CIC-Rearranged Sarcoma: A Clinical and Pathological Study of a Peculiar Entity
Abstract
1. Introduction
2. Materials and Methods
3. Results
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Antonescu, C.R.; Yoshida, A. CIC-Rearranged Sarcoma. In Undifferentiated Small Round Cell Sarcomas of Bone and Soft Tissue, 5th ed.; IARC Press: Lyon, France, 2020; pp. 330–332. [Google Scholar]
- Antonescu, C.R.; Owosho, A.A.; Zhang, L.; Chen, S.; Deniz, K.; Huryn, J.M.; Kao, Y.C.; Huang, S.C.; Singer, S.; Tap, W.; et al. Sarcomas with CIC-Rearrangements Are a Distinct Pathologic Entity with Aggressive Outcome: A Clinicopathologic and Molecular Study of 115 Cases. Am. J. Surg. Pathol. 2017, 41, 941–949. [Google Scholar] [CrossRef] [PubMed]
- Kawamura-Saito, M.; Yamazaki, Y.; Kaneko, K.; Kawaguchi, N.; Kanda, H.; Mukai, H.; Gotoh, T.; Motoi, T.; Fukayama, M.; Aburatani, H.; et al. Fusion between CIC and DUX4 Up-Regulates PEA3 Family Genes in Ewing-like Sarcomas with t(4;19)(q35;q13) Translocation. Hum. Mol. Genet. 2006, 15, 2125–2137. [Google Scholar] [CrossRef] [PubMed]
- Yoshimoto, M.; Graham, C.; Chilton-MacNeill, S.; Lee, E.; Shago, M.; Squire, J.; Zielenska, M.; Somers, G.R. Detailed Cytogenetic and Array Analysis of Pediatric Primitive Sarcomas Reveals a Recurrent CIC-DUX4 Fusion Gene Event. Cancer Genet. Cytogenet. 2009, 195, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Italiano, A.; Sung, Y.S.; Zhang, L.; Singer, S.; Maki, R.G.; Coindre, J.M.; Antonescu, C.R. High Prevalence of CIC Fusion with Double-Homeobox (DUX4) Transcription Factors in EWSR1-Negative Undifferentiated Small Blue Round Cell Sarcomas. Genes Chromosomes Cancer 2012, 51, 207–218. [Google Scholar] [CrossRef]
- Graham, C.; Chilton-MacNeill, S.; Zielenska, M.; Somers, G.R. The CIC-DUX4 Fusion Transcript Is Present in a Subgroup of Pediatric Primitive Round Cell Sarcomas. Hum. Pathol. 2012, 43, 180–189. [Google Scholar] [CrossRef]
- Choi, E.Y.; Thomas, D.G.; McHugh, J.B.; Patel, R.M.; Roulston, D.; Schuetze, S.M.; Chugh, R.; Biermann, J.S.; Lucas, D.R. Undifferentiated Small Round Cell Sarcoma with t(4;19)(q35;q13.1) CIC-DUX4 Fusion: A Novel Highly Aggressive Soft Tissue Tumor with Distinctive Histopathology. Am. J. Surg. Pathol. 2013, 37, 1379–1386. [Google Scholar] [CrossRef]
- Le Loarer, F.; Pissaloux, D.; Watson, S.; Godfraind, C.; Galmiche-Rolland, L.; Silva, K.; Mayeur, L.; Italiano, A.; Michot, A.; Pierron, G.; et al. Clinicopathologic Features of CIC-NUTM1 Sarcomas, a New Molecular Variant of the Family of CIC-Fused Sarcomas. Am. J. Surg. Pathol. 2019, 43, 268–276. [Google Scholar] [CrossRef]
- Sugita, S.; Arai, Y.; Aoyama, T.; Asanuma, H.; Mukai, W.; Hama, N.; Emori, M.; Shibata, T.; Hasegawa, T. NUTM2A-CIC Fusion Small Round Cell Sarcoma: A Genetically Distinct Variant of CIC-Rearranged Sarcoma. Hum. Pathol. 2017, 65, 225–230. [Google Scholar] [CrossRef]
- Sugita, S.; Arai, Y.; Tonooka, A.; Hama, N.; Totoki, Y.; Fujii, T.; Aoyama, T.; Asanuma, H.; Tsukahara, T.; Kaya, M.; et al. A Novel CIC-FOXO4 Gene Fusion in Undifferentiated Small Round Cell Sarcoma: A Genetically Distinct Variant of Ewing-Like Sarcoma. Am. J. Surg. Pathol. 2014, 38, 1571–1576. [Google Scholar] [CrossRef]
- Connolly, E.A.; Bhadri, V.A.; Wake, J.; Ingley, K.M.; Lewin, J.; Bae, S.; Wong, D.D.; Long, A.P.; Pryor, D.; Thompson, S.R.; et al. Systemic Treatments and Outcomes in CIC-Rearranged Sarcoma: A National Multi-Centre Clinicopathological Series and Literature Review. Cancer Med. 2022, 11, 1805–1816. [Google Scholar] [CrossRef]
- Yoshimoto, T.; Tanaka, M.; Homme, M.; Yamazaki, Y.; Takazawa, Y.; Antonescu, C.R.; Nakamura, T. CIC-DUX4 Induces Small Round Cell Sarcomas Distinct from Ewing Sarcoma. Cancer Res. 2017, 77, 2927–2937. [Google Scholar] [CrossRef]
- Specht, K.; Sung, Y.S.; Zhang, L.; Richter, G.H.; Fletcher, C.D.; Antonescu, C.R. Distinct Transcriptional Signature and Immunoprofile of CIC-DUX4 Fusion-Positive Round Cell Tumors Compared to EWSR1-Rearranged Ewing Sarcomas: Further Evidence Toward Distinct Pathologic Entities. Genes Chromosomes Cancer 2014, 53, 622–633. [Google Scholar] [CrossRef]
- Kinnaman, M.D.; Zhu, C.; Weiser, D.A.; Mohiuddin, S.; Hingorani, P.; Roth, M.; Gill, J.; Janeway, K.A.; Gorlick, R.; Lessnick, S.L.; et al. Survey of Paediatric Oncologists and Pathologists Regarding Their Views and Experiences with Variant Translocations in Ewing and Ewing-Like Sarcoma: A Report of the Children’s Oncology Group. Sarcoma 2020, 2020, 3498549. [Google Scholar] [CrossRef]
- Brahmi, M.; Gaspar, N.; Gantzer, J.; Toulmonde, M.; Boudou-Rouquette, P.; Bompas, E.; Firmin, N.; Valentin, T.; Cancel, M.; Duffaud, F.; et al. Patterns of Care and Outcome of CIC-Rearranged Sarcoma Patients: A Nationwide Study of the French Sarcoma Group. Cancer Med. 2023, 12, 7801–7807. [Google Scholar] [CrossRef]
- Palmerini, E.; Gambarotti, M.; Italiano, A.; Nathenson, M.J.; Ratan, R.; Dileo, P.; Provenzano, S.; Jones, R.L.; DuBois, S.G.; Martin-Broto, J.; et al. A Global Collaborative Study of CIC-Rearranged, BCOR::CCNB3-Rearranged and Other Ultra-Rare Unclassified Undifferentiated Small Round Cell Sarcomas (GRACefUl). Eur. J. Cancer 2023, 183, 11–23. [Google Scholar] [CrossRef]
- Makise, N.; Yoshida, A. CIC-Rearranged Sarcoma. Surg. Pathol. Clin. 2024, 17, 141–151. [Google Scholar] [CrossRef]
- Hung, Y.P.; Fletcher, C.D.; Hornick, J.L. Evaluation of NKX2-2 Expression in Round Cell Sarcomas and Other Tumors with EWSR1 Rearrangement: Imperfect Specificity for Ewing Sarcoma. Mod. Pathol. 2016, 29, 370–380. [Google Scholar] [CrossRef]
- Yoshida, A.; Arai, Y.; Kobayashi, E.; Yonemori, K.; Ogura, K.; Hama, N.; Mukai, W.; Motoi, T.; Kawai, A.; Shibata, T.; et al. CIC Break-Apart Fluorescence In-Situ Hybridization Misses a Subset of CIC-DUX4 Sarcomas: A Clinicopathological and Molecular Study. Histopathology 2017, 71, 461–469. [Google Scholar] [CrossRef]
- Cocchi, S.; Gamberi, G.; Magagnoli, G.; Maioli, M.; Righi, A.; Frisoni, T.; Gambarotti, M.; Benini, S. CIC Rearranged Sarcomas: A Single Institution Experience of the Potential Pitfalls in Interpreting CIC FISH Results. Pathol. Res. Pract. 2022, 231, 153773. [Google Scholar] [CrossRef]
- Koo, S.C.; Cardenas, M.; Stow, P.; Neary, J.; Wheeler, D.A.; Shi, Z.; Furtado, L.V. Comparison of Molecular Testing Methodologies for CIC-Rearranged Sarcomas. Arch. Pathol. Lab. Med. 2025. advance online publication. [Google Scholar] [CrossRef]
- Hung, Y.P.; Fletcher, C.D.; Hornick, J.L. Evaluation of ETV4 and WT1 expression in CIC-rearranged sarcomas and histologic mimics. Mod. Pathol. 2016, 29, 1324–1334. [Google Scholar] [CrossRef]
- Le Guellec, S.; Velasco, V.; Pérot, G.; Watson, S.; Tirode, F.; Coindre, J.M. ETV4 is a useful marker for the diagnosis of CIC-rearranged undifferentiated round-cell sarcomas: A study of 127 cases including mimicking lesions. Mod. Pathol. 2016, 29, 1523–1531. [Google Scholar] [CrossRef]
- Macedo, R.T.; Baranovska-Andrigo, V.; Pancsa, T.; Klubíčková, N.; Rubin, B.P.; Kilpatrick, S.E.; Goldblum, J.R.; Fritchie, K.J.; Billings, S.D.; Michal, M.; et al. Nuclear DUX4 immunohistochemistry is a highly sensitive and specific marker for the presence of CIC::DUX4 fusion in CIC-rearranged sarcomas: A study of 48 molecularly confirmed cases. Histopathology 2025, 86, 423–432. [Google Scholar] [CrossRef]
- Nishino, S.; Kojima, N.; Sugino, H.; Mori, T.; Yatabe, Y.; Yoshida, A. MUC5AC immunoreactivity in scattered tumor cells is useful for diagnosing CIC-rearranged sarcoma. Virchows Arch. 2024, 485, 359–363. [Google Scholar] [CrossRef]
- Murphy, J.; Resch, E.E.; Leland, C.; Meyer, C.F.; Llosa, N.J.; Gross, J.M.; Pratilas, C.A. Clinical outcomes of patients with CIC-rearranged sarcoma: A single institution retrospective analysis. J. Cancer Res. Clin. Oncol. 2024, 150, 112. [Google Scholar] [CrossRef]
- Donthi, D.; Malik, P.; Prenshaw, K.L.; Hong, H. A Rare Case of Round Cell Sarcoma with CIC-DUX4 Mutation Mimicking a Phlegmon: Review of Literature. Am. J. Case Rep. 2020, 21, e925683. [Google Scholar] [CrossRef]
- Vieira, A.C.; Xavier, C.B.; Vieira, T.D.; Carvalho, F.M.; Scaranti, M.; Munhoz, R.R.; Carvalho, J.P. CIC-DUX4 rearranged uterine cervix round-cell sarcoma exhibiting near-complete pathologic response following radiation and neoadjuvant chemotherapy: A case report. Gynecol. Oncol. Rep. 2021, 36, 100745. [Google Scholar] [CrossRef]
- Smith, S.C.; Buehler, D.; Choi, E.Y.; McHugh, J.B.; Rubin, B.P.; Billings, S.D.; Balzer, B.; Thomas, D.G.; Lucas, D.R.; Goldblum, J.R.; et al. CIC-DUX sarcomas demonstrate frequent MYC amplification and ETS-family transcription factor expression. Mod. Pathol. 2015, 28, 57–68. [Google Scholar] [CrossRef]



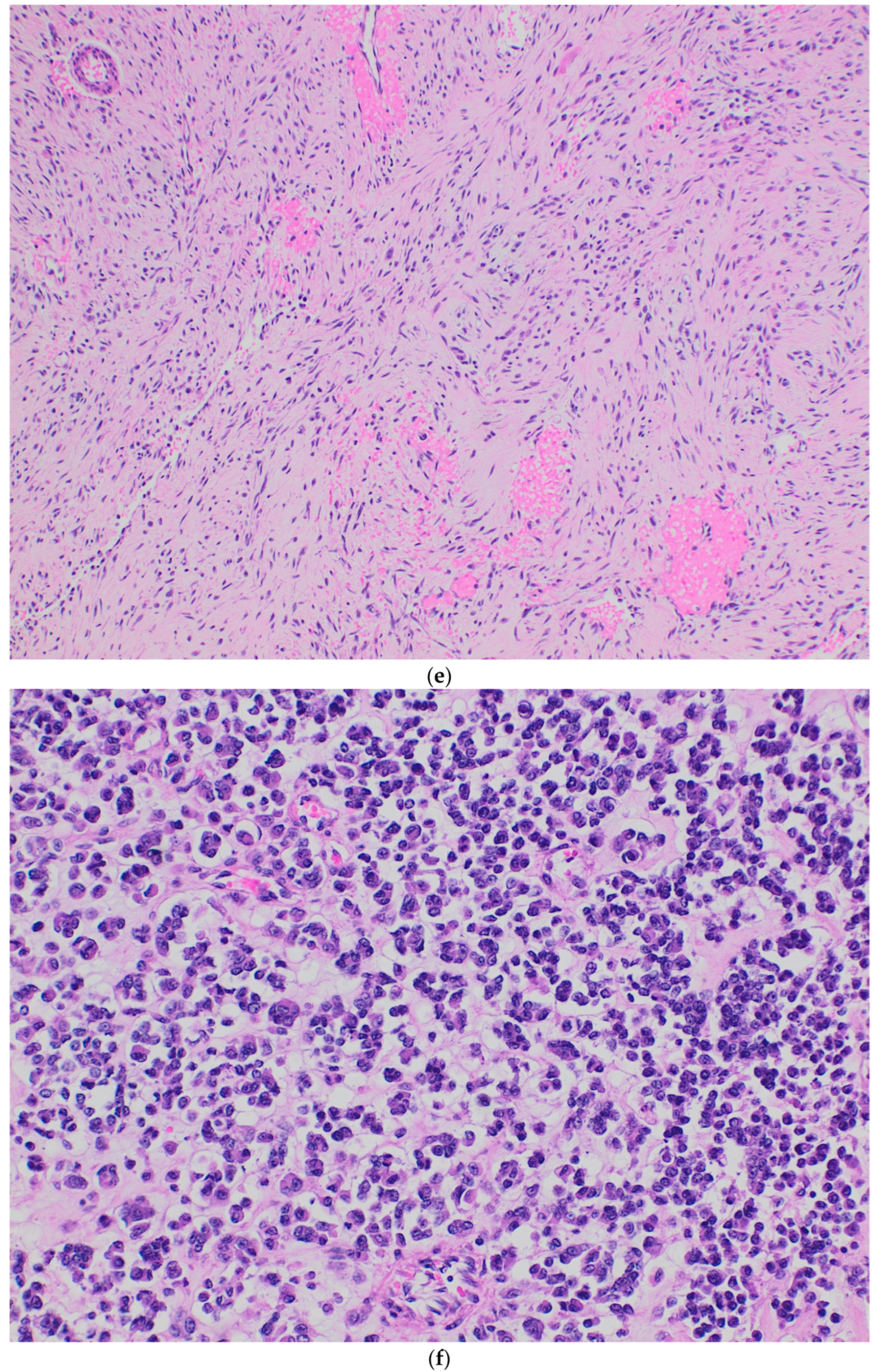
| Case | Age (Years)/Gender/Symptoms and Clinical Features | Site | Size (cm) | Radiological Features | Metastasis | Treatment | Outcome |
|---|---|---|---|---|---|---|---|
| 1 | 52/F/mass | Left thigh | 8.3 | MRI: Large subcutaneous left thigh soft tissue mass suggestive of soft tissue sarcoma. | Lung at presentation, then brain metastasis | Ifosfamide, cisplatin, and etoposide at another institution. Palliative radiotherapy. | DOD 10 months |
| 2 | 42/F/mass | Right axilla/infra clavicular | 7.0 | CT scan: Large well-defined soft tissue density mass lesion in the right upper axilla anterior to the shoulder joint, scapula, and upper ribs. | No metastasis at presentation, then lung metastasis | R2 excision, adjuvant radiotherapy. Chemotherapy (VDC-IE). | DOD 19 months |
| 3 | 25/M/pain and mass | Left thigh | 15.0 | MRI: Osseous mass at left iliac bone and acetabulum with post-gadolinium enhancement, associated with a soft tissue component at both sides of the iliac wing. | Lung at presentation, then brain metastasis | Chemotherapy (VDC-IE), radiotherapy. | DOD 8 months |
| 4 | 53/M/pain and mass | Left groin | 7.4 | CT scan: Infiltrative left iliopsoas muscle mass. | Lung at presentation, then brain metastasis | Chemotherapy (VDC-IE) followed by surgical excision and adjuvant radiotherapy. | DOD 12 months |
| 5 | 47/F/mass | Left thigh | 10.2 | MRI: A heterogeneous enhancing soft tissue mass with hemorrhagic contents at the medial anterior compartment of the left thigh encasing the superficial femoral artery. | Lung metastasis at presentation | Chemotherapy (VDC-IE), neoadjuvant radiotherapy, excision with negative margins. | DOD 10 months |
| 6 | 47/M/mass | Dorsum of the right foot | 4.8 | MRI: A soft tissue mass in the second web space with dorsal and plantar components. | Lung and bone metastasis at presentation | Chemotherapy (VDC), radiotherapy for spine metastasis. | DOD 1 month |
| 7 | 43/F/abdominal pain | Mesentery | 7.0 | CT scan: Left pelvic mesenteric mass abutting the left ovary, sigmoid, and anterior abdominal wall without definite invasion. | No | Excision (margins negative), one cycle VDC. | NED 25 months |
| 8 | 19/M/mass with restriction of shoulder movement | Right shoulder | 20.0 | MRI: An aggressive mass in the shoulder with destruction of the distal clavicle and acromion with a large extra osseous soft tissue component occupying most of the ventral and superolateral aspect of the shoulder. It showed heterogeneous enhancement with a large necrotic component. | Lung metastasis at presentation | VDC-IE. | DOD 4 months |
| 9 | 53/F/back pain | Left paraspina-l at the level of L2-L4 levels | 10.0 | MRI: A relatively large heterogeneous left paraspinal soft tissue mass invading the posterior elements of the lower lumbar vertebrae and showing intraspinal extension compressing the thecal sac with focal canal stenosis. | Lung metastasis at presentation | Chemotherapy (VDC-IE), L3-L4 spine decompression, radiotherapy. | AWD 10 months |
| 10 | 16/M/pain, rapidly progressing mass | Left gluteus maximus muscle | 15.0 | MRI: Large multilobulated mass within the left gluteus maximus, with heterogeneous enhancement and hemorrhagic components. | Lung metastasis at presentation | Chemotherapy (VDC-IE). | DOD 4 months |
| 11 | 14/M/kno-wn case of pre-B-cell ALL t (12;21) post-BMT presented with status epilepticus. On exam, the patient had left-sided upper limb weakness and diffuse skin nevi but no café au lait spots | Brain/right frontal lobe | 3.5 | MRI: Right frontal cortical and subcortical complex mass (cystic and solid). | No | Surgery, chemotherapy (ICE). | AWD 6 months |
| Case | Referred/Outside Diagnosis | Histologic Pattern | IHC Profile | Molecular Findings |
|---|---|---|---|---|
| 1 | Sarcoma, not otherwise specified | Sheets of small, undifferentiated round cells with dark nuclei. | Positive: FLI-1, BCL2, NSE, and SMA. Focally positive: CD99 and WT1. Negative: S100, desmin, CK-MNF, and myogenin. | FISH for CIC fusion is positive. |
| 2 | High-grade sarcoma with myxoid and round cell areas. | Nests, cords, and trabeculae of undifferentiated round- to oval-shaped cells with vesicular nuclei and prominent nucleoli in a fibrotic and myxoid background. Occasional cells with clear cytoplasm and a few cells with epithelioid and rhabdoid features. | Positive: FLI-1 and CD56. Focally positive: CD99 and synaptophysin. Negative: CD99, CK-MNF, desmin, S100, SOX10, TLE-1, and ERG. | CIC::DUX4 by RNA sequencing. |
| 3 | Not applicable | Sheets of small, undifferentiated round to oval-shaped and short-spindled cells with dark nuclei. | Positive: FLI-1 and WT-1. Focally positive: CD99. Negative: NKX2.2, panCK, desmin, synaptophysin, S100, SATB2 and ERG. | CIC::DUX4 by RNA sequencing. |
| 4 | Low- to intermediate-grade sarcoma | Sheets of small, undifferentiated round- to oval-shaped cells with dark nuclei. Post neoadjuvant: Biphasic pattern with spindle cell areas showing abrupt transition to atypical epithelioid cells. | Diffusely positive: CD99 and WT-1. Dot-like positivity: EMA and CK-MNF. Negative: S100, desmin, synaptophysin, NKX2.2, and STAT6. | CIC::DUX4 by RNA sequencing. |
| 5 | Extraskeletal Ewing sarcoma | Sheets of small, undifferentiated round cells with dark nuclei. | Diffusely positive: CD99, WT1, and ERG. Negative: NKX2.2, synaptophysin, and BCOR. | CIC::DUX4 by RNA sequencing. |
| 6 | Synovial sarcoma | Sheets of small, undifferentiated round- to oval-shaped and short-spindled cells with focal myxoid background. | Focally positive: CD99 and SATB2 Negative: ERG, BCOR, NKX2.2, synaptophysin, WT1, S100, panCK, SS18, desmin, STAT6, and CK-MNF. Retained nuclear expression: H3K27me3 | FISH for CIC rearrangement is positive. Methylation confirmed the diagnosis. |
| 7 | Extraskeletal Ewing sarcoma | Sheets of monotonous small, undifferentiated round cells, very similar to Ewing sarcoma. | Diffusely positive: CD99, NKX2.2, and S100. Negative: DOG-1, synaptophysin, SS18, WT-1, panCK, CK-MNF, desmin, and SALL4. Retained nuclear expression: INI1. | FISH for CIC rearrangement positive. FISH for EWSR-1 gene rearrangement is negative. |
| 8 | Not applicable | Sheets of oval to short-spindled cells with dark nuclei. | Focally positive: CD99, WT1, and ERG. Negative: PanCK, Desmin, S100, NKX2.2, BCOR, SS18, and SATB2. | FISH for CIC rearrangement positive. |
| 9 | Not applicable | Sheets, short-spindled and anastomosing cords of round- to oval-shaped cells with vesicular nuclei and prominent nucleoli. Microcyst formation and focal myxoid background. | Positive: MDM2. Dot-like positivity: CD99. Focally positive: EMA. Negative: PanCK, CK-MNF, WT-1, CD31, S100, BCOR, S100, and STAT-6. | FISH for CIC rearrangement positive. |
| 10 | Sarcoma, not otherwise specified | Sheets of small, undifferentiated round cells with dark nuclei. | Focally positive: CD99 and WT1. Negative: SATB2, MyoD1, and SOX10. | FISH for CIC rearrangement positive. |
| 11 | Not applicable | Sheets of round to oval cells, similar to Ewing sarcoma. | Diffusely positive: CD99 and FLI-1. Focally positive: Synaptophysin. Negative: NKX2.2, BCOR, Desmin, STAT-6, SATB2, SOX10, S100, and panCK. Retained nuclear expression: INI1 and BRG1. | FISH for CIC rearrangement positive. Methylation confirmed the diagnosis. |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Maaita, W.; Hasasna, N.; Yaser, S.; Saleh, Y.; Abu-Hijlih, R.; Asha, W.; Halalsheh, H.; Abdel Al, S.; Al-Hussaini, M.; Jaber, O. CIC-Rearranged Sarcoma: A Clinical and Pathological Study of a Peculiar Entity. Diagnostics 2025, 15, 1758. https://doi.org/10.3390/diagnostics15141758
Maaita W, Hasasna N, Yaser S, Saleh Y, Abu-Hijlih R, Asha W, Halalsheh H, Abdel Al S, Al-Hussaini M, Jaber O. CIC-Rearranged Sarcoma: A Clinical and Pathological Study of a Peculiar Entity. Diagnostics. 2025; 15(14):1758. https://doi.org/10.3390/diagnostics15141758
Chicago/Turabian StyleMaaita, Ward, Nabil Hasasna, Sameer Yaser, Yacob Saleh, Ramiz Abu-Hijlih, Wafa Asha, Hadeel Halalsheh, Samer Abdel Al, Maysa Al-Hussaini, and Omar Jaber. 2025. "CIC-Rearranged Sarcoma: A Clinical and Pathological Study of a Peculiar Entity" Diagnostics 15, no. 14: 1758. https://doi.org/10.3390/diagnostics15141758
APA StyleMaaita, W., Hasasna, N., Yaser, S., Saleh, Y., Abu-Hijlih, R., Asha, W., Halalsheh, H., Abdel Al, S., Al-Hussaini, M., & Jaber, O. (2025). CIC-Rearranged Sarcoma: A Clinical and Pathological Study of a Peculiar Entity. Diagnostics, 15(14), 1758. https://doi.org/10.3390/diagnostics15141758






