen, Germany) | Research use only | Multiplex polymerase chain reaction | Organism identification: twenty-three bacterial targets and four Candida species; resistance genes: fifteen targets | About 5 h | Two to twelve specimens per run; midstream, suprapubic, or fresh catheter specimens | About 4000 to 100,000 CFU/mL | Unyvero system; not designed for near-patient use | Broad panel with flexible specimen types |
| DNA/RNA hybridization | ||||||||
| CAPTURE UTI (GeneCapture, Inc., Huntsville, AL, USA) | Research use only (prototype referenced) | Unamplified ribonucleic acid expression detected by probe capture | Organism identification: six bacterial targets and one Candida species; resistance genes: none reported | About 2 h | Urine; approximately forty specimens per run | About 10,000 CFU/mL | GeneCapture benchtop instrument; not designed for near-patient use | Representative capture-hybridization approach among urine platforms in development in provided sources |
| Targeted NGS | ||||||||
| Urinary Pathogen Identification and Antimicrobial Resistance Enrichment Kit (Illumina, Inc., San Diego, CA, USA) | Research use only | Targeted deoxyribonucleic acid sequencing with enrichment | Organism identification: one hundred twenty-one bacterial targets, fourteen fungal, four parasites, and thirty-five viruses; resistance genes: three thousand seven hundred twenty-eight targets | About 1 day | Urine; up to 384 specimens per run (instrument-dependent). | Demonstrated to about 100,000 CFU/mL | Illumina next-generation sequencing instruments; not designed for near-patient use | Very broad detection with extensive resistance catalog; requires sequencing infrastructure |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Baimakhanova, B.; Sadanov, A.; Berezin, V.; Baimakhanova, G.; Trenozhnikova, L.; Orasymbet, S.; Seitimova, G.; Kalmakhanov, S.; Xetayeva, G.; Shynykul, Z.; et al. Emerging Technologies for the Diagnosis of Urinary Tract Infections: Advances in Molecular Detection and Resistance Profiling. Diagnostics 2025, 15, 2469. https://doi.org/10.3390/diagnostics15192469
Baimakhanova B, Sadanov A, Berezin V, Baimakhanova G, Trenozhnikova L, Orasymbet S, Seitimova G, Kalmakhanov S, Xetayeva G, Shynykul Z, et al. Emerging Technologies for the Diagnosis of Urinary Tract Infections: Advances in Molecular Detection and Resistance Profiling. Diagnostics. 2025; 15(19):2469. https://doi.org/10.3390/diagnostics15192469
Chicago/Turabian StyleBaimakhanova, Baiken, Amankeldi Sadanov, Vladimir Berezin, Gul Baimakhanova, Lyudmila Trenozhnikova, Saltanat Orasymbet, Gulnaz Seitimova, Sundetgali Kalmakhanov, Gulzakira Xetayeva, Zhanserik Shynykul, and et al. 2025. "Emerging Technologies for the Diagnosis of Urinary Tract Infections: Advances in Molecular Detection and Resistance Profiling" Diagnostics 15, no. 19: 2469. https://doi.org/10.3390/diagnostics15192469
APA StyleBaimakhanova, B., Sadanov, A., Berezin, V., Baimakhanova, G., Trenozhnikova, L., Orasymbet, S., Seitimova, G., Kalmakhanov, S., Xetayeva, G., Shynykul, Z., Seidakhmetova, A., & Turgumbayeva, A. (2025). Emerging Technologies for the Diagnosis of Urinary Tract Infections: Advances in Molecular Detection and Resistance Profiling. Diagnostics, 15(19), 2469. https://doi.org/10.3390/diagnostics15192469