Simultaneous Detection and Differentiation of SARS-CoV-2, Influenza A/B, and Respiratory Syncytial Viruses in Respiratory Specimens Using the VitaSIRO solo™ SARS-CoV-2/Flu/RSV Assay
Abstract
1. Introduction
2. Materials and Methods
2.1. Clinical Specimens
2.2. Reference Molecular Testing for Respiratory Viruses
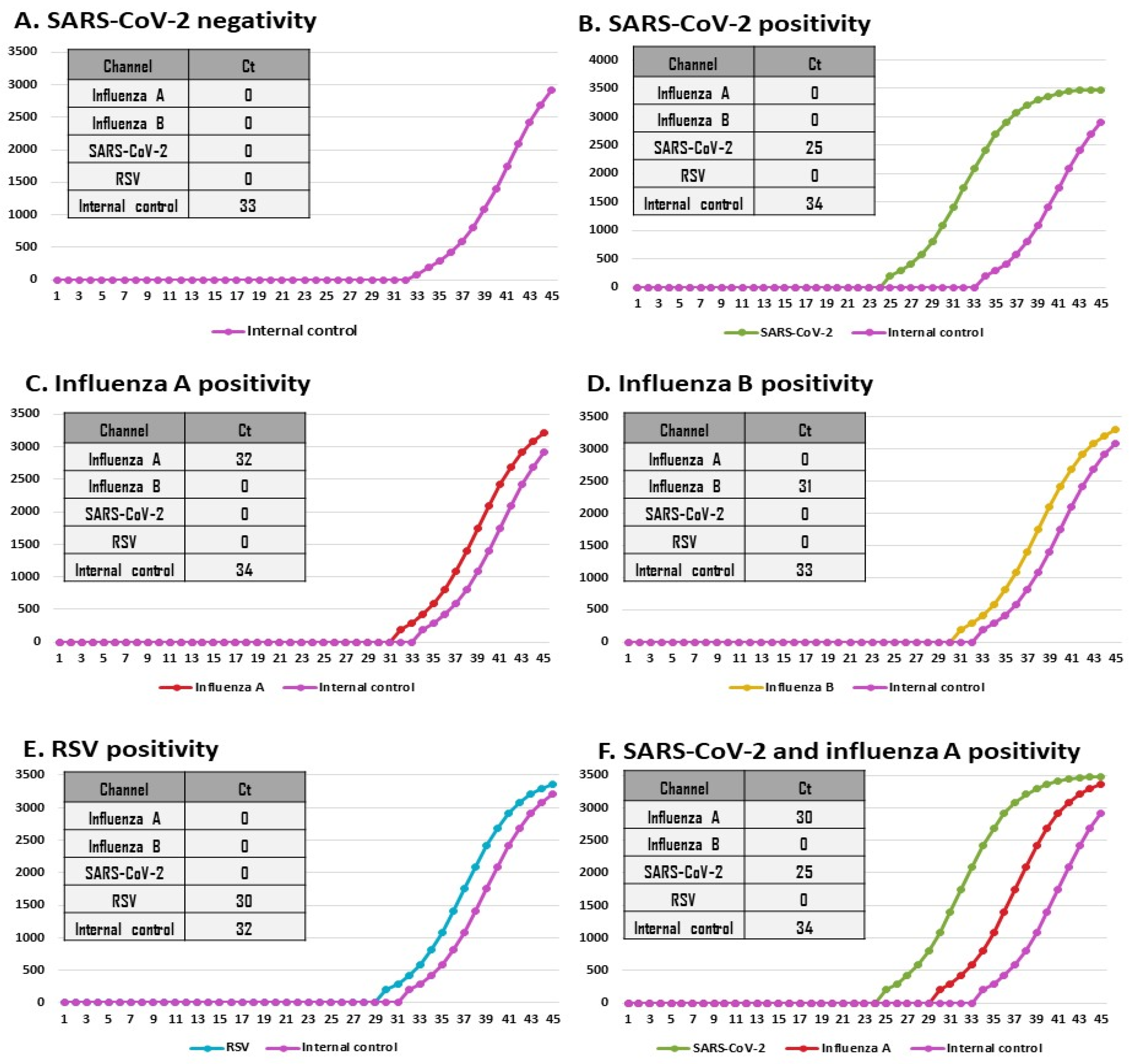
2.3. Simultaneous Detection of SARS-CoV-2, Influenza A Virus, Influenza B Virus, and RSV by VitaSIRO solo™ SARS-CoV-2/Flu/RSV Assay
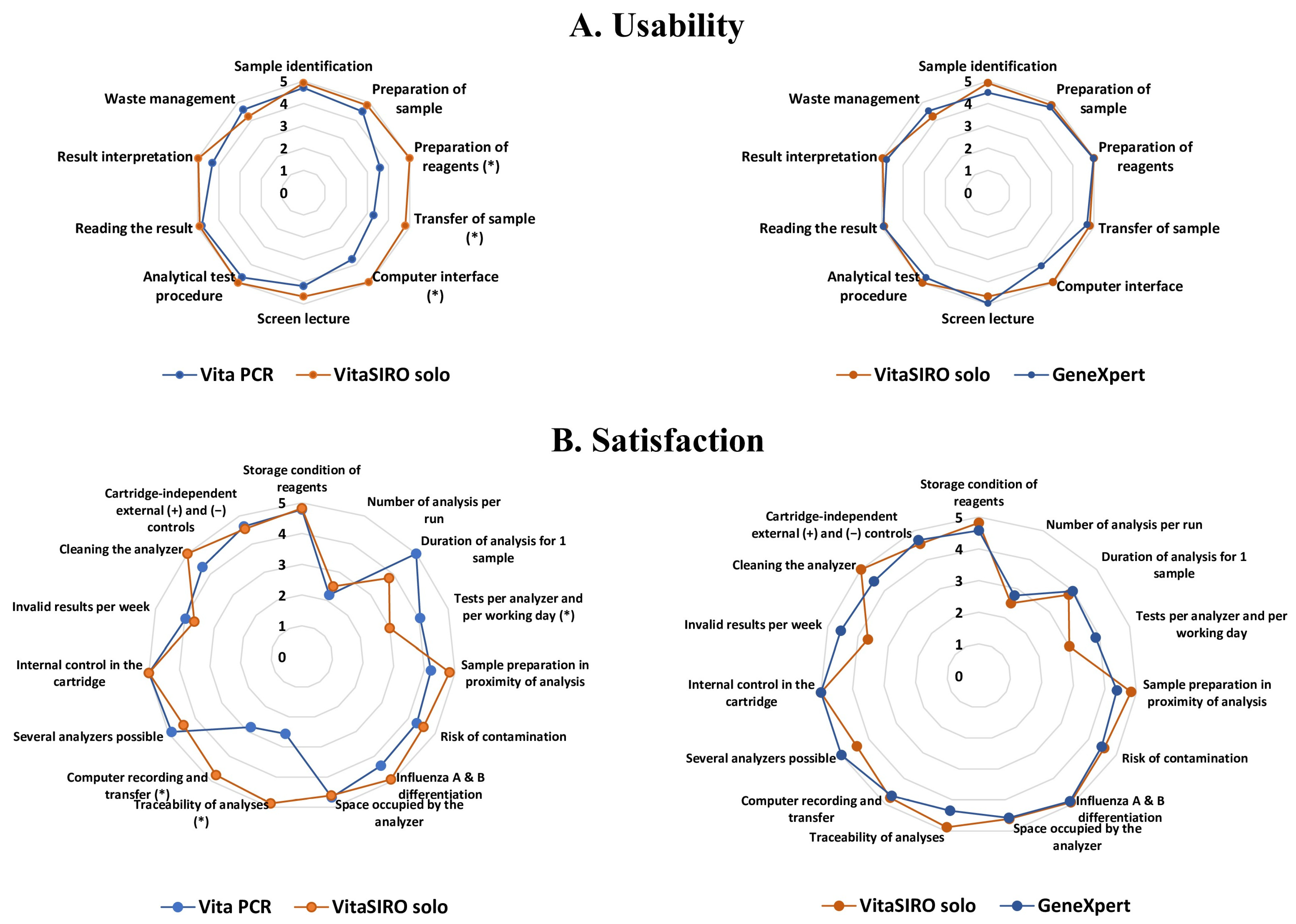
2.4. Comparative Practicability of VitaSIRO solo™ Instrument, Cepheid GeneXpert® Xpress System, and VitaPCR™ Instrument
- Substudy 1: Usability evaluation. The usability of each platform was assessed among volunteer health workers from the laboratory, including 6 laboratory technicians and 4 biologists. The participants were first trained in the three platforms, and everyone carefully read the instructions for each kit. The volunteers performed at least 5 measurements with each system, then completed the usability grid comprising 10 items (Figure 3).
- Substudy 2: Satisfaction questionnaire. Afterwards, the participants filled out the satisfaction questionnaire concerning their experiences with each platform, comprising 15 items (Figure 3).
2.5. Verification of Method According to EN ISO 15189:2022 Criteria for Accreditation
2.6. Statistical Analysis
3. Results
3.1. Repeatability, Reproducibility, and Invalid Results
3.2. Comparative Usability and Satisfaction Evaluation
3.3. Analytical Performances Using Clinical Samples
3.3.1. Results by Reference Assays
3.3.2. Analytical Performances of the VitaSIRO solo™ SARS-CoV-2/Flu/RSV Assay (Table 1)
| VitaSIRO solo™ SARS-CoV-2/Flu/RSV Assay | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| n | Positive (n) | Negative (n) | Sensitivity (% [95%CI]) µ | Specificity (% [95%CI]) | Agreement a (% [95%CI]) | Concordance b (% [95%CI]) | Youden’ J Index c (% [95%CI]) | PPV d (% [95%CI]) | NPV d (% [95%CI]) | ||||
| Reference assays | SARS-CoV-2 | Positive | All Ct $ values | 39 | 37 | 2 | 94.8 [82.2–99.4] | 100.0 [98.2–100.0] | 99.3 [97.4–99.9] | 0.970 [0.928–1.0] | 0.948 [0.822–0.994] | 100.0 [88.8–100.0] | 99.2 [97.1–99.9] |
| ≤33 | 34 | 34 | 0 | 100.0 [87.9–100.0] | 100.0 [98.2–100.0] | 100.0 [99.8–100.0] | 1.0 [0.999–1.0] | 1.0 [0.982–1.0] | 100.0 [87.9–100.0] | 100.0 [98.2–100.0] | |||
| >33 | 5 | 3 | 2 | 60.0 [22.9–88.4] | 100.0 [98.2–100.0] | 99.2 [97.1–99.9] | 0.746 [0.408–1.0] | 0.600 [0.229–0.884] | 100.0 [38.2–100.0] | 99.2 [97.1–99.9] | |||
| Negative | 262 | 0 | 262 | - | - | - | - | - | - | - | |||
| Influenza A | Positive | 77 | 75 | 2 | 97.4 [90.4–99.8] | 99.5 [97.2–99.9] | 99.0 [96.9–99.8] | 0.974 [0.944–1.0] | 0.969 [0.931–0.989] | 98.6 [92.2–99.9] | 99.1 [96.6–99.9] | ||
| Negative | 224 | 1 | 223 | ||||||||||
| Influenza B | Positive | 20 | 18 | 2 | 90.0 [68.6–98.4] | 100.0 [98.6–100.0] | 99.3 [97.4–>99.9] | 0.944 [0.866–1.0] | 0.900 [0.686–0.984] | 100.0 [79.3–100.0] | 99.2 [97.2–99.9] | ||
| Negative | 281 | 0 | 281 | ||||||||||
| Influenza A/B | Positive | 97 | 93 | 4 | 95.8 [89.5–98.7] | 99.5 [98.6–100.0] | 98.3 [96.0–99.74 | 0.962 [0.928–0.995] | 0.953 [0.914–0.977] | 98.9 [93.6–99.9] | 98.0 [94.9–99.4] | ||
| Negative | 204 | 1 | 203 | ||||||||||
| RSV | Positive | 42 | 40 | 2 | 95.2 [83.3–99.5] | 100.0 [99.9–100.0] | 99.3 [97.84–>99.9] | 0.972 [0.933–1.0] | 0.952 [0.833–0.995] | 100.0 [89.5–100.0] | 99.2 [97.0–99.9] | ||
| Negative | 259 | 0 | 259 | ||||||||||
| SARS-CoV2 + influenza A/B + RSV | Positive | 175 | 167 | 8 | 95.4 [91.1–97.8] | 99.2 [99.7–100.0] | 97.0 [94.3–98.5] | 0.939 [0.900–0.978] | 0.946 [0.906–0.969] | 99.4 [94.5–99.9] | 93.9 [88.4–97.0] | ||
| Negative | 126 | 1 | 125 | ||||||||||
3.4. Agreement Between VitaSIRO solo™ SARS-CoV-2/Flu/RSV Assay and Comparator NeuMoDx™ Flu A-B/RSV/SARS-CoV-2 Vantage Assay
3.5. Association of Ct Values Between VitaSIRO solo™ SARS-CoV-2/Flu/RSV Assay and Comparator NeuMoDx™ Flu A-B/RSV/SARS-CoV-2 Vantage Assay
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zhou, F.; Yu, T.; Du, R.; Fan, G.; Liu, Y.; Liu, Z.; Xiang, J.; Wang, Y.; Song, B.; Gu, X.; et al. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: A retrospective cohort study. Lancet 2020, 395, 1054–1062. [Google Scholar] [CrossRef]
- Zylke, J.W.; Bauchner, H. Mortality and Morbidity. The measure of a Pandemic. JAMA J. Am. Med. Assoc. 2020, 324, 458–459. [Google Scholar] [CrossRef] [PubMed]
- Groves, H.E.; Piché-Renaud, P.P.; Peci, A.; Farrar, D.S.; Buckrell, S.; Bancej, C.; Sevenhuysen, C.; Campigotto, A.; Gubbay, J.B.; Morris, S.K. The impact of the COVID-19 pandemic on influenza, respiratory syncytial virus, and other seasonal respiratory virus circulation in Canada: A population-based study. Lancet Reg. Health Am. 2021, 1, 100015. [Google Scholar] [PubMed]
- Heiskanen, A.; Galipeau, Y.; Little, J.; Mortimer, L.; Ramotar, K.; Langlois, M.A.; Cooper, C.L. Seasonal respiratory virus circulation was diminished during the COVID-19 pandemic. Influenza Other Respir. Viruses 2023, 17, e13065. [Google Scholar] [CrossRef]
- Heiskanen, A.; Galipeau, Y.; Little, J.; Langlois, M.A.; Cooper, C.L. Reduced seasonal coronavirus incidence in high-risk population groups during the COVID-19 pandemic. Immun. Inflamm. Dis. 2024, 12, e1342. [Google Scholar] [CrossRef]
- Kuitunen, I.; Artama, M.; Haapanen, M.; Renko, M. Respiratory virus circulation in children after relaxation of COVID-19 restrictions in fall 2021—A nationwide register study in Finland. J. Med. Virol. 2022, 94, 4528–4532. [Google Scholar] [CrossRef] [PubMed]
- Liu, P.; Xu, J. Resurgence of influenza virus activity during COVID-19 pandemic in Shanghai, China. J. Infect. 2023, 86, 66–117. [Google Scholar] [CrossRef]
- Li, Y.; Yu, J.; Wang, Y.; Yi, J.; Guo, L.; Wang, Q.; Zhang, G.; Xu, Y.; Zhao, Y. Cocirculation and Coinfection of Multiple Respiratory Viruses during Autumn and Winter Seasons of 2023 in Beijing, China: A Retrospective Study. J. Med. Virol. 2024, 96, e29602. [Google Scholar] [CrossRef]
- Quintero-Salgado, E.; Briseno-Ramírez, J.; Vega-Cornejo, G.; Damian-Negrete, R.; Rosales-Chavez, G.; De Arcos-Jiménez, J.C. Seasonal Shifts in Influenza, Respiratory Syncytial Virus, and Other Respiratory Viruses After the COVID-19 Pandemic: An Eight-Year Retrospective Study in Jalisco, Mexico. Viruses 2024, 16, 1892. [Google Scholar] [CrossRef]
- Contes, K.M.; Liu, B.M. Epidemiology, Clinical Significance, and Diagnosis of Respiratory Viruses and Their Co-Infections in the Post-COVID Era. Pathogens 2025, 14, 262. [Google Scholar] [CrossRef]
- Nott, R.; Fuller, T.L.; Brasil, P.; Nielsen-Saines, K. Out-of-season influenza during a COVID-19 void in the State of Rio de Janeiro, Brazil: Temperature matters. Vaccines 2022, 10, 821. [Google Scholar] [CrossRef]
- Chow, E.J.; Uyeki, T.M.; Chu, H.Y. The effects of the COVID-19 pandemic on community respiratory virus activity. Nat. Rev. Microbiol. 2023, 21, 195–210. [Google Scholar] [CrossRef]
- Treggiari, D.; Piubelli, C.; Formenti, F.; Silva, R.; Perandin, F. Resurgence of Respiratory Virus after Relaxation of COVID-19 Containment Measures: A Real-World Data Study from a Regional Hospital of Italy. Int. J. Microbiol. 2022, 2022, 4915678. [Google Scholar] [CrossRef] [PubMed]
- Treggiari, D.; Pomari, C.; Zavarise, G.; Piubelli, C.; Formenti, F.; Perandin, F. Characteristics of Respiratory Syncytial Virus Infections in Children in the Post-COVID Seasons: A Northern Italy Hospital Experience. Viruses 2024, 16, 126. [Google Scholar] [CrossRef]
- Boehm, A.B.; Wolfe, M.K.; White, B.J.; Hughes, B.; Duong, D.; Bidwell, A. More than a tripledemic: Influenza A virus, respiratory syncytial virus, SARS-CoV-2, and human metapneumovirus in wastewater during winter 2022–2023. Environ. Sci. Technol. Lett. 2023, 10, 622–627. [Google Scholar] [CrossRef] [PubMed]
- Furlow, B. Triple-demic overwhelms paediatric units in US hospitals. Lancet Child Adolesc. Health 2023, 7, 86. [Google Scholar] [CrossRef] [PubMed]
- Kandeel, A.; Fahim, M.; Deghedy, O.; Roshdy, W.H.; Khalifa, M.K.; Shesheny, R.E.; Kandeil, A.; Naguib, A.; Afifi, S.; Mohsen, A.; et al. Resurgence of influenza and respiratory syncytial virus in Egypt following two years of decline during the COVID-19 pandemic: Outpatient clinic survey of infants and children, October 2022. BMC Public Health 2023, 23, 1067. [Google Scholar] [CrossRef]
- Luo, W.; Liu, Q.; Zhou, Y.; Ran, Y.; Liu, Z.; Hou, W.; Pei, S.; Lai, S. Spatiotemporal variations of “triple-demic” outbreaks of respiratory infections in the United States in the post-COVID-19 era. BMC Public Health 2023, 23, 2452. [Google Scholar] [CrossRef]
- Quarg, C.; Jörres, R.A.; Engelhardt, S.; Alter, P.; Budweiser, S. Characteristics and outcomes of patients hospitalized for infection with influenza, SARS-CoV-2 or respiratory syncytial virus in the season 2022/2023 in a large German primary care centre. Eur. J. Med. Res. 2023, 28, 568. [Google Scholar] [CrossRef]
- Luštrek, M.; Cesar, Z.; Suljič, A.; Kogoj, R.; Knap, N.; Virant, M.J.; Uršič, T.; Petrovec, M.; Avšič-Županc, T.; Korva, M. Influenza A, Influenza B, human respiratory syncytial virus and SARS-CoV-2 molecular diagnostics and epidemiology in the post COVID-19 era. Respir. Res. 2024, 25, 234. [Google Scholar] [CrossRef]
- Wiechert, L.; Fischer, C.; Jörres, R.A.; Engelhardt, S.; Alter, P.; Kahnert, K.; Budweiser, S. Characteristics and outcomes of patients hospitalized for infection with Influenza A, SARS-CoV-2 or respiratory syncytial virus in the season 2023/2024 in a large German primary care centre. Eur. J. Med. Res. 2024, 29, 509. [Google Scholar] [CrossRef]
- Madad, S.; Salway, R.J.; Raggi, J.; Silvestri, D.; Cotter, T.; Romolt, C. The 2022–2023 USA Respiratory Viral ‘Tripledemic’: Healthcare Lessons Learned. Microbiol. Infect. Dis. AMJ 2023, 1, 35–39. [Google Scholar] [CrossRef]
- Solomon, D.A.; Sherman, A.C.; Kanjilal, S. Influenza in the COVID-19 Era. JAMA 2020, 324, 1342–1343. [Google Scholar] [CrossRef]
- Wu, D.; Lu, J.; Ma, X.; Liu, Q.; Wang, D.; Gu, Y.; Li, Y.; He, W. Coinfection of Influenza Virus and Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2). Pediatr. Infect. Dis. J. 2020, 39, e79. [Google Scholar] [CrossRef] [PubMed]
- Feng, L.; Li, Z.; Zhao, S.; Nair, H.; Lai, S.; Xu, W.; Li, M.; Wu, J.; Ren, L.; Liu, W.; et al. Viral etiologies of hospitalized acute lower respiratory infection patients in China, 2009–2013. PLoS ONE 2014, 9, e99419. [Google Scholar] [CrossRef]
- Malosh, R.E.; Martin, E.T.; Callear, A.P.; Petrie, J.G.; Lauring, A.S.; Lamerato, L.; Fry, A.M.; Ferdinands, J.; Flannery, B.; Monto, A.S. Respiratory syncytial virus hospitalization in middle-aged and older adults. J. Clin. Virol. 2017, 96, 37–43. [Google Scholar] [CrossRef]
- Rao, S.; Nyquist, A.C. Respiratory viruses and their impact in healthcare. Curr. Opin. Infect. Dis. 2014, 27, 342–347. [Google Scholar] [CrossRef] [PubMed]
- Musuuza, J.S.; Watson, L.; Parmasad, V.; Putman-Buehler, N.; Christensen, L.; Safdar, N. Prevalence and outcomes of co-infection and superinfection with SARS-CoV-2 and other pathogens: A systematic review and meta-analysis. PLoS ONE 2021, 16, e0251170. [Google Scholar] [CrossRef] [PubMed]
- Cong, B.; Deng, S.; Wang, X.; Li, Y. The role of respiratory co-infection with influenza or respiratory syncytial virus in the clinical severity of COVID-19 patients: A systematic review and meta-analysis. J. Glob. Health 2022, 12, 05040. [Google Scholar] [CrossRef]
- Zhang, N.; Wang, L.; Deng, X.; Liang, R.; Su, M.; He, C.; Hu, L.; Su, Y.; Ren, J.; Yu, F.; et al. Recent advances in the detection of respiratory virus infection in humans. J. Med. Virol. 2020, 92, 408–417. [Google Scholar] [CrossRef]
- Ursic, T.; Kogoj, R.; Erzar, K.; Virant, M.J.; Petrovec, M. Respiratory viruses in hospitalized patients before and after SARS-CoV-2. J. Biomed. Res. Environ. Sci. 2022, 23, 703–711. [Google Scholar] [CrossRef]
- Onwuchekwa, C.; Moreo, L.M.; Menon, S.; Machado, B.; Curcio, D.; Kalina, W.; Atwell, J.E.; Gessner, B.D.; Siapka, M.; Agarwal, N.; et al. Underascertainment of Respiratory Syncytial Virus Infection in Adults Due to Diagnostic Testing Limitations: A Systematic Literature Review and Meta-analysis. J. Infect. Dis. 2023, 228, 173–184. [Google Scholar] [CrossRef] [PubMed]
- Basile, K.; Kok, J.; Dwyer, D.E. Point-of-care diagnostics for respiratory viral infections. Expert Rev. Mol. Diagn. 2018, 18, 75–83. [Google Scholar] [CrossRef] [PubMed]
- Elnifro, E.M.; Ashshi, A.M.; Cooper, R.J.; Klapper, P.E. Multiplex PCR: Optimization and application in diagnostic virology. Clin. Microbiol. Rev. 2000, 13, 559–570. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Li, D.; Wang, J.; Zhang, R.; Li, J. Design, optimization, and application of multiplex rRT-PCR in the detection of respiratory viruses. Crit. Rev. Clin. Lab. Sci. 2022, 59, 555–572. [Google Scholar] [CrossRef]
- Clark, T.W.; Lindsley, K.; Wigmosta, T.B.; Bhagat, A.; Hemmert, R.B.; Uyei, J.; Timbrook, T.T. Rapid multiplex PCR for respiratory viruses reduces time to result and improves clinical care: Results of a systematic review and meta-analysis. J. Infect. 2023, 86, 462–475. [Google Scholar] [CrossRef]
- Chan, M.; Koo, S.H.; Jiang, B.; Lim, P.Q.; Tan, T.Y. Comparison of the Biofire FilmArray Respiratory Panel, Seegene AnyplexII RV16, and Argene for the detection of respiratory viruses. J. Clin. Virol. 2018, 106, 13–17. [Google Scholar] [CrossRef]
- Leber, A.L.; Everhart, K.; Daly, J.A.; Hopper, A.; Harrington, A.; Schreckenberger, P.; McKinley, K.; Jones, M.; Holmberg, K.; Kensinger, B. Multicenter Evaluation of BioFire FilmArray Respiratory Panel 2 for Detection of Viruses and Bacteria in Nasopharyngeal Swab Samples. J. Clin. Microbiol. 2018, 56, e01945-17. [Google Scholar] [CrossRef]
- Creager, H.M.; Cabrera, B.; Schnaubelt, A.; Cox, J.L.; Cushman-Vokoun, A.M.; Shakir, S.M.; Tardif, K.D.; Huang, M.L.; Jerome, K.R.; Greninger, A.L.; et al. Clinical evaluation of the BioFire® Respiratory Panel 2.1 and detection of SARS-CoV-2. J. Clin. Virol. 2020, 129, 104538. [Google Scholar] [CrossRef]
- Chung, H.Y.; Jian, M.J.; Chang, C.K.; Lin, J.C.; Yeh, K.M.; Chen, C.W.; Chiu, S.K.; Wang, Y.H.; Liao, S.J.; Li, S.Y.; et al. Novel dual multiplex real-time RT-PCR assays for the rapid detection of SARS-CoV-2, influenza A/B, and respiratory syncytial virus using the BD MAX open system. Emerg. Microbes. Infect. 2021, 10, 161–166. [Google Scholar] [CrossRef]
- Lade, H.; Kim, J.M.; Chung, Y.; Han, M.; Mo, E.K.; Kim, J.S. Comparative Evaluation of Allplex Respiratory Panels 1, 2, 3, and BioFire FilmArray Respiratory Panel for the Detection of Respiratory Infections. Diagnostics 2021, 12, 9. [Google Scholar] [CrossRef]
- Leung, E.C.; Chow, V.C.; Lee, M.K.; Tang, K.P.; Li, D.K.; Lai, R.W. Evaluation of the Xpert Xpress SARS-CoV-2/Flu/RSV Assay for Simultaneous Detection of SARS-CoV-2, Influenza A and B Viruses, and Respiratory Syncytial Virus in Nasopharyngeal Specimens. J. Clin. Microbiol. 2021, 59, e02965-20. [Google Scholar] [CrossRef]
- Mostafa, H.H.; Carroll, K.C.; Hicken, R.; Berry, G.J.; Manji, R.; Smith, E.; Rakeman, J.L.; Fowler, R.C.; Leelawong, M.; Butler-Wu, S.M.; et al. Multicenter Evaluation of the Cepheid Xpert Xpress SARS-CoV-2/Flu/RSV Test. J. Clin. Microbiol. 2021, 59, e02955-20. [Google Scholar] [CrossRef]
- Nörz, D.; Hoffmann, A.; Aepfelbacher, M.; Pfefferle, S.; Lütgehetmann, M. Clinical evaluation of a fully automated, laboratory-developed multiplex RT-PCR assay integrating dual-target SARS-CoV-2 and influenza A/B detection on a high-throughput platform. J. Med. Microbiol. 2021, 70, 001295. [Google Scholar] [CrossRef]
- Wang, C.; Liu, M.; Wang, Z.; Li, S.; Deng, Y.; He, N. Point-of-care diagnostics for infectious diseases: From methods to devices. Nano Today 2021, 37, 101092. [Google Scholar] [CrossRef] [PubMed]
- Calderaro, A.; Buttrini, M.; Farina, B.; Montecchini, S.; De Conto, F.; Chezzi, C. Respiratory tract infections and laboratory diagnostic methods: A review with A focus on syndromic Panel-Based assays. Microorganisms 2022, 10, 1856. [Google Scholar] [CrossRef] [PubMed]
- Garzarelli, V.; Chiriacò, M.S.; Cereda, M.; Autuori, I.; Ferrara, F. Miniaturized Real-Time PCR systems for SARS-CoV-2 detection at the Point-of-Care. Clin. Chim. Acta 2022, 536, 104–111. [Google Scholar] [CrossRef] [PubMed]
- Gradisteanu Pircalabioru, G.; Iliescu, F.S.; Mihaescu, G.; Cucu, A.I.; Ionescu, O.N.; Popescu, M.; Simion, M.; Burlibasa, L.; Tica, M.; Chifiriuc, M.C.; et al. Advances in the Rapid Diagnostic of Viral Respiratory Tract Infections. Front. Cell. Infect. Microbiol. 2022, 12, 807253. [Google Scholar] [CrossRef]
- Meletis, G.; Tychala, A.; Gkeka, I.; Gkotzia, A.; Triantafyllou, A.; Pappa, S.; Exindari, M.; Gioula, G.; Papa, A.; Skoura, L. Clinical Performance Evaluation of the NeuMoDx Flu A-B/RSV/SARS-CoV-2 Vantage Assay. Diagnostics 2022, 12, 3201. [Google Scholar] [CrossRef]
- Noble, L.D.; Scott, L.E.; Munir, R.; Du Plessis, M.; Steegen, K.; Hans, L.; Marokane, P.; Da Silva, P.; Stevens, W.S. Rapid Evaluation of the Xpert® Xpress CoV-2 plus and Xpert® Xpress CoV-2/Flu/RSV plus Tests. Diagnostics 2022, 13, 34. [Google Scholar] [CrossRef]
- Quinton, M.; Geahr, M.; Gluck, L.; Jarrett, J.; Mostafa, H.H. Evaluation of the respiratory NeuMoDx™ Flu A-B/RSV/SARS-CoV-2 Vantage and Alinity m Resp-4-Plex assays. J. Clin. Virol. 2022, 150-151, 105164. [Google Scholar] [CrossRef]
- Mboumba Bouassa, R.S.; Tonen-Wolyec, S.; Veyer, D.; Péré, H.; Bélec, L. Analytical performances of the AMPLIQUICK® Respiratory Triplex assay for simultaneous detection and differentiation of SARS-CoV-2, influenza A/B and respiratory syncytial viruses in respiratory specimens. PLoS ONE 2022, 17, e0262258. [Google Scholar] [CrossRef]
- Yang, S.M.; Lv, S.; Zhang, W.; Cui, Y. Microfluidic Point-of-Care (POC) Devices in Early Diagnosis: A Review of Opportunities and Challenges. Sensors 2022, 22, 1620. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Zhou, X.; Wang, Q.; Liu, W.; Chen, C. Microfluidics for COVID-19, From Current Work to Future Perspective. Biosensors 2023, 13, 163. [Google Scholar] [CrossRef] [PubMed]
- Lübke, N.; Repges, K.; Menne, C.; Walker, A.; Jensen, B.O.; Freise, N.F.; Gliga, S.; Eickhoff, S.B.; Bosse, H.M.; Adams, O.; et al. Quantitative analysis of different respiratory specimens on two automated test systems for detection of SARS-CoV-2 RNA. Diagn. Microbiol. Infect. Dis. 2023, 105, 115800. [Google Scholar] [CrossRef]
- Maltezou, H.C.; Papanikolopoulou, A.; Vassiliu, S.; Theodoridou, K.; Nikolopoulou, G.; Sipsas, N.V. COVID-19 and Respiratory Virus Co-Infections: A Systematic Review of the Literature. Viruses 2023, 15, 865. [Google Scholar] [CrossRef]
- Domnich, A.; Massaro, E.; Icardi, G.; Orsi, A. Multiplex molecular assays for the laboratory-based and point-of-care diagnosis of infections caused by seasonal influenza, COVID-19, and RSV. Expert Rev. Mol. Diagn. 2024, 24, 997–1008. [Google Scholar] [CrossRef]
- Van Der Pol, B. Opportunities and challenges of point of care testing paradigms in the post-COVID era. Expert Rev. Mol. Diagn. 2024, 24, 135–137. [Google Scholar] [CrossRef]
- Caldwell, J.M.; Espinosa, C.M.; Banerjee, R.; Domachowske, J.B. Rapid diagnosis of acute pediatric respiratory infections with Point-of-Care and multiplex molecular testing. Infection 2025, 53 (Suppl. 1), 1–14. [Google Scholar] [CrossRef] [PubMed]
- Komu, J.G.; Jamsransuren, D.; Matsuda, S.; Ogawa, H.; Yohei Takeda, Y. Performance Evaluation of a Fully Automated Molecular Diagnostic System for Multiplex Detection of SARS-CoV-2, Influenza A/B Viruses, and Respiratory Syncytial Virus. Diagnostics 2025, 15, 1791. [Google Scholar] [CrossRef]
- Lakshmanan, K.; Liu, B.M. Impact of Point-of-Care Testing on Diagnosis, Treatment, and Surveillance of Vaccine-Preventable Viral Infections. Diagnostics 2025, 15, 123. [Google Scholar] [CrossRef]
- Fitoussi, F.; Dupont, R.; Tonen-Wolyec, S.; Bélec, L. Performances of the VitaPCR™ SARS-CoV-2 Assay during the second wave of the COVID-19 epidemic in France. J. Med. Virol. 2021, 93, 4351–4357. [Google Scholar] [CrossRef]
- Mboumba Bouassa, R.S.; Tonen-Wolyec, S.; Rodary, J.; Bélec, L. Comparative practicability and analytical performances of Credo VitaPCR Flu A&B and Cepheid Xpert Xpress Flu/RSV platforms. Diagn. Microbiol. Infect. Dis. 2021, 100, 115381. [Google Scholar]
- Oliver, P.; Fernandez-Calle, P.; Buno, A. POCT Accreditation ISO 15189 and ISO 22870, Making the Point. Electron. J. Int. Fed. Clin. Chem. Lab. Med. 2021, 32, 131–139. [Google Scholar]
- ISO 15189; International Organization for Standardization. 2022 Medical Laboratories—Requirements for Quality and Competence. ISO: Geneva, Switzerland, 2022. Available online: https://www.iacld.com/UpFiles/Documents/2e096ce5-485b-4f22-b7be-e557fb7d06f8.pdf (accessed on 11 July 2025).
- Likert, R.A. Technique for the Measurement of Attitudes. Arch. Psychol. 1932, 140, 5–55. [Google Scholar]
- Bonett, D.G.; Price, R.M. Confidence intervals for a ratio of binomial proportions based on paired data. Stat. Med. 2006, 25, 3039–3047. [Google Scholar] [CrossRef] [PubMed]
- Cohen, J. A coefficient of agreement for nominal scales. Educ. Psychol. Meas. 1960, 20, 37–46. [Google Scholar] [CrossRef]
- Landis, J.R.; Koch, G.G. The measurement of observer agreement for categorical data. Biometrics 1977, 33, 159–174. [Google Scholar] [CrossRef] [PubMed]
- Youden, W.J. Index for rating diagnostic tests. Cancer 1950, 3, 32–35. [Google Scholar] [CrossRef] [PubMed]
- U.S. Food & Drug Administration. March 2007. Statistical Guidance on Reporting Results from Studies Evaluating Diagnostic Tests-Guidance for Industry and FDA Staff. Available online: https://www.fda.gov/regulatory-information/search-fda-guidance-documents/statistical-guidance-reporting-results-studies-evaluating-diagnostic-tests-guidance-industry-and-fda (accessed on 14 July 2025).
- Bland, J.M.; Altman, D.G. Statistical methods for assessing agreement between two methods of clinical measurement. Lancet 1986, 1, 307–310. [Google Scholar] [CrossRef]
- Bland, J.M.; Altman, D.G. Measuring agreement in method comparison studies. Stat. Methods Med. Res. 1999, 8, 135–160. [Google Scholar] [CrossRef]
- Passing, H.; Bablok, W. A new biometrical procedure for testing the equality of measurements from two different analytical methods. Application of linear regression procedures for method comparison studies in clinical chemistry, Part I. J. Clin. Chem. Clin. Biochem. 1983, 21, 709–720. [Google Scholar] [CrossRef] [PubMed]
- Yu, F.; Yan, L.; Wang, N.; Yang, S.; Wang, L.; Tang, Y.; Gao, G.; Wang, S.; Ma, C.; Xie, R.; et al. Quantitative Detection and Viral Load Analysis of SARS-CoV-2 in Infected Patients. Clin. Infect. Dis. 2020, 71, 793–798. [Google Scholar] [CrossRef]
- Jefferson, T.; Spencer, E.A.; Brassey, J.; Heneghan, C. Viral Cultures for Coronavirus Disease 2019 Infectivity Assessment: A Systematic Review. Clin. Infect. Dis. 2021, 73, e3884–e3899. [Google Scholar] [CrossRef] [PubMed]
- Ebentier, D.L.; Hanley, K.T.; Cao, Y.; Badgley, B.D.; Boehm, A.B.; Ervin, J.S.; Goodwin, K.D.; Gourmelon, M.; Griffith, J.F.; Holden, P.A.; et al. Evaluation of the repeatability and reproducibility of a suite of qPCR-based microbial source tracking methods. Water Res. 2013, 47, 6839–6848. [Google Scholar]
- Schulten, L.; Boehm, A.B.; Mieszkin, S.; Rosado, V.; Van De Werfhorst, L.C. Evaluation of the repeatability and reproducibility of a suite of qPCR-based microbial source tracking methods. Water Res. 2014, 67, 270–281. [Google Scholar]
- Mabey, D.; Peeling, R.W.; Ustianowski, A.; Perkins, M.D. Diagnostics for the developing world. Nat. Rev. Microbiol. 2004, 2, 231–240. [Google Scholar] [CrossRef]
- Naseri, M.; Ziora, Z.M.; Simon, G.P.; Batchelor, W. ASSURED-compliant point-of-care diagnostics for the detection of human viral infections. Rev. Med. Virol. 2022, 32, e2263. [Google Scholar]
- Douze, L.; Schiro, J.; Heyndels, L.; Pazart, L.; Pelayo, S. Evaluations of medical device usability during clinical investigations: A scoping review of clinical study protocols. Expert Rev. Med. Devices 2024, 21, 781–788. [Google Scholar] [CrossRef]
- Venner, A.A.; Beach, L.A.; Shea, J.L.; Knauer, M.J.; Huang, Y.; Fung, A.W.S.; Dalton, J.; Provencal, M.; Shaw, J.L.V. Quality assurance practices for point of care testing programs: Recommendations by the Canadian society of clinical chemists point of care testing interest group. Clin. Biochem. 2021, 88, 11–17. [Google Scholar] [CrossRef] [PubMed]
- Chokkalla, A.K.; Recio, B.D.; Devaraj, S. Best Practices for Effective Management of Point of Care Testing. Electron. J. Int. Fed. Clin. Chem. Lab. Med. 2023, 34, 245–249. [Google Scholar]
- Brendish, N.J.; Malachira, A.K.; Armstrong, L.; Houghton, R.; Aitken, S.; Nyimbili, E.; Ewings, S.; Lillie, P.J.; Clark, T.W. Routine molecular point-of-care testing for respiratory viruses in adults presenting to hospital with acute respiratory illness (ResPOC): A pragmatic, open-label, randomised controlled trial. Lancet Respir. Med. 2017, 5, 401–411. [Google Scholar] [CrossRef]
- Brendish, N.J.; Davis, C.; Chapman, M.E.; Borca, F.; Waddington, D.; Hill, C.; White, N.; Clark, T.W. Emergency Department point-of-care antiviral host response testing is accurate during periods of multiple respiratory virus co-circulation. J. Infect. 2024, 88, 41–47. [Google Scholar] [CrossRef]
- Tan, C.; Chan, C.K.; Ofner, M.; O’Brien, J.; Thomas, N.R.; Callahan, J.; Pascual, B.; Palmer, S.J.; Serapion, V.; Fabro, H.; et al. Implementation of point-of-care molecular testing for respiratory viruses in congregate living settings. Infect. Control Hosp. Epidemiol. 2024, 45, 1085–1089. [Google Scholar] [CrossRef]
- Nichols, J.H. Laboratory quality control based on risk management. Ann. Saudi. Med. 2011, 31, 223–228. [Google Scholar] [CrossRef][Green Version]
- Martínez-Murcia, A.; Navarro, A.; Garcia-Sirera, A.; Pérez, L.; Bru, G. Internal Validation of a Real-Time qPCR Kit following the UNE/EN ISO/IEC 17025,2005 for Detection of the Re-Emerging Monkeypox virus. Diagnostics 2023, 13, 1560. [Google Scholar] [CrossRef] [PubMed]
- Ali, A.E.; Hassan, T.; Elamim, M.; Hamza, A.M. Investigating the Performance of Unacceptable Results in Proficiency Testing Among Medical Laboratories. J. BioMed. Res. Rep. 2024, 5, 1–18. [Google Scholar][Green Version]
- Kazmierczak, S.C.; Morosyuk, S.; Rajkumar, R. Evaluation of Preanalytical Point-of-Care Testing Errors and Their Impact on Productivity in the Emergency Department in the United States. J. Appl. Lab. Med. 2022, 7, 650–660. [Google Scholar] [CrossRef]
- Hansen, G.; Marino, J.; Wang, Z.X.; Beavis, K.G.; Rodrigo, J.; Labog, K.; Westblade, L.F.; Jin, R.; Love, N.; Ding, K.; et al. Clinical Performance of the Point-of-Care cobas Liat for Detection of SARS-CoV-2 in 20 Minutes: A Multicenter Study. J. Clin. Microbiol. 2021, 59, e02811–e02820. [Google Scholar] [CrossRef] [PubMed]
- Vemula, S.V.; Zhao, J.; Liu, J.; Wang, X.; Biswas, S.; Hewlett, I. Current approaches for diagnosis of influenza virus infections in humans. Viruses 2016, 8, 96. [Google Scholar] [CrossRef]
- Corman, V.M.; Eckerle, I.; Bleicker, T.; Zaki, A.; Landt, O.; Eschbach-Bludau, M.; van Boheemen, S.; Gopal, R.; Ballhause, M.; Bestebroer, T.M.; et al. Detection of a novel human coronavirus by real-time reverse-transcription polymerase chain reaction. Euro. Surveill. 2012, 17, 20285. [Google Scholar] [CrossRef]
- Yu, K.; Xing, J.; Zhang, J.; Zhao, R.; Zhang, Y.; Zhao, L. Effect of multiple cycles of freeze thawing on the RNA quality of lung cancer tissues. Cell Tissue Bank. 2017, 18, 433–440. [Google Scholar] [CrossRef] [PubMed]
- Brukner, I.; Eintracht, S.; Papadakis, A.I.; Faucher, D.; Lamontagne, B.; Spatz, A.; Oughton, M. Maximizing confidence in a negative result: Quantitative sample adequacy control. J. Infect. Public Health 2020, 13, 991–993. [Google Scholar] [CrossRef]
- Bouzid, D.; Vila, J.; Hansen, G.; Manissero, D.; Pareja, J.; Rao, S.N.; Visseaux, B. Systematic review on the association between respiratory virus real-time PCR cycle threshold values and clinical presentation or outcomes. J. Antimicrob. Chemother. 2021, 76 (Suppl. 3), iii33–iii49. [Google Scholar] [CrossRef] [PubMed]
- Bentahar, I.; Loubet, P.; Salipante, F.; Choquet, C.; Descamps, D.; Visseaux, B.; Smadja, N.P.; Le Hingrat, Q.; Bouzid, D. Respiratory viruses Ct values and association with clinical outcomes among adults visiting the ED with lower respiratory tract infections. PLoS ONE 2025, 20, e0320503. [Google Scholar] [CrossRef]
- Santé Publique France. Infections Respiratoires Aiguës (Grippe, Bronchiolite, COVID-19). Bilan de la Saison 2024–2025. Publié le 16 Avril 2025. Mis à Jour le 16 Avril 2025. Available online: https://www.santepubliquefrance.fr/maladies-et-traumatismes/maladies-et-infections-respiratoires/grippe/documents/bulletin-national/infections-respiratoires-aigues-grippe-bronchiolite-covid-19-.-bilan-de-la-saison-2024-2025 (accessed on 17 July 2025).
- Ali, S.T.; Lau, Y.C.; Shan, S.; Ryu, S.; Du, Z.; Wang, L.; Xu, X.K.; Chen, D.; Xiong, J.; Tae, J.; et al. Prediction of upcoming global infection burden of influenza seasons after relaxation of public health and social measures during the COVID-19 pandemic: A modelling study. Lancet Glob. Health 2022, 10, e1612–e1622. [Google Scholar] [CrossRef]
- Sciuto, E.L.; Leonardi, A.A.; Calabrese, G.; Luca, G.; Coniglio, M.A.; Irrera, A.; Conoci, S. Nucleic Acids Analytical Methods for Viral Infection Diagnosis: State-of-the-Art and Future Perspectives. Biomolecules 2021, 11, 1585. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Bai, Y.; You, M.; Hu, J.; Yao, C.; Cao, L.; Xu, F. Fully integrated microfluidic devices for qualitative, quantitative and digital nucleic acids testing at point of care. Biosens. Bioelectron. 2021, 177, 112952. [Google Scholar] [CrossRef]
- Journal Officiel de la République Française. Ordonnance n 2010-49 du 13 Janvier 2010 Relative à la Biologie Médicale. Available online: https://www.legifrance.gouv.fr/jorf/id/JORFTEXT000021683301/ (accessed on 14 July 2025).
- Bourdin, J.; Salmona, M.; Fidouh, N.; Fouéré, S.; LeGoff, J.; Maylin, S. Evaluation of the analytical performances of the Alinity-i HSV-1 IgG and HSV-2 IgG chemiluminescent immunoassays. J. Clin. Virol. 2025, 177, 105776. [Google Scholar] [CrossRef]
- Timsit, S.; Mellon, G.; Forgeard, N.; Mercier-Delarue, S.; Mahjoub, N.; Euzen, V.; Harel, S.; Elessa, D.; Osinski, N.; Diaz, E.; et al. Clinical and Epidemiological Insights into a Parainfluenza Virus Type 3 Outbreak in Multiple Myeloma Patients. J. Med. Virol. 2025, 97, e70411. [Google Scholar] [CrossRef]
- Carré, J.; Guérineau, H.; Le Beller, C.; Mauge, L.; Huynh, B.; Nili, R.; Planquette, B.; Clauser, S.; Smadja, D.M.; Helley, D.; et al. Direct Oral Anticoagulants as Successful Treatment of Heparin-Induced Thrombocytopenia: A Parisian Retrospective Case Series. Front. Med. 2021, 8, 713649. [Google Scholar] [CrossRef] [PubMed]
- Souche, R.; Mas, S.; Scatton, O.; Fabre, J.M.; Gimeno, L.; Herrero, A.; Gaujoux, S. French legislation on retrospective clinical research: What to know and what to do. J. Visc. Surg. 2022, 159, 222–228. [Google Scholar] [CrossRef] [PubMed]
- GIRCI–La Réutilisation D’échantillons Biologiques Humains à des Fins de Recherche–Webinaire 17 Janvier 2023–T. Roche, Avocat. Available online: https://girci-idf.fr/wp-content/uploads/2023/10/20230117_Webinaire-Juridique_Reutilisation-EBH-Recherche.pdf (accessed on 3 August 2025).

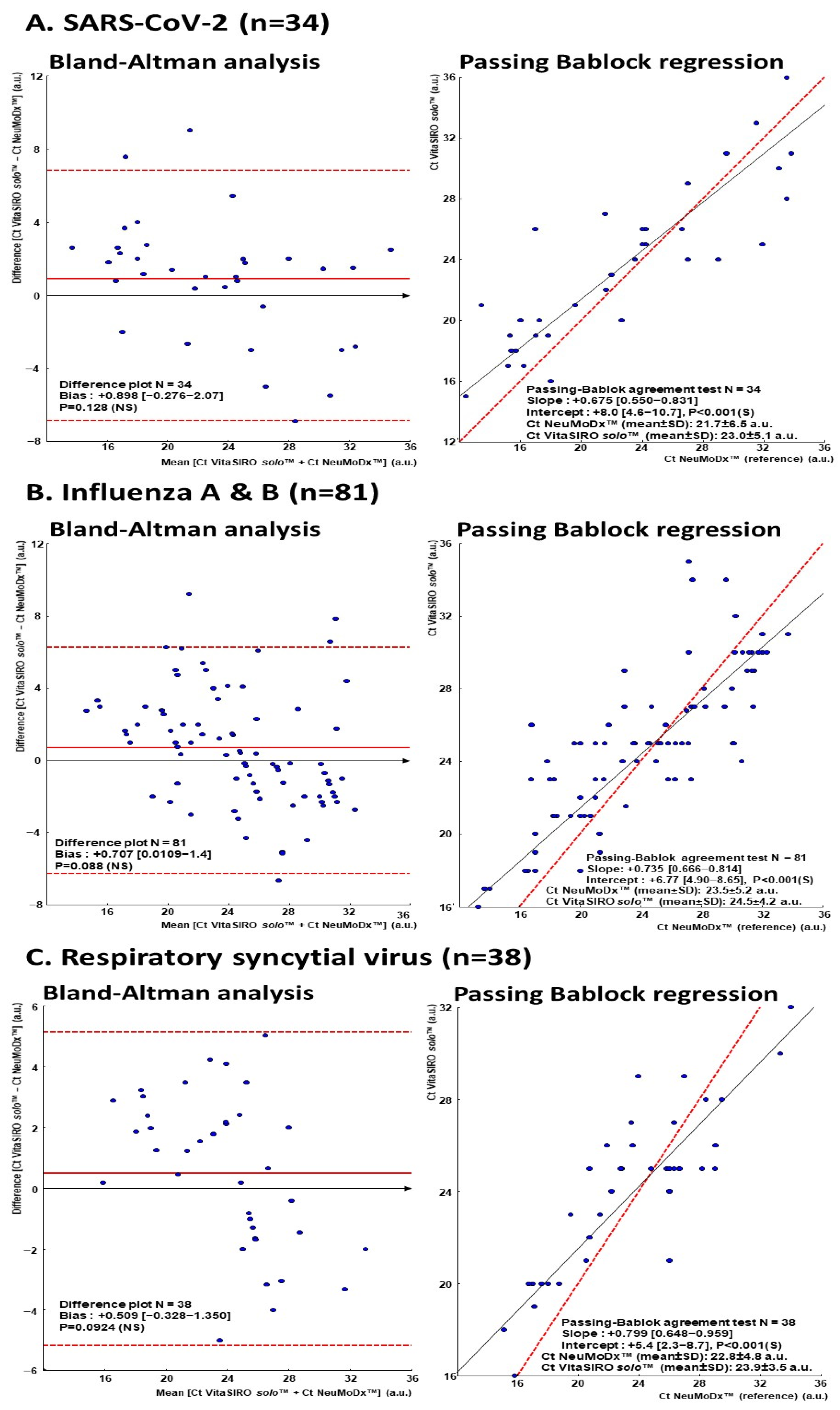
| SARS-CoV-2 (n = 34) | Influenza A and B (n = 81) | RSV (n = 38) | |
|---|---|---|---|
| VitaSIRO solo™ SARS-CoV-2/Flu/RSV assay | 23.01 ± 5.17 (15 µ–36) µµ | 24.55 ± 4.21 (16–35) | 23.90 ± 3.49 (16–32) |
| NeuMoDx™ Flu A-B/RSV/SARS-CoV-2 Vantage Assay | 21.74 ± 6.55 (12.4–33.8) | 23.59 ± 5.29 (13.2–33.7) | 22.88 ± 4.85 (15.1–34.0) |
| P µµµ | 0.128 | 0.088 | 0.092 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mboumba Bouassa, R.-S.; Lukumbisa, S.; Bélec, L. Simultaneous Detection and Differentiation of SARS-CoV-2, Influenza A/B, and Respiratory Syncytial Viruses in Respiratory Specimens Using the VitaSIRO solo™ SARS-CoV-2/Flu/RSV Assay. Diagnostics 2025, 15, 2249. https://doi.org/10.3390/diagnostics15172249
Mboumba Bouassa R-S, Lukumbisa S, Bélec L. Simultaneous Detection and Differentiation of SARS-CoV-2, Influenza A/B, and Respiratory Syncytial Viruses in Respiratory Specimens Using the VitaSIRO solo™ SARS-CoV-2/Flu/RSV Assay. Diagnostics. 2025; 15(17):2249. https://doi.org/10.3390/diagnostics15172249
Chicago/Turabian StyleMboumba Bouassa, Ralph-Sydney, Sarah Lukumbisa, and Laurent Bélec. 2025. "Simultaneous Detection and Differentiation of SARS-CoV-2, Influenza A/B, and Respiratory Syncytial Viruses in Respiratory Specimens Using the VitaSIRO solo™ SARS-CoV-2/Flu/RSV Assay" Diagnostics 15, no. 17: 2249. https://doi.org/10.3390/diagnostics15172249
APA StyleMboumba Bouassa, R.-S., Lukumbisa, S., & Bélec, L. (2025). Simultaneous Detection and Differentiation of SARS-CoV-2, Influenza A/B, and Respiratory Syncytial Viruses in Respiratory Specimens Using the VitaSIRO solo™ SARS-CoV-2/Flu/RSV Assay. Diagnostics, 15(17), 2249. https://doi.org/10.3390/diagnostics15172249






