RadiomiX for Radiomics Analysis: Automated Approaches to Overcome Challenges in Replicability
Abstract
1. Introduction
2. Materials and Methods
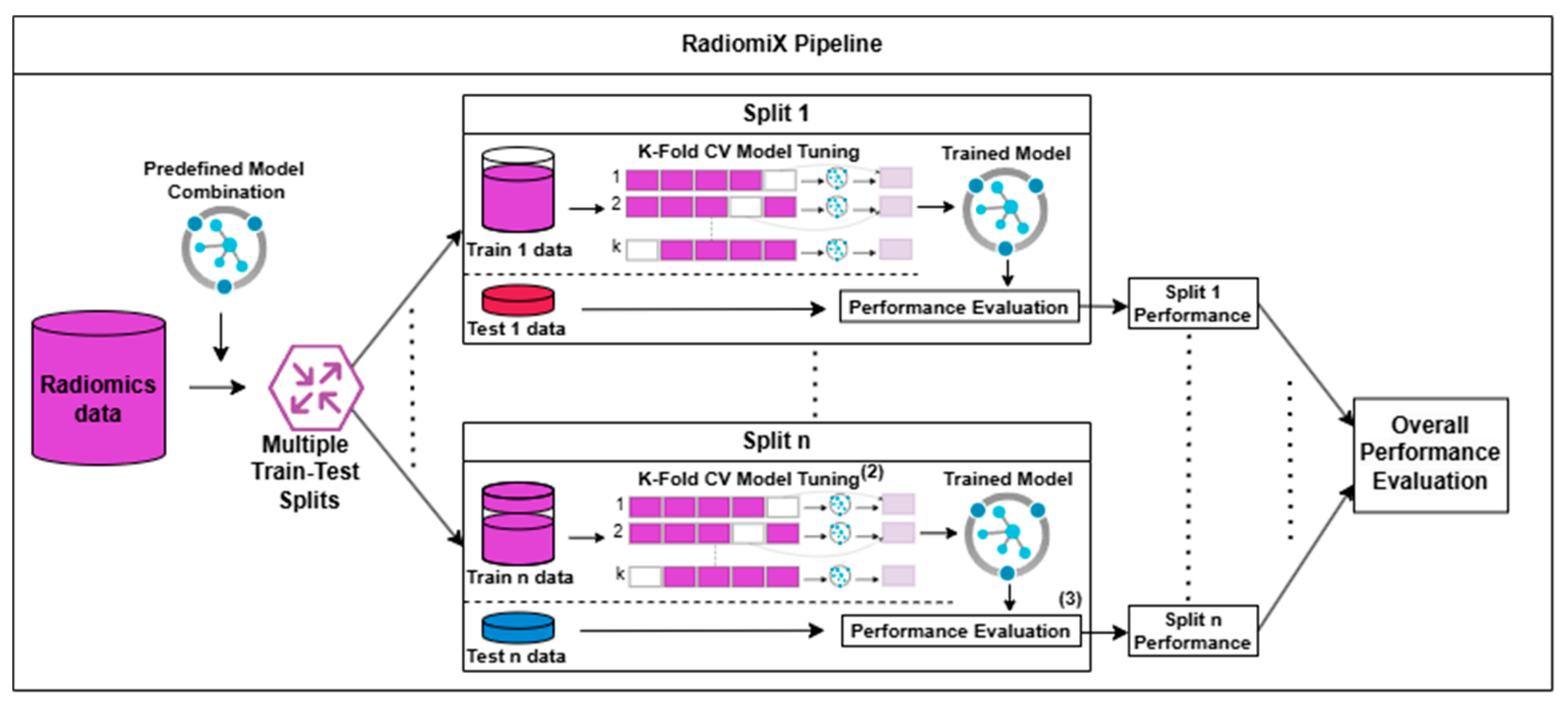
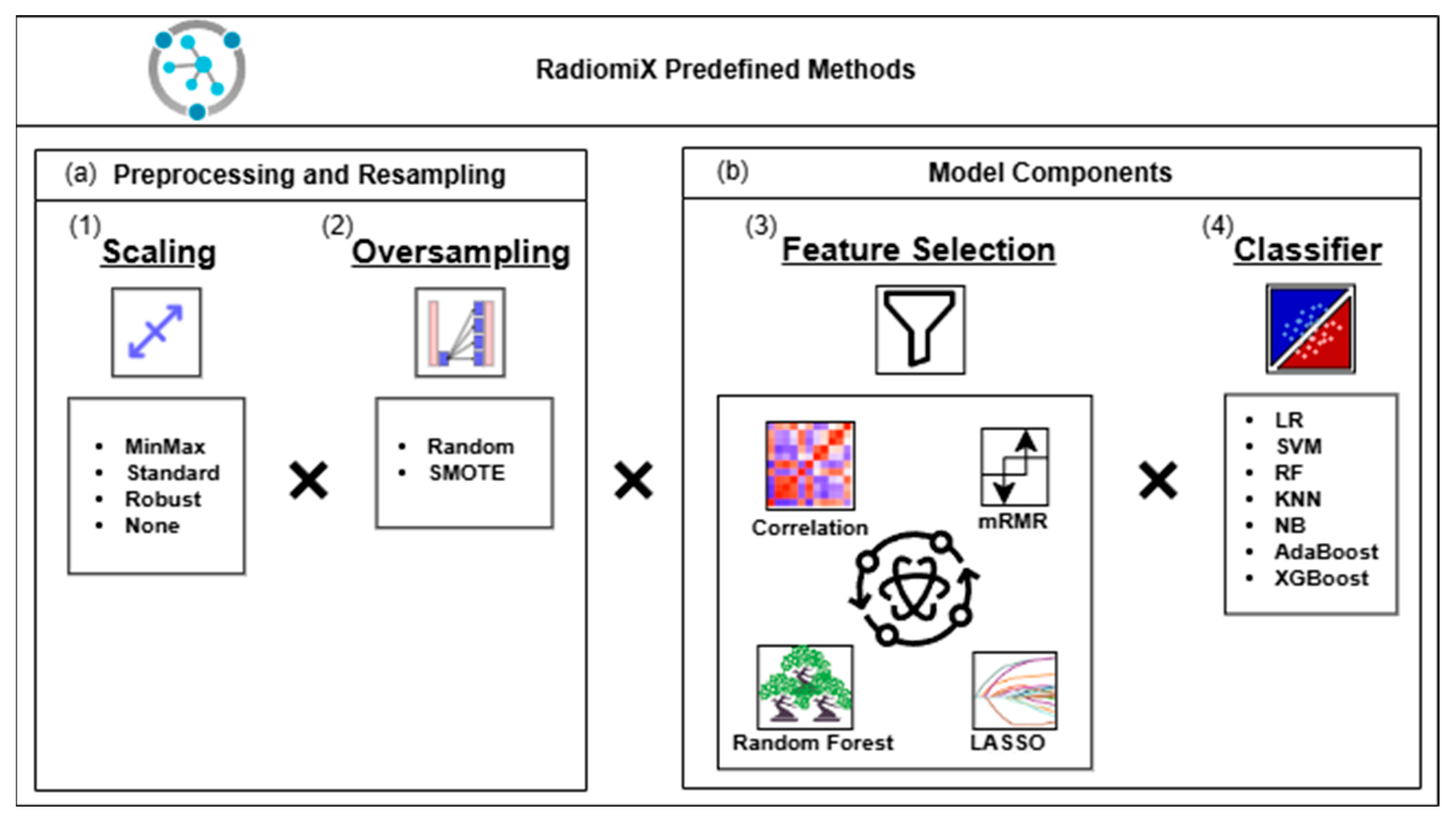
2.1. Algorithm Description
2.2. Datasets
2.3. Statistical Analysis
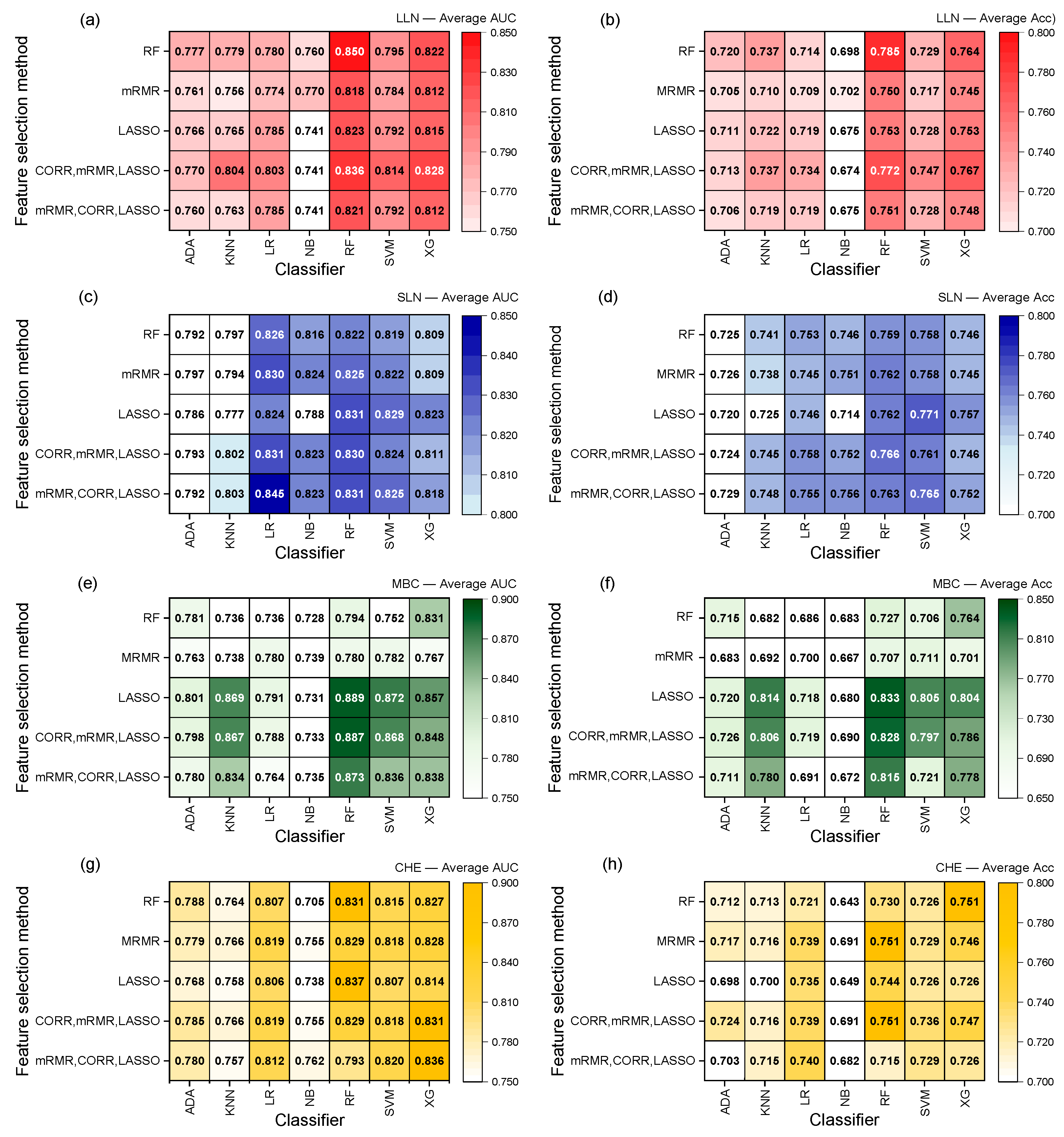
3. Results
3.1. Dataset Characteristics
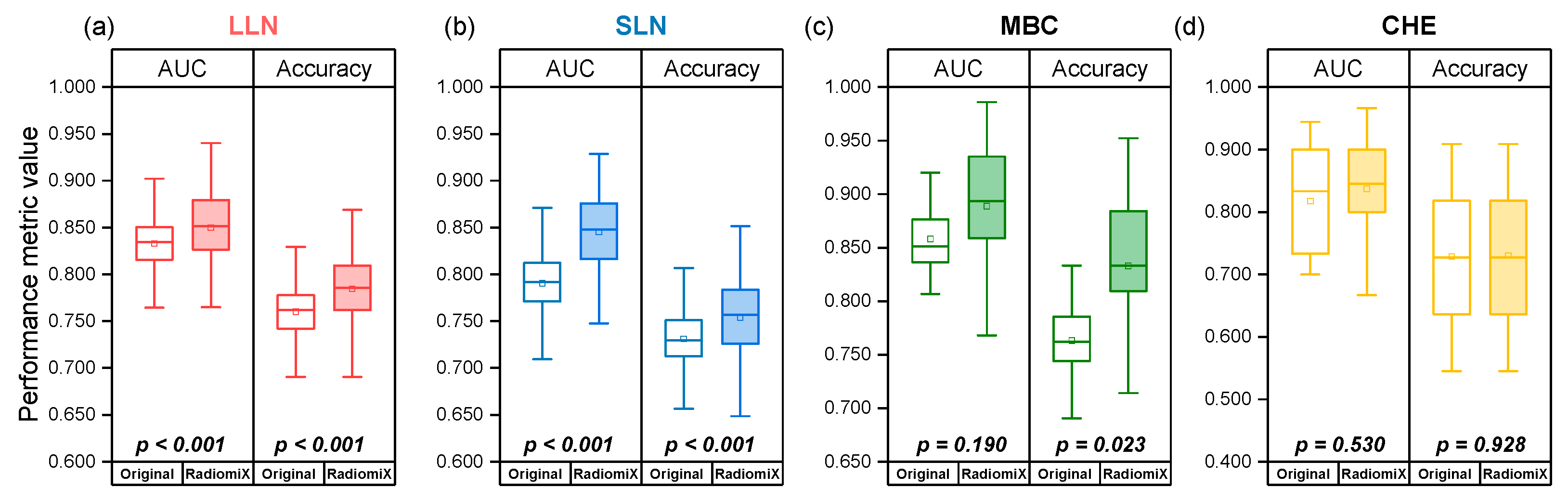
3.2. RadiomiX Performance
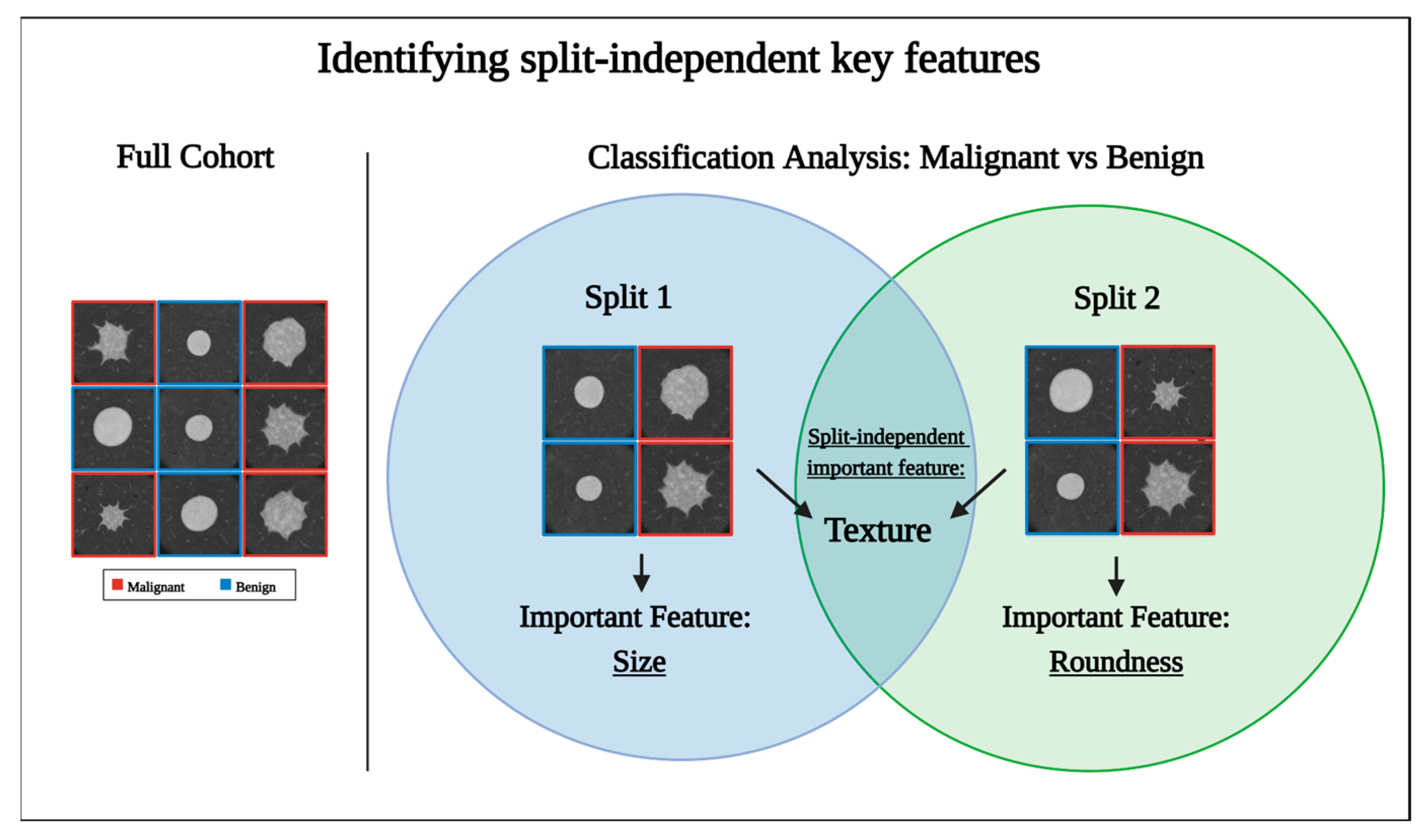
3.3. The Model’s Performance Is Dependent on the Dataset
3.4. Performance Dependence on Dataset Train–Test Splitting
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Lambin, P.; Rios-Velazquez, E.; Leijenaar, R.; Carvalho, S.; van Stiphout, R.G.P.M.; Granton, P.; Zegers, C.M.L.; Gillies, R.; Boellard, R.; Dekker, A.; et al. Radiomics: Extracting more information from medical images using advanced feature analysis. Eur. J. Cancer 2012, 48, 441–446. [Google Scholar] [CrossRef]
- Zwanenburg, A.; Vallières, M.; Abdalah, M.A.; Aerts, H.J.W.L.; Andrearczyk, V.; Apte, A.; Ashrafinia, S.; Bakas, S.; Beukinga, R.J.; Boellaard, R.; et al. The Image Biomarker Standardization Initiative: Standardized Quantitative Radiomics for High-Throughput Image-based Phenotyping. Radiology 2020, 295, 328–338. [Google Scholar] [CrossRef] [PubMed]
- Shur, J.D.; Doran, S.J.; Kumar, S.; Ap Dafydd, D.; Downey, K.; O’cOnnor, J.P.B.; Papanikolaou, N.; Messiou, C.; Koh, D.-M.; Orton, M.R. Radiomics in Oncology: A Practical Guide. RadioGraphics 2021, 41, 1717–1732. [Google Scholar] [CrossRef]
- Sugai, Y.; Kadoya, N.; Tanaka, S.; Tanabe, S.; Umeda, M.; Yamamoto, T.; Takeda, K.; Dobashi, S.; Ohashi, H.; Takeda, K.; et al. Impact of feature selection methods and subgroup factors on prognostic analysis with CT-based radiomics in non-small cell lung cancer patients. Radiat. Oncol. 2021, 16, 80. [Google Scholar] [CrossRef] [PubMed]
- Chang, C.; Sun, X.; Wang, G.; Yu, H.; Zhao, W.; Ge, Y.; Duan, S.; Qian, X.; Wang, R.; Lei, B.; et al. A Machine Learning Model Based on PET/CT Radiomics and Clinical Characteristics Predicts ALK Rearrangement Status in Lung Adenocarcinoma. Front. Oncol. 2021, 11, 603882. [Google Scholar] [CrossRef]
- Kirienko, M.; Sollini, M.; Corbetta, M.; Voulaz, E.; Gozzi, N.; Interlenghi, M.; Gallivanone, F.; Castiglioni, I.; Asselta, R.; Duga, S.; et al. Radiomics and gene expression profile to characterise the disease and predict outcome in patients with lung cancer. Eur. J. Nucl. Med. Mol. Imaging 2021, 48, 3643–3655. [Google Scholar] [CrossRef] [PubMed]
- Mu, W.; Tunali, I.; Gray, J.E.; Qi, J.; Schabath, M.B.; Gillies, R.J. Radiomics of 18F-FDG PET/CT images predicts clinical benefit of advanced NSCLC patients to checkpoint blockade immunotherapy. Eur. J. Nucl. Med. Mol. Imaging 2019, 47, 1168–1182. [Google Scholar] [CrossRef]
- Faghani, S.; Khosravi, B.; Zhang, K.; Moassefi, M.; Jagtap, J.M.; Nugen, F.; Vahdati, S.; Kuanar, S.P.; Rassoulinejad-Mousavi, S.M.; Singh, Y.; et al. Mitigating Bias in Radiology Machine Learning: 3. Performance Metrics. Radiol. Artif. Intell. 2022, 4, e220061. [Google Scholar] [CrossRef]
- Demircioğlu, A. Benchmarking Feature Selection Methods in Radiomics. Investig. Radiol. 2022, 57, 433–443. [Google Scholar] [CrossRef]
- Rouzrokh, P.; Khosravi, B.; Faghani, S.; Moassefi, M.; Garcia, D.V.V.; Singh, Y.; Zhang, K.; Conte, G.M.; Erickson, B.J. Mitigating Bias in Radiology Machine Learning: 1. Data Handling. Radiol. Artif. Intell. 2022, 4, e210290. [Google Scholar] [CrossRef]
- Sollini, M.; Antunovic, L.; Chiti, A.; Kirienko, M. Towards clinical application of image mining: A systematic review on artificial intelligence and radiomics. Eur. J. Nucl. Med. Mol. Imaging 2019, 46, 2656–2672. [Google Scholar] [CrossRef]
- Park, J.E.; Park, S.Y.; Kim, H.J.; Kim, H.S. Reproducibility and Generalizability in Radiomics Modeling: Possible Strategies in Radiologic and Statistical Perspectives. Korean J. Radiol. 2019, 20, 1124–1137. [Google Scholar] [CrossRef]
- An, C.; Park, Y.W.; Ahn, S.S.; Han, K.; Kim, H.; Lee, S.-K.; Le, K.N. Radiomics machine learning study with a small sample size: Single random training-test set split may lead to unreliable results. PLoS ONE 2021, 16, e0256152. [Google Scholar] [CrossRef] [PubMed]
- Zhang, K.; Khosravi, B.; Vahdati, S.; Faghani, S.; Nugen, F.; Rassoulinejad-Mousavi, S.M.; Moassefi, M.; Jagtap, J.M.M.; Singh, Y.; Rouzrokh, P.; et al. Mitigating Bias in Radiology Machine Learning: 2. Model Development. Radiol. Artif. Intell. 2022, 4, e220010. [Google Scholar] [CrossRef]
- Demircioğlu, A. The effect of data resampling methods in radiomics. Sci. Rep. 2024, 14, 2858. [Google Scholar] [CrossRef]
- Uddin, S.; Khan, A.; Hossain, E.; Moni, M.A. Comparing different supervised machine learning algorithms for disease prediction. BMC Med. Inform. Decis. Mak. 2019, 19, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Decoux, A.; Duron, L.; Habert, P.; Roblot, V.; Arsovic, E.; Chassagnon, G.; Arnoux, A.; Fournier, L. Comparative performances of machine learning algorithms in radiomics and impacting factors. Sci. Rep. 2023, 13, 14069. [Google Scholar] [CrossRef]
- Husso, M.; Afara, I.O.; Nissi, M.J.; Kuivanen, A.; Halonen, P.; Tarkia, M.; Teuho, J.; Saunavaara, V.; Vainio, P.; Sipola, P.; et al. Quantification of Myocardial Blood Flow by Machine Learning Analysis of Modified Dual Bolus MRI Examination. Ann. Biomed. Eng. 2020, 49, 653–662. [Google Scholar] [CrossRef]
- Molinaro, A.M.; Simon, R.; Pfeiffer, R.M. Prediction error estimation: A comparison of resampling methods. Bioinformatics 2005, 21, 3301–3307. [Google Scholar] [CrossRef] [PubMed]
- Tsamardinos, I. Don’t lose samples to estimation. Patterns 2022, 3, 100612. [Google Scholar] [CrossRef]
- Bouckaert, R.R.; Frank, E. Evaluating the Replicability of Significance Tests for Comparing Learning Algorithms. In Advances in Knowledge Discovery and Data Mining; Dai, H., Srikant, R., Zhang, C., Eds.; Springer: Berlin/Heidelberg, Germany, 2004; pp. 3–12. [Google Scholar]
- Hunter, B.; Chen, M.; Ratnakumar, P.; Alemu, E.; Logan, A.; Linton-Reid, K.; Tong, D.; Senthivel, N.; Bhamani, A.; Bloch, S.; et al. A radiomics-based decision support tool improves lung cancer diagnosis in combination with the Herder score in large lung nodules. EBioMedicine 2022, 86, 104344. [Google Scholar] [CrossRef]
- Hunter, B.; Argyros, C.; Inglese, M.; Linton-Reid, K.; Pulzato, I.; Nicholson, A.G.; Kemp, S.V.; Shah, P.L.; Molyneaux, P.L.; McNamara, C.; et al. Radiomics-based decision support tool assists radiologists in small lung nodule classification and improves lung cancer early diagnosis. Br. J. Cancer 2023, 129, 1949–1955. [Google Scholar] [CrossRef]
- Van Gómez, O.; Herraiz, J.L.; Udías, J.M.; Haug, A.; Papp, L.; Cioni, D.; Neri, E. Analysis of Cross-Combinations of Feature Selection and Machine-Learning Classification Methods Based on [18F]F-FDG PET/CT Radiomic Features for Metabolic Response Prediction of Metastatic Breast Cancer Lesions. Cancers 2022, 14, 2922. [Google Scholar] [CrossRef]
- Sparacia, G.; Parla, G.; Cannella, R.; Mamone, G.; Petridis, I.; Maruzzelli, L.; Re, V.L.; Shahriari, M.; Iaia, A.; Comelli, A.; et al. Brain magnetic resonance imaging radiomics features associated with hepatic encephalopathy in adult cirrhotic patients. Neuroradiology 2022, 64, 1969–1978. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Xu, Z.; Liu, G.; Jiang, B.; de Bock, G.H.; Groen, H.J.M.; Vliegenthart, R.; Xie, X. Simultaneous Identification of EGFR,KRAS,ERBB2, and TP53 Mutations in Patients with Non-Small Cell Lung Cancer by Machine Learning-Derived Three-Dimensional Radiomics. Cancers 2021, 13, 1814. [Google Scholar] [CrossRef] [PubMed]
- Xiong, Z.; Jiang, Y.; Che, S.; Zhao, W.; Guo, Y.; Li, G.; Liu, A.; Li, Z. Use of CT radiomics to differentiate minimally invasive adenocarcinomas and invasive adenocarcinomas presenting as pure ground-glass nodules larger than 10 mm. Eur. J. Radiol. 2021, 141, 109772. [Google Scholar] [CrossRef]
- Zhao, W.; Xiong, Z.; Jiang, Y.; Wang, K.; Zhao, M.; Lu, X.; Liu, A.; Qin, D.; Li, Z. Radiomics based on enhanced CT for differentiating between pulmonary tuberculosis and pulmonary adenocarcinoma presenting as solid nodules or masses. J. Cancer Res. Clin. Oncol. 2022, 149, 3395–3408. [Google Scholar] [CrossRef] [PubMed]
- Chen, Q.; Shao, J.; Xue, T.; Peng, H.; Li, M.; Duan, S.; Feng, F. Intratumoral and peritumoral radiomics nomograms for the preoperative prediction of lymphovascular invasion and overall survival in non-small cell lung cancer. Eur. Radiol. 2022, 33, 947–958. [Google Scholar] [CrossRef]
- Li, Y.; Mansmann, U.; Du, S.; Hornung, R. Benchmark study of feature selection strategies for multi-omics data. BMC Bioinform. 2022, 23, 412. [Google Scholar] [CrossRef]
- Hemphill, E.; Lindsay, J.; Lee, C.; Măndoiu, I.I.; Nelson, C.E. Feature selection and classifier performance on diverse bio- logical datasets. BMC Bioinform. 2014, 15, S4. [Google Scholar] [CrossRef]
- Le, T.T.; Fu, W.; Moore, J.H.; Kelso, J. Scaling tree-based automated machine learning to biomedical big data with a feature set selector. Bioinformatics 2019, 36, 250–256. [Google Scholar] [CrossRef] [PubMed]
- Imrie, F.; Cebere, B.; McKinney, E.F.; van der Schaar, M.; Guillot, G. AutoPrognosis 2.0: Democratizing Diagnostic and Prognostic Modeling in Healthcare with Automated Machine Learning. PLOS Digit. Health 2023, 2, e0000276. [Google Scholar] [CrossRef] [PubMed]
- Rácz, A.; Bajusz, D.; Héberger, K. Effect of Dataset Size and Train/Test Split Ratios in QSAR/QSPR Multiclass Classification. Molecules 2021, 26, 1111. [Google Scholar] [CrossRef] [PubMed]
- Feurer, M.; Eggensperger, K.; Falkner, S.; Lindauer, M.; Hutter, F. Auto-Sklearn 2.0: Hands-free AutoML via Meta-Learning. J. Mach. Learn. Res. 2022, 23, 1–61. [Google Scholar] [CrossRef]

| Dataset | Classification Label | N° Samples | N° Features | N° Train-Test Splits (Ratio) |
|---|---|---|---|---|
| LLN | Malignant/ benign | Total: 838 Malignant: 524 Benign: 314 | 666 * | 1 (70:30) |
| SLN | Malignant/ benign | Total: 736 Malignant: 377 Benign: 359 | 1998 | 1 (70:30) |
| MBC | Responders/non-responders | Total: 228 Responders: 127 Non-responders: 101 | 222 | 1 (80:20) |
| CHE | HE present/ HE absent | Total: 124 HE present: 38 HE absent: 86 | 43 | 1 (Ratio undisclosed) |
| Dataset | Performance Metric | Originally Published Performance (95% CI) | RadiomiX’s Best Model Performance (95% CI) | p-Value |
|---|---|---|---|---|
| LLN | AUC | 0.83 (0.77, 0.88) * | 0.850 (0.734, 0.919) | <0.001 |
| Accuracy | 0.76 (0.70, 0.81) * | 0.785 (0.694, 0.863) | <0.001 | |
| SLN | AUC | 0.78 (0.70, 0.86) * | 0.845 (0.772, 0.915) | <0.001 |
| Accuracy | 0.73 (0.65, 0.81) * | 0.754 (0.653, 0.830) | <0.001 | |
| MBC | AUC | 0.85 (0.73, 0.95) * | 0.889 (0.768, 0.979) | 0.190 |
| Accuracy | 0.763 (0.696, 0.829) | 0.833 (0.714, 0.952) | 0.023 | |
| CHE | AUC | 0.82 (0.73-0.90) * | 0.837 (0.649, 0.967) | 0.530 |
| Accuracy | 0.729 (0.566, 0.892) | 0.730 (0.584-0.909) | 0.928 |
| Dataset | Performance Metric | Originally Published Performance Based on a Single Split (95% CI) | Recalculated Performance Based on 10 Splits (95% CI) | p-Value |
|---|---|---|---|---|
| LLN | AUC | 0.83 (0.77, 0.88) * | 0.783 (0.717, 0.846) | <0.001 |
| Accuracy | 0.76 (0.70, 0.81) * | 0.731 (0.667, 0.794) | <0.001 | |
| SLN | AUC | 0.78 (0.70, 0.86) * | 0.748 (0.668, 0.821) | <0.001 |
| Accuracy | 0.73 (0.65, 0.81) * | 0.714 (0.644, 0.781) | <0.001 | |
| MBC | AUC | 0.85 (0.73, 0.95) * | 0.764 (0.626, 0.871) | 0.005 |
| Accuracy | 0.763 (0.696, 0.829) | 0.711 (0.600, 0.805) | 0.064 | |
| CHE | AUC | 0.82 (0.73, 0.90) * | 0.755 (0.533, 0.933) | 0.003 |
| Accuracy | 0.729 (0.566, 0.892) | 0.677 (0.500, 0.833) | 0.252 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kotler, H.; Bergamin, L.; Aiolli, F.; Scagliori, E.; Grassi, A.; Pasello, G.; Ferro, A.; Caumo, F.; Gennaro, G. RadiomiX for Radiomics Analysis: Automated Approaches to Overcome Challenges in Replicability. Diagnostics 2025, 15, 1968. https://doi.org/10.3390/diagnostics15151968
Kotler H, Bergamin L, Aiolli F, Scagliori E, Grassi A, Pasello G, Ferro A, Caumo F, Gennaro G. RadiomiX for Radiomics Analysis: Automated Approaches to Overcome Challenges in Replicability. Diagnostics. 2025; 15(15):1968. https://doi.org/10.3390/diagnostics15151968
Chicago/Turabian StyleKotler, Harel, Luca Bergamin, Fabio Aiolli, Elena Scagliori, Angela Grassi, Giulia Pasello, Alessandra Ferro, Francesca Caumo, and Gisella Gennaro. 2025. "RadiomiX for Radiomics Analysis: Automated Approaches to Overcome Challenges in Replicability" Diagnostics 15, no. 15: 1968. https://doi.org/10.3390/diagnostics15151968
APA StyleKotler, H., Bergamin, L., Aiolli, F., Scagliori, E., Grassi, A., Pasello, G., Ferro, A., Caumo, F., & Gennaro, G. (2025). RadiomiX for Radiomics Analysis: Automated Approaches to Overcome Challenges in Replicability. Diagnostics, 15(15), 1968. https://doi.org/10.3390/diagnostics15151968






