Performance Evaluation of a Fully Automated Molecular Diagnostic System for Multiplex Detection of SARS-CoV-2, Influenza A/B Viruses, and Respiratory Syncytial Virus
Abstract
1. Introduction
2. Materials and Methods
2.1. Viruses and Preparation of Virus Stocks
2.2. Viral Gene Detection Using geneLEAD/VIASURE
2.3. Reference Assays
2.4. Determination of the Analytical Reactivity and the Viral Titer Detection Limit of geneLEAD/VIASURE
2.5. Analysis of Single-, Double-, and Triple-Infection Models
2.6. Correlation Between geneLEAD/VIASURE and the Manual Assays
2.7. Data Analysis
3. Results
3.1. Analytical Reactivity and the Viral Titer Detection Limit of geneLEAD/VIASURE
3.2. Simultaneous Detection of Single, Double, and Triple Viral Genes
3.3. Agreement Between geneLEAD/VIASURE and the Manual Assays
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Groves, H.E.; Piché-Renaud, P.-P.; Peci, A.; Farrar, D.S.; Buckrell, S.; Bancej, C.; Sevenhuysen, C.; Campigotto, A.; Gubbay, J.B.; Morris, S.K. The Impact of the COVID-19 Pandemic on Influenza, Respiratory Syncytial Virus, and Other Seasonal Respiratory Virus Circulation in Canada: A Population-Based Study. Lancet Reg. Health Am. 2021, 1, 100015. [Google Scholar] [CrossRef] [PubMed]
- Heiskanen, A.; Galipeau, Y.; Little, J.; Mortimer, L.; Ramotar, K.; Langlois, M.; Cooper, C.L. Seasonal Respiratory Virus Circulation Was Diminished during the COVID-19 Pandemic. Influenza Other Respir. Viruses 2023, 17, e13065. [Google Scholar] [CrossRef] [PubMed]
- Cong, B.; Deng, S.; Wang, X.; Li, Y. The Role of Respiratory Co-Infection with Influenza or Respiratory Syncytial Virus in the Clinical Severity of COVID-19 Patients: A Systematic Review and Meta-Analysis. J. Glob. Health 2022, 12, 05040. [Google Scholar] [CrossRef] [PubMed]
- Musuuza, J.S.; Watson, L.; Parmasad, V.; Putman-Buehler, N.; Christensen, L.; Safdar, N. Prevalence and Outcomes of Co-Infection and Superinfection with SARS-CoV-2 and Other Pathogens: A Systematic Review and Meta-Analysis. PLoS ONE 2021, 16, e0251170. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Yu, J.; Wang, Y.; Yi, J.; Guo, L.; Wang, Q.; Zhang, G.; Xu, Y.; Zhao, Y. Cocirculation and Coinfection of Multiple Respiratory Viruses during Autumn and Winter Seasons of 2023 in Beijing, China: A Retrospective Study. J. Med. Virol. 2024, 96, e29602. [Google Scholar] [CrossRef] [PubMed]
- Luo, W.; Liu, Q.; Zhou, Y.; Ran, Y.; Liu, Z.; Hou, W.; Pei, S.; Lai, S. Spatiotemporal Variations of “Triple-Demic” Outbreaks of Respiratory Infections in the United States in the Post-COVID-19 Era. BMC Public Health 2023, 23, 2452. [Google Scholar] [CrossRef] [PubMed]
- Quarg, C.; Jörres, R.A.; Engelhardt, S.; Alter, P.; Budweiser, S. Characteristics and Outcomes of Patients Hospitalized for Infection with Influenza, SARS-CoV-2 or Respiratory Syncytial Virus in the Season 2022/2023 in a Large German Primary Care Centre. Eur. J. Med. Res. 2023, 28, 568. [Google Scholar] [CrossRef] [PubMed]
- Madad, S.; Salway, R.J.; Raggi, J.; Silvestri, D.; Cotter, T.; Romolt, C. The 2022–2023 USA Respiratory Viral ‘Tripledemic’: Healthcare Lessons Learned. Microbiol. Infect. Dis. AMJ 2023, 1, 35–39. [Google Scholar] [CrossRef]
- Lippi, G. Impact on Laboratory Medicine of Transitioning from COVID-19 Pandemic to “Tripledemic”. Biochim. Clin. 2023, 47, 68–73. [Google Scholar] [CrossRef]
- Eftekhari, A.; Alipour, M.; Chodari, L.; Maleki Dizaj, S.; Ardalan, M.; Samiei, M.; Sharifi, S.; Zununi Vahed, S.; Huseynova, I.; Khalilov, R.; et al. A Comprehensive Review of Detection Methods for SARS-CoV-2. Microorganisms 2021, 9, 232. [Google Scholar] [CrossRef] [PubMed]
- Vemula, S.V.; Zhao, J.; Liu, J.; Wang, X.; Biswas, S.; Hewlett, I. Current Approaches for Diagnosis of Influenza Virus Infections in Humans. Viruses 2016, 8, 96. [Google Scholar] [CrossRef] [PubMed]
- Onwuchekwa, C.; Moreo, L.M.; Menon, S.; Machado, B.; Curcio, D.; Kalina, W.; Atwell, J.E.; Gessner, B.D.; Siapka, M.; Agarwal, N.; et al. Underascertainment of Respiratory Syncytial Virus Infection in Adults Due to Diagnostic Testing Limitations: A Systematic Literature Review and Meta-Analysis. J. Infect. Dis. 2023, 228, 173–184. [Google Scholar] [CrossRef] [PubMed]
- Moreira, V.M.; Mascarenhas, P.; Machado, V.; Botelho, J.; Mendes, J.J.; Taveira, N.; Almeida, M.G. Diagnosis of SARS-Cov-2 Infection by RT-PCR Using Specimens Other Than Naso- and Oropharyngeal Swabs: A Systematic Review and Meta-Analysis. Diagnostics 2021, 11, 363. [Google Scholar] [CrossRef] [PubMed]
- To, K.K.W.; Yip, C.C.Y.; Lai, C.Y.W.; Wong, C.K.H.; Ho, D.T.Y.; Pang, P.K.P.; Ng, A.C.K.; Leung, K.-H.; Poon, R.W.S.; Chan, K.-H.; et al. Saliva as a Diagnostic Specimen for Testing Respiratory Virus by a Point-of-Care Molecular Assay: A Diagnostic Validity Study. Clin. Microbiol. Infect. 2019, 25, 372–378. [Google Scholar] [CrossRef] [PubMed]
- Clark, T.W.; Lindsley, K.; Wigmosta, T.B.; Bhagat, A.; Hemmert, R.B.; Uyei, J.; Timbrook, T.T. Rapid Multiplex PCR for Respiratory Viruses Reduces Time to Result and Improves Clinical Care: Results of a Systematic Review and Meta-Analysis. J. Infect. 2023, 86, 462–475. [Google Scholar] [CrossRef] [PubMed]
- Chung, H.-Y.; Jian, M.-J.; Chang, C.-K.; Lin, J.-C.; Yeh, K.-M.; Yang, Y.-S.; Chen, C.-W.; Hsieh, S.-S.; Tang, S.-H.; Perng, C.-L.; et al. Multicenter Study Evaluating One Multiplex RT-PCR Assay to Detect SARS-CoV-2, Influenza A/B, and Respiratory Syncytia Virus Using the LabTurbo AIO Open Platform: Epidemiological Features, Automated Sample-to-Result, and High-Throughput Testing. Aging 2021, 13, 24931–24942. [Google Scholar] [CrossRef] [PubMed]
- Brouwer, C.W.E.; Russcher, A.; Rezek, Y.; Nijhuis, R.H.T. Evaluation of the Fully Automated, Sample-to-Result Seegene STARlet-AIOS Platform for Detection of SARS-CoV-2, Influenza Virus A, Influenza Virus B, and RSV. Eur. J. Clin. Microbiol. Infect. Dis. 2024, 43, 87–93. [Google Scholar] [CrossRef] [PubMed]
- Quinton, M.; Geahr, M.; Gluck, L.; Jarrett, J.; Mostafa, H.H. Evaluation of the Respiratory NeuMoDxTM Flu A-B/RSV/SARS-CoV-2 Vantage and Alinity m Resp-4-Plex Assays. J. Clin. Virol. 2022, 150–151, 105164. [Google Scholar] [CrossRef] [PubMed]
- Sluimer, J.; Goderski, G.; van den Brink, S.; Broeders, M.; Rahamat-Langendoen, J.; Then, E.; Wijsman, L.; Wolters, F.; van de Bovenkamp, J.; Melchers, W.J.; et al. Multi-Center Evaluation of Cepheid Xpert® Xpress SARS-CoV-2/Flu/RSV Molecular Point-of-Care Test. J. Clin. Virol. Plus. 2021, 1, 100042. [Google Scholar] [CrossRef] [PubMed]
- Komu, J.G.; Nguyen, H.D.; Takeda, Y.; Fukumoto, S.; Imai, K.; Takemae, H.; Mizutani, T.; Ogawa, H. Challenges for Precise Subtyping and Sequencing of a H5N1 Clade 2.3.4.4b Highly Pathogenic Avian Influenza Virus Isolated in Japan in the 2022–2023 Season Using Classical Serological and Molecular Methods. Viruses 2023, 15, 2274. [Google Scholar] [CrossRef] [PubMed]
- Mohamed, I.M.A.; Ogawa, H.; Takeda, Y. In Vitro Virucidal Activity of the Theaflavin-Concentrated Tea Extract TY-1 against Influenza A Virus. J. Nat. Med. 2022, 76, 152–160. [Google Scholar] [CrossRef] [PubMed]
- Nao, N.; Sato, K.; Yamagishi, J.; Tahara, M.; Nakatsu, Y.; Seki, F.; Katoh, H.; Ohnuma, A.; Shirogane, Y.; Hayashi, M.; et al. Consensus and Variations in Cell Line Specificity among Human Metapneumovirus Strains. PLoS ONE 2019, 14, e0215822. [Google Scholar] [CrossRef] [PubMed]
- Takeda, Y.; Jamsransuren, D.; Matsuda, S.; Crea, R.; Ogawa, H. The SARS-CoV-2-Inactivating Activity of Hydroxytyrosol-Rich Aqueous Olive Pulp Extract (HIDROX®) and Its Use as a Virucidal Cream for Topical Application. Viruses 2021, 13, 232. [Google Scholar] [CrossRef] [PubMed]
- Komu, J.G.; Jamsransuren, D.; Matsuda, S.; Ogawa, H.; Takeda, Y. Efficacy Validation of SARS-CoV-2-Inactivation and Viral Genome Stability in Saliva by a Guanidine Hydrochloride and Surfactant-Based Virus Lysis/Transport Buffer. Viruses 2023, 15, 509. [Google Scholar] [CrossRef] [PubMed]
- National Institute of Infectious Diseases. Influenza Diagnostic Manual, 5th Edition (In Japanese). Available online: https://id-info.jihs.go.jp/relevant/manual/010/influenza20230829.pdf (accessed on 11 July 2025).
- National Institute of Infectious Diseases. Manual for the Detection of Pathogen 2019-NCoV Ver.2.9.1. Available online: https://id-info.jihs.go.jp/relevant/manual/010/2019-nCoV20200319.pdf (accessed on 11 July 2025).
- National Institute of Infectious Diseases. Human Orthopneumovirus (RS Virus) Pathogen Detection Manual Version 4.0. Available online: https://id-info.jihs.go.jp/relevant/manual/010/RSVirus20230807.pdf (accessed on 11 July 2025).
- Caraguel, C.G.B.; Stryhn, H.; Gagné, N.; Dohoo, I.R.; Hammell, K.L. Selection of a Cutoff Value for Real-Time Polymerase Chain Reaction Results to Fit a Diagnostic Purpose: Analytical and Epidemiologic Approaches. J. Vet. Diagn. Investig. 2011, 23, 2–15. [Google Scholar] [CrossRef] [PubMed]
- Boers, S.A.; Mourik, B.C.; van Bussel, M.J.A.W.M.; de Brouwer, C.S.; Wessels, E.; Claas, E.C.J. Increasing Diagnostic Possibilities Using the GeneLEAD VIII Platform for Detection of SARS-CoV-2. J. Virol. Methods 2021, 298, 114291. [Google Scholar] [CrossRef] [PubMed]
- Matsumura, Y.; Noguchi, T.; Shinohara, K.; Yamamoto, M.; Nagao, M. Development and Evaluation of Three Automated Media Pooling and Molecular Diagnostic Systems for the Detection of SARS-CoV-2. Microbiol. Spectr. 2024, 12, e0368423. [Google Scholar] [CrossRef] [PubMed]
- Sydenham, T.V.; Bek-Thomsen, M.; Andersen, S.D.; Kolmos, B.; Marmolin, E.S.; Trebbien, R.; Møller, J.K. Comparative Evaluation of the CerTest VIASURE Flu A, B & RSV Real Time RT-PCR Detection Kit on the BD MAX System versus a Routine in-House Assay for Detection of Influenza A and B Virus during the 2016/17 Influenza Season. J. Clin. Virol. 2018, 99–100, 35–37. [Google Scholar] [CrossRef] [PubMed]
- Bosworth, A.; Whalley, C.; Poxon, C.; Wanigasooriya, K.; Pickles, O.; Aldera, E.L.; Papakonstantinou, D.; Morley, G.L.; Walker, E.M.; Zielinska, A.E.; et al. Rapid Implementation and Validation of a Cold-Chain Free SARS-CoV-2 Diagnostic Testing Workflow to Support Surge Capacity. J. Clin. Virol. 2020, 128, 104469. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization Cumulative Number of Confirmed Human Cases for Avian Influenza A(H5N1) Reported to WHO, 2003–2024, 26 February 2024. Available online: https://www.who.int/publications/m/item/cumulative-number-of-confirmed-human-cases-for-avian-influenza-a(h5n1)-reported-to-who--2003-2024-26-february-2024 (accessed on 19 April 2024).
- Elnifro, E.M.; Ashshi, A.M.; Cooper, R.J.; Klapper, P.E. Multiplex PCR: Optimization and Application in Diagnostic Virology. Clin. Microbiol. Rev. 2000, 13, 559–570. [Google Scholar] [CrossRef] [PubMed]
- Grohmann, L.; Barbante, A.; Eriksson, R.; Gatto, F.; Georgieva, T.; Huber, I.; Hulin, J.; Köppel, R.; Marchesi, U.; Marmin, L.; et al. Guidance Document on Multiplex Real-Time PCR Methods, EUR 30708 EN, 2021. Available online: https://gmo-crl.jrc.ec.europa.eu/doc/KJNA30708ENN.en.pdf (accessed on 17 February 2024).
- An, S.-H.; Kim, N.-Y.; Heo, G.-B.; Kang, Y.-M.; Lee, Y.-J.; Lee, K.-N. Development and Evaluation of a Multiplex Real-Time RT-PCR Assay for Simultaneous Detection of H5, H7, and H9 Subtype Avian Influenza Viruses. J. Virol. Methods 2024, 327, 114942. [Google Scholar] [CrossRef] [PubMed]
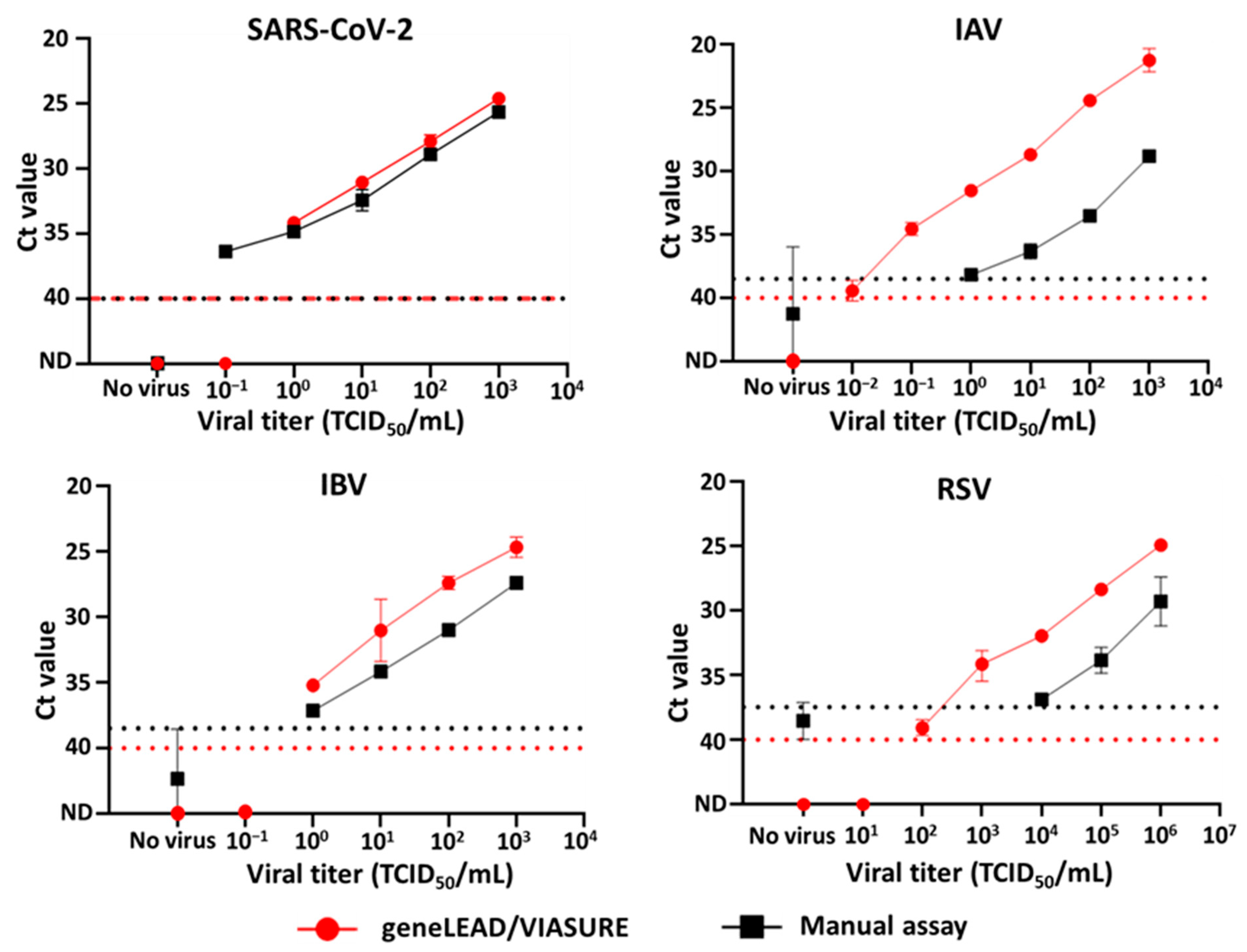
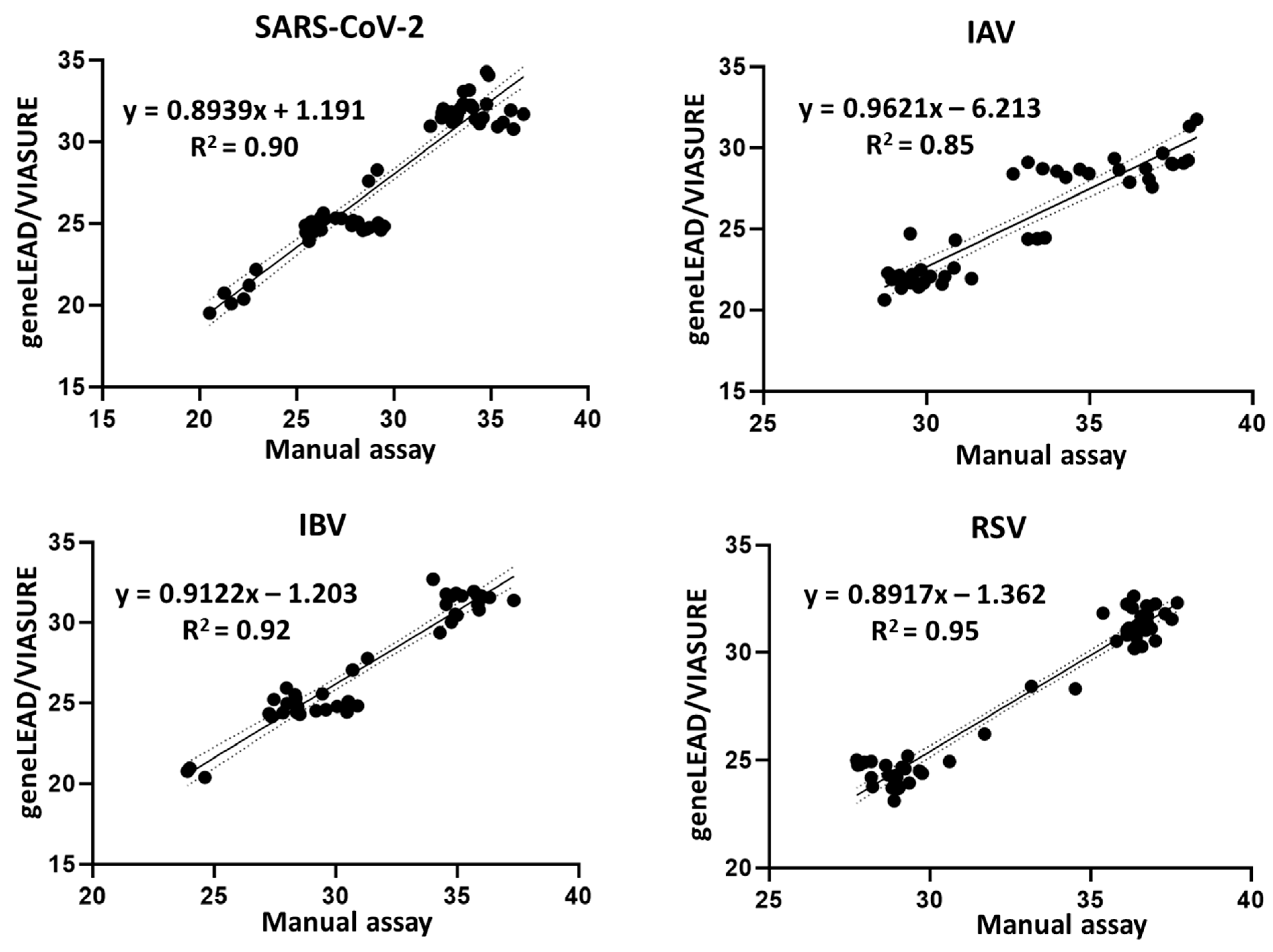
| Virus | Variant, Subtype, or Lineage | Strain Name | GISAID ID/NCBI Accession No. | Source |
|---|---|---|---|---|
| SARS-CoV-2 | Ancestral (A a) | 2019-nCoV/Japan/TY/WK-521/2020 | EPI_ISL_408667 c | NIID |
| Alpha (B.1.1.7 a) | hCoV-19/Japan/QHN001/2020 | EPI_ISL_804007 c | NIID | |
| Delta (B.1.716.2 a) | hCoV-19/Japan/TY11-927/2021 | EPI_ISL_2158617 c | NIID | |
| Gamma (P.1 a) | hCoV-19/Japan/TY7-501/2021 | EPI_ISL_833366 c | NIID | |
| Beta (B.1.351 a) | hCoV-19/Japan/TY8-612/2021 | EPI_ISL_1123289 c | NIID | |
| Omicron (BA.5 a) | hCoV-19/Japan/TY41-702/2022 | EPI_ISL_13241867 c | NIID | |
| IAV | H1N1 | A/Narita/1/2009 | EPI_ISL_30176 c | NIID |
| H3N2 | A/Hokkaido/19/1998 | Not available | HIPH | |
| H5N1 (HPAIV, 2022 b) | A/white-tailed eagle/Japan/OU-1/2022 | LC775579 d | Our lab [20] | |
| H5N1 (HPAIV, 2004 b) | A/chicken/Yamaguchi/7/04 | AB166862 d | NIAH | |
| IBV | Yamagata-lineage | B/Hokkaido/25/2018 | Not available | HIPH |
| Victoria-lineage | B/Hokkaido/1/2019 | EPI_ISL_363734 c | HIPH | |
| RSV | Type A | RSV/A/NIID/2370/14 | LC474558 e | NIID |
| Type B | RSV/B/NIID/2472/14 | LC474559 e | NIID |
| Virus (n = 2) | Manual Assays (Ct ± SD) | geneLEAD/VIASURE (Ct ± SD) | |
|---|---|---|---|
| Virus-Spiked Saliva | Virus-Spiked Saliva | Virus-Spiked Nasal Swab Solutions | |
| SARS-CoV-2 (Ancestral) | 22.92 ± 0.18 | 22.18 ± 0.34 | 23.85 ± 0.24 |
| SARS-CoV-2 (Alpha) | 21.65 ± 0.28 | 20.09 ± 0.00 | 23.32 ± 0.04 |
| SARS-CoV-2 (Delta) | 22.29 ± 0.54 | 20.38 ± 0.14 | 22.29 ± 0.01 |
| SARS-CoV-2 (Gamma) | 21.28 ± 0.24 | 20.74 ± 0.52 | 21.67 ± 0.41 |
| SARS-CoV-2 (Beta) | 22.56 ± 0.66 | 21.21 ± 0.35 | 22.15 ± 0.35 |
| SARS-CoV-2 (Omicron) | 20.53 ± 0.49 | 19.50 ± 0.21 | 21.25 ± 0.47 |
| IAV (H1N1) | 28.83 ± 0.16 | 22.27 ± 0.42 | 23.10 ± 0.64 |
| IAV (H3N2) | 30.49 ± 0.01 | 21.61 ± 1.08 | 20.97 ± 0.09 |
| AIV (H5N1, 2022 a) | 30.90 ± 0.09 | 24.30 ± 0.06 | 24.61 ± 0.25 |
| AIV (H5N1, 2004 a) | 33.12 ± 0.04 | 24.37 ± 0.01 | 25.36 ± 0.21 |
| IBV (Yamagata lineage) | 24.63 ± 0.21 | 20.40 ± 0.03 | 20.54 ± 0.17 |
| IBV (Victoria lineage) | 28.49 ± 0.24 | 27.99 ± 0.42 | 29.41 ± 0.39 |
| RSV (Type A) | 31.71 ± 0.16 | 26.21 ± 0.45 | 28.37 ± 0.43 |
| RSV (Type B) | 35.40 ± 0.21 | 31.82 ± 0.67 | 34.24 ± 0.30 |
| Viruses Spiked in Saliva Samples and Their Viral Titers (TCID50/mL) | Ct Value (±SD) Obtained Using geneLEAD/VIASURE | ||||||
|---|---|---|---|---|---|---|---|
| SARS-CoV-2 | IAV | IBV | RSV | SARS-CoV-2 a (FAM) | IAV a (ROX) | IBV a (ROX) | RSV a (Cy5) |
| 103 (high) | - | - | - | 24.90 ± 0.25 | n/d | n/d | n/d |
| - | 103 (high) | - | - | n/d b | 22.66 ± 1.38 | n/d | n/d |
| - | - | 103 (high) | - | n/d | n/d | 24.90 ± 0.74 | n/d |
| - | - | - | 106 (high) | n/d | n/d | n/d | 24.67 ± 0.26 |
| 103 (high) | 103 (high) | - | - | 25.03 ± 0.33 | 21.91 ± 0.46 | n/d | n/d |
| 103 (high) | - | 103 (high) | - | 24.84 ± 0.27 | n/d | 24.76 ± 0.52 | n/d |
| 103 (high) | - | - | 106 (high) | 25.02 ± 0.30 | n/d | n/d | 24.56 ± 0.40 |
| - | 103 (high) | - | 106 (high) | n/d | 22.17 ± 0.26 | n/d | 24.12 ± 0.15 |
| - | - | 103 (high) | 106 (high) | n/d | n/d | 24.96 ± 0.52 | 24.81 ± 0.26 |
| 103 (high) | 103 (high) | - | 106 (high) | 24.47 ± 0.40 | 21.78 ± 0.32 | n/d | 23.57 ± 0.30 |
| 103 (high) | - | 103 (high) | 106 (high) | 25.27 ± 0.26 | n/d | 25.27 ± 0.27 | 23.83 ± 0.25 |
| 101 (low) | - | - | - | 31.88 ± 1.02 | n/d | n/d | n/d |
| - | 101 (low) | - | - | n/d | 29.23 ± 0.31 | n/d | n/d |
| - | - | 101 (low) | - | n/d | n/d | 30.60 ± 0.45 | n/d |
| - | - | - | 104 (low) | n/d | n/d | n/d | 32.03 ± 0.65 |
| 101 (low) | 101 (low) | - | - | 31.68 ± 0.15 | 28.55 ± 0.40 | n/d | n/d |
| 101 (low) | - | 101 (low) | - | 31.24 ± 0.39 | n/d | 31.66 ± 0.22 | n/d |
| 101 (low) | - | - | 104 (low) | 31.95 ± 0.35 | n/d | n/d | 31.55 ± 0.53 |
| - | 101 (low) | - | 104 (low) | n/d | 28.14 ± 0.64 | n/d | 31.31 ± 0.53 |
| - | - | 101 (low) | 104 (low) | n/d | n/d | 31.30 ± 0.60 | 31.74 ± 0.27 |
| 101 (low) | 101 (low) | - | 104 (low) | 31.74 ± 0.31 | 28.78 ± 0.39 | n/d | 30.44 ± 0.22 |
| 101 (low) | - | 101 (low) | 104 (low) | 32.21 ± 0.65 | n/d | 31.55 ± 0.29 | 30.65 ± 0.32 |
| Viruses Spiked in Saliva Samples and Their Viral Titers (TCID50/mL) | Ct Value (±SD) Obtained Using geneLEAD/VIASURE | ||||||
|---|---|---|---|---|---|---|---|
| SARS-CoV-2 | IAV | IBV | RSV | SARS-CoV-2 a (FAM) | IAV a (ROX) | IBV a (ROX) | RSV a (Cy5) |
| 103 (high) | 101 (low) | - | 104 (low) | 24.89 ± 0.38 | 30.17 ± 0.46 | n/d | 30.43 ± 1.53 |
| 103 (high) | - | 101 (low) | 104 (low) | 25.17 ± 0.38 | n/d b | 32.16 ± 1.31 | 33.28 ± 0.27 |
| 103 (high) | 103 (high) | - | 104 (low) | 24.82 ± 0.39 | 23.50 ± 0.42 | n/d | 35.46 ± 1.21 |
| 103 (high) | - | 103 (high) | 104 (low) | 24.91 ± 0.28 | n/d | 23.88 ± 0.26 | 36.09 ± 2.03 |
| 101 (low) | 103 (high) | - | 104 (low) | 32.18 ± 0.85 | 22.4 ± 1.29 | n/d | 32.84 ± 0.03 |
| 101 (low) | - | 103 (high) | 104 (low) | 33.06 ± 0.38 | n/d | 23.68 ± 0.94 | 34.65 ± 0.30 |
| 101 (low) | 101 (low) | - | 106 (high) | 31.25 ± 0.15 | 27.72 ± 0.22 | n/d | 23.77 ± 0.14 |
| 101 (low) | - | 101 (low) | 106 (high) | 31.36 ± 0.21 | n/d | 32.65 ± 0.76 | 23.63 ± 0.43 |
| 101 (low) | 103 (high) | - | 106 (high) | 31.44 ± 0.39 | 24.02 ± 0.42 | n/d | 23.88 ± 0.16 |
| 101 (low) | - | 103 (high) | 106 (high) | 32.51 ± 0.85 | n/d | 24.72 ± 1.68 | 24.73 ± 0.67 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Komu, J.G.; Jamsransuren, D.; Matsuda, S.; Ogawa, H.; Takeda, Y. Performance Evaluation of a Fully Automated Molecular Diagnostic System for Multiplex Detection of SARS-CoV-2, Influenza A/B Viruses, and Respiratory Syncytial Virus. Diagnostics 2025, 15, 1791. https://doi.org/10.3390/diagnostics15141791
Komu JG, Jamsransuren D, Matsuda S, Ogawa H, Takeda Y. Performance Evaluation of a Fully Automated Molecular Diagnostic System for Multiplex Detection of SARS-CoV-2, Influenza A/B Viruses, and Respiratory Syncytial Virus. Diagnostics. 2025; 15(14):1791. https://doi.org/10.3390/diagnostics15141791
Chicago/Turabian StyleKomu, James G., Dulamjav Jamsransuren, Sachiko Matsuda, Haruko Ogawa, and Yohei Takeda. 2025. "Performance Evaluation of a Fully Automated Molecular Diagnostic System for Multiplex Detection of SARS-CoV-2, Influenza A/B Viruses, and Respiratory Syncytial Virus" Diagnostics 15, no. 14: 1791. https://doi.org/10.3390/diagnostics15141791
APA StyleKomu, J. G., Jamsransuren, D., Matsuda, S., Ogawa, H., & Takeda, Y. (2025). Performance Evaluation of a Fully Automated Molecular Diagnostic System for Multiplex Detection of SARS-CoV-2, Influenza A/B Viruses, and Respiratory Syncytial Virus. Diagnostics, 15(14), 1791. https://doi.org/10.3390/diagnostics15141791





