Artificial Intelligence in Primary Malignant Bone Tumor Imaging: A Narrative Review
Abstract
1. Introduction
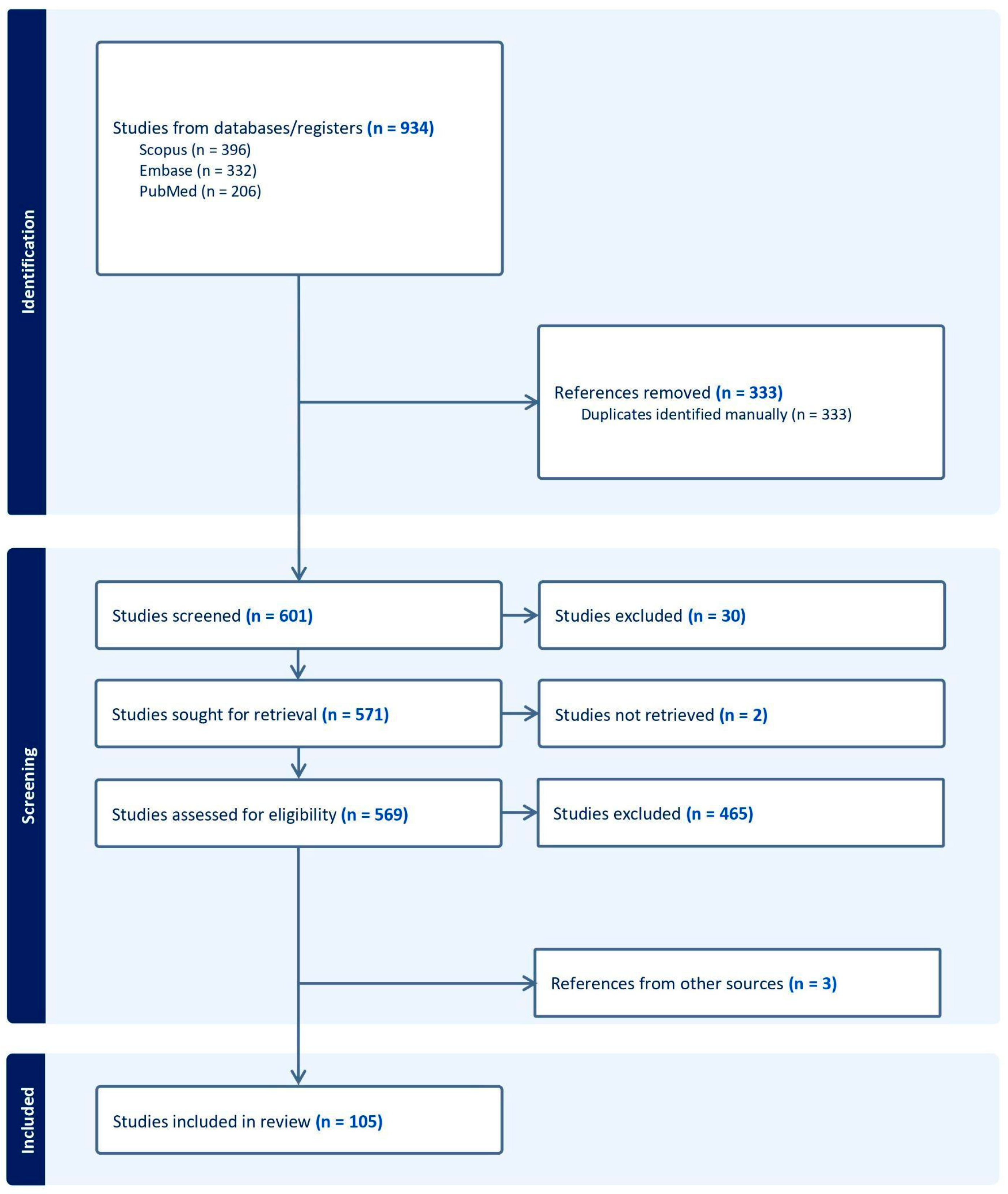
2. Materials and Methods
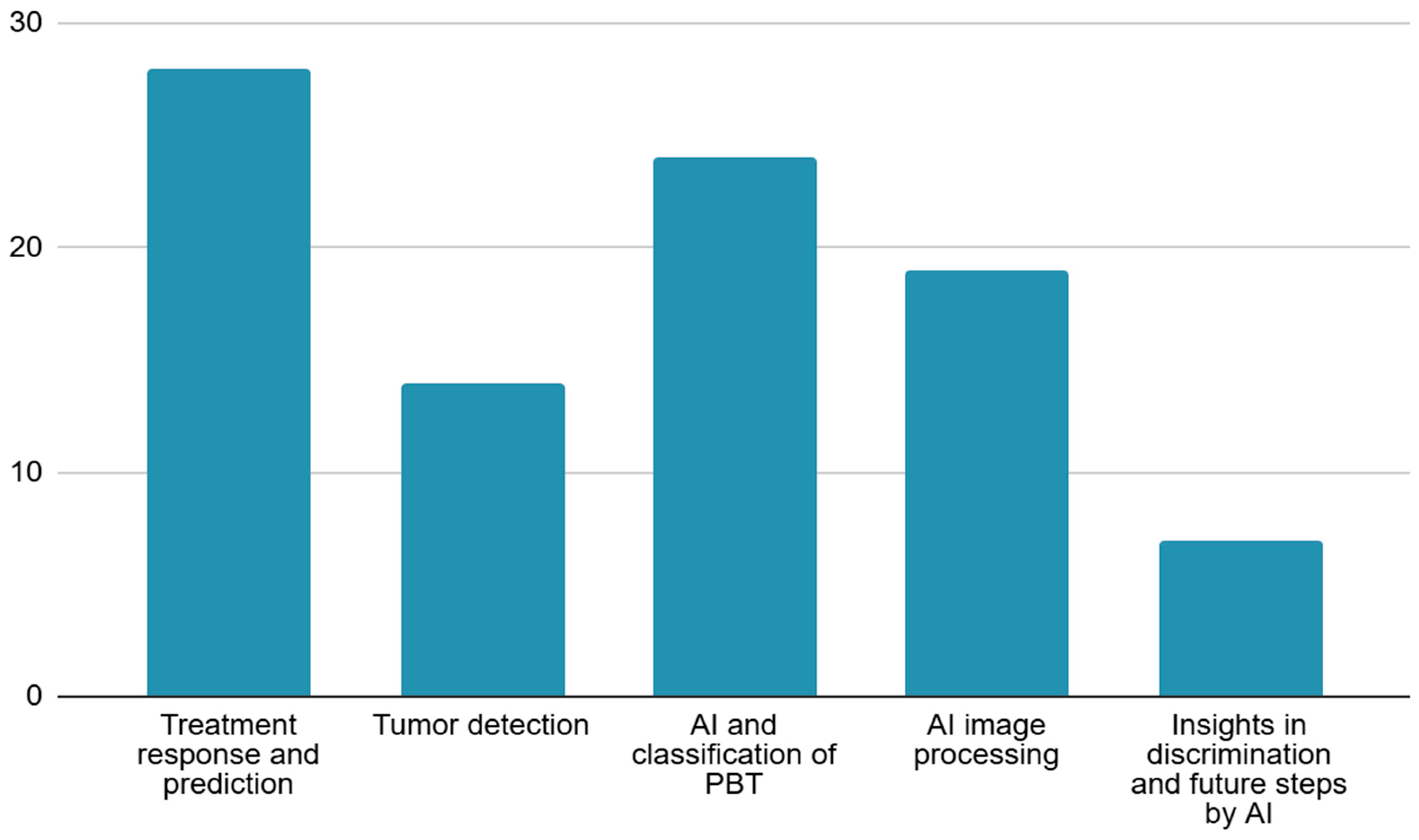
3. Results
4. Discussion
4.1. Treatment Response and Prediction
4.2. Tumor Detection
4.3. AI and Classification of PBT
4.4. Tumor Segmentation
4.5. Insights into Discrimination and Future Steps by AI
4.6. Limitations of the Study
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| AI | Artificial Intelligence |
| PBT | Primary Bone Tumors |
| NAC | Neoadjuvant Chemotherapy |
| ML | Machine Learning |
| DL | Deep Learning |
| RS | Radiomics Signature |
| RBF | Radial Basis Function |
| CNN | Convolutional Neural Networks |
| MRI | Magnetic Resonance Imaging |
| AUC | Area Under the Curve |
| DT | Decision Tree |
| LR | Logic Recession |
| SVM | Support Vector Machine |
| DCA | Decision Curve Analysis |
| DLRM | Deep Learning Radiomics Model |
| DIaL | Deep Learning Interactive Model |
| DS-Net | Deep Supervision Network |
| H&E | Hematoxylin and Eosin |
| 18F-FDG | Fluorine 18 Fluorodeoxyglucose |
| PET | Positron Emission Tomography |
| DWI | Diffusion-Weighted Imaging |
| PSNR | Peak Signal-to-Noise Ratio |
| MSE | Mean Squared Error |
| EPI | Edge Presence Index |
| LASSO | Least Absolute Shrinkage and Selection Operator |
| ROI | Region Of Interest |
| T2WI | T2 Weighted Imaging |
| T1CE | T1 Weighted Contrast-Enhanced Imaging |
| KNN | K Nearest Neighbor |
| NAC | Neoadjuvant Chemotherapy |
| DCE-MRI | Dynamic Contrast-Enhanced Magnetic Resonance Imaging |
| SUVmax | Maximum Standardized Uptake Value |
| CT | Computed Tomography |
| VGG16 | Visual Geometry Group 16-layer Network |
| VGG19 | Visual Geometry Group 19-layer Network |
| DenseNet201 | Densely Connected Convolutional Network 201 Layers |
| ResNet101 | Residual Network 101 Layers |
| NASNetLarge | Neural Architecture Search Network Large |
| EfficientNetV2L | Efficient Network Version 2 Large |
| IF-FSM-C | Inception Framework with Feature Selection Mechanism for Classification |
| BCDNet | Bone Cancer Detection Network |
| GCT | Giant Cell Tumor |
| ALP | Alkaline Phosphatase |
| LDH | Lactate Dehydrogenase |
| ChatGPT-4 | Chat Generative Pre-trained Transformer 4 |
| U-net | U-shaped Convolutional Network |
| DUconViT | Dual Convolutional Vision Transformer |
| Mask R-CNN | Mask Region-Based Convolutional Neural Network |
| PCA-IPSO | Principal Component Analysis Improved Particle Swarm Optimization |
| DECIDE | Deep Ensemble Classifier with Integration of Dual Enhancers |
| Grad-CAM | Gradient-weighted Class Activation Mapping |
| CATS | Computer-Assisted Tumor Surgery |
References
- Vogrin, M.; Trojner, T.; Kelc, R. Artificial Intelligence in Musculoskeletal Oncological Radiology. Radiol. Oncol. 2020, 55, 1–6. [Google Scholar] [CrossRef]
- Erdem, F.; Tamsel, İ.; Demirpolat, G. The Use of Radiomics and Machine Learning for the Differentiation of Chondrosarcoma from Enchondroma. J. Clin. Ultrasound 2023, 51, 1027–1035. [Google Scholar] [CrossRef]
- Meng, Y.; Yang, Y.; Hu, M.; Zhang, Z.; Zhou, X. Artificial Intelligence-Based Radiomics in Bone Tumors: Technical Advances and Clinical Application. Semin. Cancer Biol. 2023, 95, 75–87. [Google Scholar] [CrossRef]
- Ye, Q.; Yang, H.; Lin, B.; Wang, M.; Song, L.; Xie, Z.; Lu, Z.; Feng, Q.; Zhao, Y. Automatic Detection, Segmentation, and Classification of Primary Bone Tumors and Bone Infections Using an Ensemble Multi-Task Deep Learning Framework on Multi-Parametric MRIs: A Multi-Center Study. Eur. Radiol. 2024, 34, 4287–4299. [Google Scholar] [CrossRef]
- Emil, N.S.; Sibbitt, R.R.; Sibbitt, W.L., Jr. Machine Learning and Magnetic Resonance Imaging: Differentiating Benign from Malignant Osseous Tumors. J. Clin. Ultrasound 2023, 51, 1036–1038. [Google Scholar] [CrossRef]
- Yildirim, M.; Yildirim, H. CT Radiomics-Based Machine Learning Model for Differentiating between Enchondroma and Low-Grade Chondrosarcoma. Med. Baltim. 2024, 103, e39311. [Google Scholar] [CrossRef]
- Zheng, F.; Yin, P.; Liang, K.; Wang, Y.; Hao, W.; Hao, Q.; Hong, N. Fusion Radiomics-Based Prediction of Response to Neoadjuvant Chemotherapy for Osteosarcoma. Acad. Radiol. 2024, 31, 2444–2455. [Google Scholar] [CrossRef]
- Avery, E.; Sanelli, P.C.; Aboian, M.; Payabvash, S. Radiomics: A Primer on Processing Workflow and Analysis. Semin. Ultrasound CT MRI 2022, 43, 142–146. [Google Scholar] [CrossRef]
- Li, M.D.; Ahmed, S.R.; Choy, E.; Lozano-Calderon, S.A.; Kalpathy-Cramer, J.; Chang, C.Y. Artificial Intelligence Applied to Musculoskeletal Oncology: A Systematic Review. Skelet. Radiol. 2022, 51, 245–256. [Google Scholar] [CrossRef]
- Gitto, S.; Corino, V.; Bologna, M.; Marzorati, L.; Milazzo Machado, E.; Albano, D.; Messina, C.; Mainardi, L.; Sconfienza, L.M. MRI Radiomics-Based Machine Learning to Predict Neoadjuvant Chemotherapy Response in Ewing Sarcoma. Insights Imaging 2022, 14, 77–78. [Google Scholar] [CrossRef]
- Gitto, S.; Corino, V.D.A.; Annovazzi, A.; Milazzo Machado, E.; Bologna, M.; Marzorati, L.; Albano, D.; Messina, C.; Serpi, F.; Anelli, V.; et al. 3D vs. 2D MRI Radiomics in Skeletal Ewing Sarcoma: Feature Reproducibility and Preliminary Machine Learning Analysis on Neoadjuvant Chemotherapy Response Prediction. Front. Oncol. 2022, 12, 1016123. [Google Scholar] [CrossRef]
- Lin, P.; Yang, P.F.; Chen, S.; Shao, Y.Y.; Xu, L.; Wu, Y.; Teng, W.; Zhou, X.Z.; Li, B.H.; Luo, C.; et al. A Delta-Radiomics Model for Preoperative Evaluation of Neoadjuvant Chemotherapy Response in High-Grade Osteosarcoma. Cancer Imaging 2020, 20, 7. [Google Scholar] [CrossRef]
- He, F.; Xie, L.; Sun, X.; Xu, J.; Li, Y.; Liu, R.; Sun, K.; Shen, D.; Gu, J.; Ji, T.; et al. A Scoring System for Predicting Neoadjuvant Chemotherapy Response in Primary High-Grade Bone Sarcomas: A Multicenter Study. Orthop. Surg. 2022, 14, 2499–2509. [Google Scholar] [CrossRef]
- Zhong, J.; Zhang, C.; Hu, Y.; Zhang, J.; Liu, Y.; Si, L.; Xing, Y.; Ding, D.; Geng, J.; Jiao, Q.; et al. Automated Prediction of the Neoadjuvant Chemotherapy Response in Osteosarcoma with Deep Learning and an MRI-Based Radiomics Nomogram. Eur. Radiol. 2022, 32, 6196–6206. [Google Scholar] [CrossRef]
- Nie, P.; Zhao, X.; Ma, J.; Wang, Y.; Li, B.; Li, X.; Li, Q.; Xu, Y.; Dai, Z.; Wu, J.; et al. Can the Preoperative CT-Based Deep Learning Radiomics Model Predict Histologic Grade and Prognosis of Chondrosarcoma? Eur. J. Radiol. 2024, 181, 111719. [Google Scholar] [CrossRef]
- Teo, K.Y.; Daescu, O.; Cederberg, K.; Sengupta, A.; Leavey, P.J. Correlation of Histopathology and Multi-Modal Magnetic Resonance Imaging in Childhood Osteosarcoma: Predicting Tumor Response to Chemotherapy. PLoS ONE 2022, 17, e0259564. [Google Scholar] [CrossRef]
- Ho, D.J.; Agaram, N.P.; Schüffler, P.J.; Vanderbilt, C.M.; Jean, M.-H.; Hameed, M.R.; Fuchs, T.J. Deep Interactive Learning: An Efficient Labeling Approach for Deep Learning-Based Osteosarcoma Treatment Response Assessment. In Medical Image Computing and Computer Assisted Intervention–MICCAI 2020, Proceedings of the 23rd International Conference, Lima, Peru, 4–8 October 2020; Springer Science and Business Media Deutschland GmbH: Cham, Switzerland, 2020; Volume 12265 LNCS, pp. 540–549. [Google Scholar]
- Fu, Y.; Xue, P.; Ji, H.; Cui, W.; Dong, E. Deep Model with Siamese Network for Viable and Necrotic Tumor Regions Assessment in Osteosarcoma. Med. Phys. 2020, 47, 4895–4905. [Google Scholar] [CrossRef]
- Kim, W.; Park, J.; Sheen, H.; Byun, B.H.; Lim, I.; Kong, C.-B.; Lim, S.M.; Woo, S.-K. Development of Deep Learning Model for Prediction of Chemotherapy Response Using PET Images and Radiomics Features. In Proceedings of the 2018 IEEE Nuclear Science Symposium and Medical Imaging Conference Proceedings (NSS/MIC), Sydney, Australia, 10–17 November 2018; Institute of Electrical and Electronics Engineers Inc.: Piscataway, NJ, USA, 2018. [Google Scholar]
- Hu, Y.; Tang, J.; Zhao, S.; Li, Y. Diffusion-Weighted Imaging-Magnetic Resonance Imaging Information under Class-Structured Deep Convolutional Neural Network Algorithm in the Prognostic Chemotherapy of Osteosarcoma. Sci. Program. 2021, 2021, 1–12. [Google Scholar] [CrossRef]
- Djuričić, G.J.; Ahammer, H.; Rajković, S.; Kovač, J.D.; Milošević, Z.; Sopta, J.P.; Radulovic, M. Directionally Sensitive Fractal Radiomics Compatible With Irregularly Shaped Magnetic Resonance Tumor Regions of Interest: Association With Osteosarcoma Chemoresistance. J. Magn. Reson. Imaging 2023, 57, 248–258. [Google Scholar] [CrossRef]
- Zhang, L.; Gao, Q.; Dou, Y.; Cheng, T.; Xia, Y.; Li, H.; Gao, S. Evaluation of the Neoadjuvant Chemotherapy Response in Osteosarcoma Using the MRI DWI-Based Machine Learning Radiomics Nomogram. Front. Oncol. 2024, 14, 1345576. [Google Scholar] [CrossRef]
- Huang, B.; Wang, J.; Sun, M.; Chen, X.; Xu, D.; Li, Z.P.; Ma, J.; Feng, S.T.; Gao, Z. Feasibility of Multi-Parametric Magnetic Resonance Imaging Combined with Machine Learning in the Assessment of Necrosis of Osteosarcoma after Neoadjuvant Chemotherapy: A Preliminary Study. BMC Cancer 2020, 20, 322. [Google Scholar] [CrossRef]
- Zhang, L.; Ge, Y.; Gao, Q.; Zhao, F.; Cheng, T.; Li, H.; Xia, Y. Machine Learning-Based Radiomics Nomogram With Dynamic Contrast-Enhanced MRI of the Osteosarcoma for Evaluation of Efficacy of Neoadjuvant Chemotherapy. Front. Oncol. 2021, 11, 758921. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhi, L.; Li, J.; Wang, M.; Chen, G.; Yin, S. Magnetic Resonance Imaging Radiomics Predicts Histological Response to Neoadjuvant Chemotherapy in Localized High-Grade Osteosarcoma of the Extremities. Acad. Radiol. 2024, 31, 5100–5107. [Google Scholar] [CrossRef]
- Mori, Y.; Ren, H.; Mori, N.; Watanuki, M.; Hitachi, S.; Watanabe, M.; Mugikura, S.; Takase, K. Magnetic Resonance Imaging Texture Analysis Based on Intraosseous and Extraosseous Lesions to Predict Prognosis in Patients with Osteosarcoma. Diagnostics 2024, 14, 2562. [Google Scholar] [CrossRef]
- Chen, H.; Zhang, X.; Wang, X.; Quan, X.; Deng, Y.; Lu, M.; Wei, Q.; Ye, Q.; Zhou, Q.; Xiang, Z.; et al. MRI-Based Radiomics Signature for Pretreatment Prediction of Pathological Response to Neoadjuvant Chemotherapy in Osteosarcoma: A Multicenter Study. Eur. Radiol. 2021, 31, 7913–7924. [Google Scholar] [CrossRef]
- Miedler, J.; Schaal, M.; Götz, M.; Cario, H.; Beer, M. Potential Role of MRI-Based Radiomics in Prediction of Chemotherapy Response in Pediatric Patients with Ewing-Sarcoma. Pediatr. Radiol. 2023, 53, S163–S164. [Google Scholar] [CrossRef]
- Chaber, R.; Arthur, C.J.; Łach, K.; Raciborska, A.; Michalak, E.; Bilska, K.; Drabko, K.; Depciuch, J.; Kaznowska, E.; Cebulski, J. Predicting Ewing Sarcoma Treatment Outcome Using Infrared Spectroscopy and Machine Learning. Molecules 2019, 24, 1075. [Google Scholar] [CrossRef]
- Dufau, J.; Bouhamama, A.; Leporq, B.; Malaureille, L.; Beuf, O.; Gouin, F.; Pilleul, F.; Marec-Berard, P. Prediction of Chemotherapy Response in Primary Osteosarcoma Using the Machine Learning Technique on Radiomic Data. Bull. Cancer 2019, 106, 983–999. [Google Scholar] [CrossRef]
- Jeong, S.Y.; Kim, W.; Byun, B.H.; Kong, C.B.; Song, W.S.; Lim, I.; Lim, S.M.; Woo, S.K. Prediction of Chemotherapy Response of Osteosarcoma Using Baseline (18)F-FDG Textural Features Machine Learning Approaches with PCA. Contrast Media Mol. Imaging 2019, 2019, 3515080. [Google Scholar] [CrossRef]
- Bouhamama, A.; Leporq, B.; Khaled, W.; Nemeth, A.; Brahmi, M.; Dufau, J.; Marec-Bérard, P.; Drapé, J.L.; Gouin, F.; Bertrand-Vasseur, A.; et al. Prediction of Histologic Neoadjuvant Chemotherapy Response in Osteosarcoma Using Pretherapeutic MRI Radiomics. Radiol. Imaging Cancer 2022, 4, e210107. [Google Scholar] [CrossRef]
- Kim, J.; Jeong, S.Y.; Kim, B.C.; Byun, B.H.; Lim, I.; Kong, C.B.; Song, W.S.; Lim, S.M.; Woo, S.K. Prediction of Neoadjuvant Chemotherapy Response in Osteosarcoma Using Convolutional Neural Network of Tumor Center (18)F-FDG PET Images. Diagnostics 2021, 11, 1976. [Google Scholar] [CrossRef]
- Helen, R.; Gurumoorthy, G.; Thennarasu, S.R.; Sakthivel, P.R. Prediction of Osteosarcoma Using Binary Convolutional Neural Network: A Machine Learning Approach. In Proceedings of the 2024 Second International Conference on Emerging Trends in Information, Vellore, India, 22–23 February 2024; Institute of Electrical and Electronics Engineers Inc.: Piscataway, NJ, USA, 2024. [Google Scholar]
- Im, H.-J.; McIlwain, S.; Ong, I.; Lee, I.; Song, C.; Shulkin, B.; Cho, S. Prediction of Response to Neoadjuvant Chemotherapy Using Machine Learning Algorithm Trained by Baseline FDG-PET Textural Parameters in Osteosarcoma. J. Nucl. Med. 2017, 58, 44. [Google Scholar]
- Sheen, H.; Kim, W.; Byun, B.H.; Kong, C.-B.; Lim, I.; Lim, S.M.; Woo, S.-K. Prognostic and Predictive Logistic Model for Osteosarcoma Using Metabolic Imaging Phenotypes. J. Nucl. Med. 2019, 60, 58. [Google Scholar]
- White, L.M.; Atinga, A.; Naraghi, A.M.; Lajkosz, K.; Wunder, J.S.; Ferguson, P.; Tsoi, K.; Griffin, A.; Haider, M. T2-Weighted MRI Radiomics in High-Grade Intramedullary Osteosarcoma: Predictive Accuracy in Assessing Histologic Response to Chemotherapy, Overall Survival, and Disease-Free Survival. Skelet. Radiol. 2023, 52, 553–564. [Google Scholar] [CrossRef]
- Sampath, K.; Rajagopal, S.; Chintanpalli, A. A Comparative Analysis of CNN-Based Deep Learning Architectures for Early Diagnosis of Bone Cancer Using CT Images. Sci. Rep. 2024, 14, 2144. [Google Scholar] [CrossRef]
- Sun, W.; Liu, S.; Guo, J.; Hao, D.; Hou, F.; Wang, H.; Xu, W. A CT-Based Radiomics Nomogram for Distinguishing between Benign and Malignant Bone Tumours. Cancer Imaging 2021, 21, 20. [Google Scholar] [CrossRef]
- Sanmartín, J.; Azuero, P.; Hurtado, R. A Modern Approach to Osteosarcoma Tumor Identification Through Integration of FP-Growth, Transfer Learning and Stacking Model. In International Conference on Information Technology & Systems; Springer Science and Business Media Deutschland GmbH: Cham, Switzerland, 2024; Volume 932 LNNS, pp. 298–307. [Google Scholar]
- Gawade, S.; Bhansali, A.; Patil, K.; Shaikh, D. Application of the Convolutional Neural Networks and Supervised Deep-Learning Methods for Osteosarcoma Bone Cancer Detection. Healthc. Anal. 2023, 3, 100153. [Google Scholar] [CrossRef]
- Bansal, P.; Gehlot, K.; Singhal, A.; Gupta, A. Automatic Detection of Osteosarcoma Based on Integrated Features and Feature Selection Using Binary Arithmetic Optimization Algorithm. Multimed. Tools Appl. 2022, 81, 8807–8834. [Google Scholar] [CrossRef]
- Deng, S.; Huang, Y.; Li, C.; Qian, J.; Wang, X. Auxiliary Diagnosis of Primary Bone Tumors Based on Machine Learning Model. J. Bone Oncol. 2024, 49, 100648. [Google Scholar] [CrossRef]
- Rao, B.D.; Madhavi, K. BCDNet: A Deep Learning Model with Improved Convolutional Neural Network for Efficient Detection of Bone Cancer Using Histology Images. Int. J. Comput. Exp. Sci. Eng. 2024, 10, 988–998. [Google Scholar] [CrossRef]
- Shao, J.; Lin, H.; Ding, L.; Li, B.; Xu, D.; Sun, Y.; Guan, T.; Dai, H.; Liu, R.; Deng, D.; et al. Deep Learning for Differentiation of Osteolytic Osteosarcoma and Giant Cell Tumor around the Knee Joint on Radiographs: A Multicenter Study. Insights Imaging 2024, 15, 35. [Google Scholar] [CrossRef]
- Wang, S.; Shen, Y.; Zeng, F.; Wang, M.; Li, B.; Shen, D.; Tang, X.; Wang, B. Exploiting Biochemical Data to Improve Osteosarcoma Diagnosis with Deep Learning. Health Inf. Sci. Syst. 2024, 12, 31. [Google Scholar] [CrossRef]
- Yang, P.; Jiang, L.; Xiang, Y.; Wei, J.; Zhao, Z.; Cai, H.; Yi, Z.; Li, L. Deep-Learning Model for Differentiation of Pediatric Bone Diseases by Bone Scintigraphy: A Feasibility Study. Eur. J. Nucl. Med. Mol. Imaging 2023, 50, S727. [Google Scholar] [CrossRef]
- Ren, Y.; Guo, Y.; He, Q.; Cheng, Z.; Huang, Q.; Yang, L. Exploring Whether ChatGPT-4 with Image Analysis Capabilities Can Diagnose Osteosarcoma from X-Ray Images. Exp. Hematol. Oncol. 2024, 13, 71. [Google Scholar] [CrossRef]
- Loraksa, C.; Mongkolsomlit, S.; Nimsuk, N.; Uscharapong, M.; Kiatisevi, P. Effectiveness of Learning Systems from Common Image File Types to Detect Osteosarcoma Based on Convolutional Neural Networks (CNNs) Models. J. Imaging 2022, 8, 2. [Google Scholar] [CrossRef]
- Hasei, J.; Nakahara, R.; Otsuka, Y.; Nakamura, Y.; Hironari, T.; Kahara, N.; Miwa, S.; Ohshika, S.; Nishimura, S.; Ikuta, K.; et al. High-Quality Expert Annotations Enhance Artificial Intelligence Model Accuracy for Osteosarcoma X-Ray Diagnosis. Cancer Sci. 2024, 115, 3695–3704. [Google Scholar] [CrossRef]
- Ling, Z.; Yang, S.; Gou, F.; Dai, Z.; Wu, J. Intelligent Assistant Diagnosis System of Osteosarcoma MRI Image Based on Transformer and Convolution in Developing Countries. IEEE J. Biomed. Health Inf. 2022, 26, 5563–5574. [Google Scholar] [CrossRef]
- Xia, G.; Ran, T.; Wu, H.; Wang, M.; Pan, J. The Development of Mask R-CNN to Detect Osteosarcoma and Oste-Ochondroma in X-Ray Radiographs. Comput. Methods Biomech. Biomed. Eng. Imaging Vis. 2023, 11, 1869–1875. [Google Scholar] [CrossRef]
- Song, L.; Li, C.; Tan, L.; Wang, M.; Chen, X.; Ye, Q.; Li, S.; Zhang, R.; Zeng, Q.; Xie, Z.; et al. A Deep Learning Model to Enhance the Classification of Primary Bone Tumors Based on Incomplete Multimodal Images in X-Ray, CT, and MRI. Cancer Imaging 2024, 24, 135. [Google Scholar] [CrossRef]
- Xie, Z.; Zhao, H.; Song, L.; Ye, Q.; Zhong, L.; Li, S.; Zhang, R.; Wang, M.; Chen, X.; Lu, Z.; et al. A Radiograph-Based Deep Learning Model Improves Radiologists’ Performance for Classification of Histological Types of Primary Bone Tumors: A Multicenter Study. Eur. J. Radiol. 2024, 176, 111496. [Google Scholar] [CrossRef]
- He, Y.; Pan, I.; Bao, B.; Halsey, K.; Chang, M.; Liu, H.; Peng, S.; Sebro, R.A.; Guan, J.; Yi, T.; et al. Deep Learning-Based Classification of Primary Bone Tumors on Radiographs: A Preliminary Study. EBioMedicine 2020, 62, 103121. [Google Scholar] [CrossRef]
- Obaid, M.K.; Abed, H.A.; Abdullah, S.B.; Al-Jawahry, H.M.; Majed, S.; Hassan, A.R. Automated Osteosarcoma Detection and Classification Using Advanced Deep Learning with Remora Optimization Algorithm. In Proceedings of the 2023 6th International Conference on Engineering Technology and its Applications (IICETA), Al-Najaf, Iraq, 15–16 July 2023; Institute of Electrical and Electronics Engineers Inc.: Piscataway, NJ, USA, 2023; pp. 122–128. [Google Scholar]
- He, J.; Bi, X. Automatic Classification of Spinal Osteosarcoma and Giant Cell Tumor of Bone Using Optimized DenseNet. J. Bone Oncol. 2024, 46, 100606. [Google Scholar] [CrossRef]
- Malibari, A.A.; Alzahrani, J.S.; Obayya, M.; Negm, N.; Al-Hagery, M.A.; Salama, A.S.; Hilal, A.M. Biomedical Osteosarcoma Image Classification Using Elephant Herd Optimization and Deep Learning. Comput. Mater. Contin. 2022, 73, 6443–6459. [Google Scholar] [CrossRef]
- Rahouma, K.H.; Abdellatif, A.S. Bone Osteosarcoma Tumor Classification. Indones. J. Electr. Eng. Comput. Sci. 2023, 31, 582–587. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, Z.; Zhang, B.; Yang, F. Comprehensive Diagnostic Model for Osteosarcoma Classification Using CT Imaging Features. J. Bone Oncol. 2024, 47, 100622. [Google Scholar] [CrossRef]
- Georgeanu, V.; Mamuleanu, M.-L.; Selisteanu, D. Convolutional Neural Networks for Automated Detection and Classification of Bone Tumors in Magnetic Resonance Imaging. In Proceedings of the 2021 IEEE International Conference on Artificial Intelligence, Robotics, and Communication (ICAIRC), Fuzhou, China, 25–27 June 2021; Institute of Electrical and Electronics Engineers Inc.: Piscataway, NJ, USA, 2021; pp. 5–7. [Google Scholar]
- Sagar, C.V.; Bhan, A. Machine Learning Approach to Classify and Predict Osteosarcoma Grading. In Proceedings of the 2024 International Conference on Automation and Computation (AUTOCOM), Dehradun, India, 14–16 March 2024; Institute of Electrical and Electronics Engineers Inc.: Piscataway, NJ, USA, 2024; pp. 470–474. [Google Scholar]
- Gitto, S.; Albano, D.; Chianca, V.; Cuocolo, R.; Ugga, L.; Messina, C.; Sconfienza, L.M. Machine Learning Classification of Low-Grade and High-Grade Chondrosarcomas Based on MRI-Based Texture Analysis. Semin. Musculoskelet. Radiol. 2019, 23, A025. [Google Scholar] [CrossRef]
- Gitto, S.; Cuocolo, R.; van Langevelde, K.; van de Sande, M.A.J.; Parafioriti, A.; Luzzati, A.; Imbriaco, M.; Sconfienza, L.M.; Bloem, J.L. MRI Radiomics-Based Machine Learning Classification of Atypical Cartilaginous Tumour and Grade II Chondrosarcoma of Long Bones. EBioMedicine 2022, 75, 103757. [Google Scholar] [CrossRef]
- Gitto, S.; Cuocolo, R.; Albano, D.; Chianca, V.; Messina, C.; Gambino, A.; Ugga, L.; Cortese, M.C.; Lazzara, A.; Ricci, D.; et al. MRI Radiomics-Based Machine-Learning Classification of Bone Chondrosarcoma. Eur. J. Radiol. 2020, 128, 109043. [Google Scholar] [CrossRef]
- Vaiyapuri, T.; Jothi, A.; Narayanasamy, K.; Kamatchi, K.; Kadry, S.; Kim, J. Design of a Honey Badger Optimization Algorithm with a Deep Transfer Learning-Based Osteosarcoma Classification Model. Cancers 2022, 14, 6066. [Google Scholar] [CrossRef]
- Jha, A.K.; Nayak, P.; Mithun, S.; Sherkhane, U.; Jaiswar, V.; Nath, B.; Tripathi, A.; Mehta, G.M.; Panchal, S.; Purandare, N.; et al. Development and Validation of Radiomic Signature for Classification of High and Low-Grade Chondrosarcoma: A Pilot Study. Mol. Imaging Biol. 2022, 24, S218. [Google Scholar] [CrossRef]
- Shen, R.; Li, Z.; Zhang, L.; Hua, Y.; Mao, M.; Cai, Z.; Qiu, Y.; Gryak, J.; Najarian, K. Osteosarcoma Patients Classification Using Plain X-Rays and Metabolomic Data. Annu. Int. Conf. IEEE Eng. Med. Biol. Soc. 2018, 2018, 690–693. [Google Scholar] [CrossRef]
- Li, J.; Li, S.; Li, X.; Miao, S.; Dong, C.; Gao, C.; Liu, X.; Hao, D.; Xu, W.; Huang, M.; et al. Primary Bone Tumor Detection and Classification in Full-Field Bone Radiographs via YOLO Deep Learning Model. Eur. Radiol. 2023, 33, 4237–4248. [Google Scholar] [CrossRef]
- Hadi, M.R.; Hassan, A.R.; Mohammed, I.H.; Alazzai, W.K.; Alzubaidi, L.H.; Ai Sadi, H.I. Integrated Design of Artificial Neural Network with Bald Eagle Search Optimization for Osteosarcoma Classification. In Proceedings of the 2023 6th International Conference on Engineering Technology and its Applications (IICETA), Al-Najaf, Iraq, 15–16 July 2023; Institute of Electrical and Electronics Engineers Inc.: Piscataway, NJ, USA, 2023; pp. 552–558. [Google Scholar]
- Guo, C.; Chen, Y.; Li, J. Radiographic Imaging and Diagnosis of Spinal Bone Tumors: AlexNet and ResNet for the Classification of Tumor Malignancy. J. Bone Oncol. 2024, 48, 100629. [Google Scholar] [CrossRef]
- Li, Y.; Dong, B.; Yuan, P. The Diagnostic Value of Machine Learning for the Classification of Malignant Bone Tumor: A Systematic Evaluation and Meta-Analysis. Front. Oncol. 2023, 13, 1207175. [Google Scholar] [CrossRef]
- Gitto, S.; Cuocolo, R.; Annovazzi, A.; Anelli, V.; Acquasanta, M.; Cincotta, A.; Albano, D.; Chianca, V.; Ferraresi, V.; Messina, C.; et al. CT Radiomics-Based Machine Learning Classification of Atypical Cartilaginous Tumours and Appendicular Chondrosarcomas. EBioMedicine 2021, 68, 103407. [Google Scholar] [CrossRef]
- Pan, D.; Liu, R.; Zheng, B.; Yuan, J.; Zeng, H.; He, Z.; Luo, Z.; Qin, G.; Chen, W. Using Machine Learning to Unravel the Value of Radiographic Features for the Classification of Bone Tumors. BioMed Res. Int. 2021, 2021, 8811056. [Google Scholar] [CrossRef]
- von Schacky, C.E.; Wilhelm, N.J.; Schäfer, V.S.; Leonhardt, Y.; Jung, M.; Jungmann, P.M.; Russe, M.F.; Foreman, S.C.; Gassert, F.G.; Gassert, F.T.; et al. Development and Evaluation of Machine Learning Models Based on X-Ray Radiomics for the Classification and Differentiation of Malignant and Benign Bone Tumors. Eur. Radiol. 2022, 32, 6247–6257. [Google Scholar] [CrossRef]
- Gitto, S.; Annovazzi, A.; Nulle, K.; Interlenghi, M.; Salvatore, C.; Anelli, V.; Baldi, J.; Messina, C.; Albano, D.; Di Luca, F.; et al. X-Rays Radiomics-Based Machine Learning Classification of Atypical Cartilaginous Tumour and High-Grade Chondrosarcoma of Long Bones. EBioMedicine 2024, 101, 105018. [Google Scholar] [CrossRef]
- von Schacky, C.E.; Wilhelm, N.J.; Schäfer, V.S.; Leonhardt, Y.; Gassert, F.G.; Foreman, S.C.; Gassert, F.T.; Jung, M.; Jungmann, P.M.; Russe, M.F.; et al. Multitask Deep Learning for Segmentation and Classification of Primary Bone Tumors on Radiographs. Radiology 2021, 301, 398–406. [Google Scholar] [CrossRef]
- Zhong, J.; Hu, Y.; Ge, X.; Xing, Y.; Ding, D.; Zhang, G.; Zhang, H.; Yang, Q.; Yao, W. A Systematic Review of Radiomics in Chondrosarcoma: Assessment of Study Quality and Clinical Value Needs Handy Tools. Eur. Radiol. 2023, 33, 1433–1444. [Google Scholar] [CrossRef]
- Wu, J.; Xiao, P.; Huang, H.; Gou, F.; Zhou, Z.; Dai, Z. An Artificial Intelligence Multiprocessing Scheme for the Diagnosis of Osteosarcoma MRI Images. IEEE J. Biomed. Health Inf. 2022, 26, 4656–4667. [Google Scholar] [CrossRef]
- Zhan, X.; Liu, J.; Long, H.; Zhu, J.; Tang, H.; Gou, F.; Wu, J. An Intelligent Auxiliary Framework for Bone Malignant Tumor Lesion Segmentation in Medical Image Analysis. Diagnostics 2023, 13, 223. [Google Scholar] [CrossRef]
- Zhong, X.; Gou, F.; Wu, J. An Intelligent MRI Assisted Diagnosis and Treatment System for Osteosarcoma Based on Super-Resolution. Complex Intell. Syst. 2024, 10, 6031–6050. [Google Scholar] [CrossRef]
- Lv, B.; Liu, F.; Li, Y.; Nie, J.; Gou, F.; Wu, J. Artificial Intelligence-Aided Diagnosis Solution by Enhancing the Edge Features of Medical Images. Diagnostics 2023, 13, 1063. [Google Scholar] [CrossRef]
- Wang, L.; Yu, L.; Zhu, J.; Tang, H.; Gou, F.; Wu, J. Auxiliary Segmentation Method of Osteosarcoma in MRI Images Based on Denoising and Local Enhancement. Healthcare 2022, 10, 1468. [Google Scholar] [CrossRef]
- Liu, F.; Zhu, J.; Lv, B.; Yang, L.; Sun, W.; Dai, Z.; Gou, F.; Wu, J. Auxiliary Segmentation Method of Osteosarcoma MRI Image Based on Transformer and U-Net. Comput. Intell. Neurosci. 2022, 2022, 9990092. [Google Scholar] [CrossRef]
- Wu, J.; Liu, Z.; Gou, F.; Zhu, J.; Tang, H.; Zhou, X.; Xiong, W. BA-GCA Net: Boundary-Aware Grid Contextual Attention Net in Osteosarcoma MRI Image Segmentation. Comput. Intell. Neurosci. 2022, 2022, 3881833. [Google Scholar] [CrossRef]
- Lim, C.C.; Ling, A.H.W.; Chong, Y.F.; Mashor, M.Y.; Alshantti, K.; Aziz, M.E. Comparative Analysis of Image Processing Techniques for Enhanced MRI Image Quality: 3D Reconstruction and Segmentation Using 3D U-Net Architecture. Diagnostics 2023, 13, 2377. [Google Scholar] [CrossRef]
- Wu, Y.; Li, J.; Wang, X.; Zhang, Z.; Zhao, S. DECIDE: A Decoupled Semantic and Boundary Learning Network for Precise Osteosarcoma Segmentation by Integrating Multi-Modality MRI. Comput. Biol. Med. 2024, 174, 108308. [Google Scholar] [CrossRef]
- Wu, J.; Yang, S.; Gou, F.; Zhou, Z.; Xie, P.; Xu, N.; Dai, Z. Intelligent Segmentation Medical Assistance System for MRI Images of Osteosarcoma in Developing Countries. Comput. Math. Methods Med. 2022, 2022, 7703583. [Google Scholar] [CrossRef]
- Dionísio, F.C.F.; Oliveira, L.S.; Hernandes, M.A.; Engel, E.E.; Rangayyan, R.M.; Azevedo-Marques, P.M.; Nogueira-Barbosa, M.H. Manual and Semiautomatic Segmentation of Bone Sarcomas on MRI Have High Similarity. Braz. J. Med. Biol. Res. 2020, 53, e8962. [Google Scholar] [CrossRef]
- Zhang, R.; Huang, L.; Xia, W.; Zhang, B.; Qiu, B.; Gao, X. Multiple Supervised Residual Network for Osteosarcoma Segmentation in CT Images. Comput. Med. Imaging Graph. 2018, 63, 1–8. [Google Scholar] [CrossRef]
- Shen, Y.; Gou, F.; Dai, Z. Osteosarcoma MRI Image-Assisted Segmentation System Base on Guided Aggregated Bilateral Network. Mathematics 2022, 10, 1090. [Google Scholar] [CrossRef]
- Ørum, L.; Banke, K.; Borgwardt, L.; Hansen, A.; Højgaard, L.; Andersen, F.; Ladefoged, C. Pediatric Sarcoma Segmentation Using Deep Learning. J. Nucl. Med. 2019, 60, 1208. [Google Scholar]
- Kaur, C.; Grag, U. Preprocessing and Segmentation of MRI Images for Bone Cancer Detection Using Aurous Spatial Pooling With Deeplabv3. Grenze Sci. Soc. 2024, 2, 2374–2383. [Google Scholar]
- Ouyang, T.; Yang, S.; Gou, F.; Dai, Z.; Wu, J. Rethinking U-Net from an Attention Perspective with Transformers for Osteosarcoma MRI Image Segmentation. Comput. Intell. Neurosci. 2022, 2022, 7973404. [Google Scholar] [CrossRef]
- Zou, B.; Chen, Y.; Chen, Z.; Sun, Y.; Huang, Y.; Qin, F.; Wang, C. RTUNet++: Assessment of Osteosarcoma MRI Image Segmentation Leveraging Hybrid CNN-Transformer Approach with Dense Skip Connection. In Proceedings of the 2023 8th International Conference on Signal and Image Processing (ICSIP), Wuxi, China, 8–10 July 2023; Institute of Electrical and Electronics Engineers Inc.: Piscataway, NJ, USA, 2023; pp. 217–223. [Google Scholar]
- Baidya Kayal, E.; Kandasamy, D.; Sharma, R.; Bakhshi, S.; Mehndiratta, A. Segmentation of Osteosarcoma Tumor Using Diffusion Weighted MRI: A Comparative Study Using Nine Segmentation Algorithms. Signal Image Video Process 2020, 14, 727–735. [Google Scholar] [CrossRef]
- Zhou, Z.; Xie, P.; Dai, Z.; Wu, J. Self-Supervised Tumor Segmentation and Prognosis Prediction in Osteosarcoma Using Multiparametric MRI and Clinical Characteristics. Comput. Methods Programs Biomed. 2024, 244, 107974. [Google Scholar] [CrossRef]
- Cilengir, A.H.; Evrimler, S.; Serel, T.A.; Uluc, E.; Tosun, O. The Diagnostic Value of Magnetic Resonance Imaging-Based Texture Analysis in Differentiating Enchondroma and Chondrosarcoma. Skelet. Radiol. 2023, 52, 1039–1049. [Google Scholar] [CrossRef]
- Yin, P.; Wang, W.; Wang, S.; Liu, T.; Sun, C.; Liu, X.; Chen, L.; Hong, N. The Potential for Different Computed Tomography-Based Machine Learning Networks to Automatically Segment and Differentiate Pelvic and Sacral Osteosarcoma from Ewing’s Sarcoma. Quant. Imaging Med. Surg. 2023, 13, 3174–3184. [Google Scholar] [CrossRef]
- Consalvo, S.; Hinterwimmer, F.; Neumann, J.; Steinborn, M.; Salzmann, M.; Seidl, F.; Lenze, U.; Knebel, C.; Rueckert, D.; Burgkart, R.H.H. Two-Phase Deep Learning Algorithm for Detection and Differentiation of Ewing Sarcoma and Acute Osteomyelitis in Paediatric Radiographs. Anticancer Res. 2022, 42, 4371–4380. [Google Scholar] [CrossRef]
- Arunachalam, H.B.; Mishra, R.; Daescu, O.; Cederberg, K.; Rakheja, D.; Sengupta, A.; Leonard, D.; Hallac, R.; Leavey, P. Viable and Necrotic Tumor Assessment from Whole Slide Images of Osteosarcoma Using Machine-Learning and Deep-Learning Models. PLoS ONE 2019, 14, e0210706. [Google Scholar] [CrossRef]
- Prexler, C.; Kesper, M.S.; Mustafa, M.; Seemann, W.; Schmidt, O.; Gall, K.; Specht, K.; Rechl, H.; Knebel, C.; Woertler, K.; et al. Radiogenomics in Ewing Sarcoma: Integration of Functional Imaging and Transcriptomics Characterizes Tumor Glucose Uptake. Eur. J. Nucl. Med. Mol. Imaging 2019, 46, S694. [Google Scholar] [CrossRef]
- Bhatt, S.; Butola, A.; Kumar, A.; Thapa, P.; Joshi, A.; Jadhav, S.; Singh, N.; Prasad, D.K.; Agarwal, K.; Mehta, D.S. Single-Shot Multispectral Quantitative Phase Imaging of Biological Samples Using Deep Learning. Appl. Opt. 2023, 62, 3989–3999. [Google Scholar] [CrossRef]
- McCulloch, R.A.; Frisoni, T.; Kurunskal, V.; Donati, D.M.; Jeys, L. Computer Navigation and 3d Printing in the Surgical Management of Bone Sarcoma. Cells 2021, 10, 195. [Google Scholar] [CrossRef]
- Shrivastava, A.; Nag, M.K. Enhancing Bone Cancer Diagnosis Through Image Extraction and Machine Learning: A State-of-the-Art Approach. Surg. Innov. 2024, 31, 58–70. [Google Scholar] [CrossRef]
| Author | Year | Study Type | Imaging Modality | AI Model | Performance Metrics | Tumor Type |
|---|---|---|---|---|---|---|
| Gitto et al. [10] | 2022 | Retrospective | MRI | 2D vs. 3D Radiomics | 3D is superior in reproducibility | Ewing Sarcoma |
| Gitto et al. [11] | 2022 | Retrospective | MRI | 3D Radiomics | Feature reproducibility in predicting NAC response | Ewing Sarcoma |
| Lin et al. [12] | 2020 | Retrospective | MRI | Delta-Radiomics | AUC 0.871 (train), 0.843 (validation) | Osteosarcoma |
| He et al. [13] | 2022 | Multicenter | MRI | LR, DT, SVM, NN | AUC 0.893 | High-Grade Bone Sarcoma |
| Zhong et al. [14] | 2022 | Retrospective | MRI | DL + Radiomics Nomogram | AUC 0.793 (95% CI 0.610–0.975) | Osteosarcoma |
| Nie et al. [15] | 2024 | Retrospective | CT | DLRM | AUC 0.879 (95% CI 0.802–0.956) | Chondrosarcoma |
| Teo et al. [16] | 2022 | Retrospective | MRI | SVM (RBF) | Accuracy improved >95% with DCE-MRI | Osteosarcoma (Pediatric) |
| Ho et al. [17] | 2020 | Retrospective | MRI | Deep Interactive Learning (DIaL) | CNN training in 7 h | Osteosarcoma |
| Fu et al. [18] | 2020 | Retrospective | Histology (H&E) | Siamese Network (DS-Net) | Accuracy 95.1% | Osteosarcoma |
| Kim et al. [19] | 2018 | Retrospective | PET | DL + Radiomics | Higher prediction accuracy | Osteosarcoma |
| Hu et al. [20] | 2021 | Retrospective | DWI-MRI | CSDCNN | Better PSNR, MSE, EPI, accuracy, recall, F1, ADC stats | Osteosarcoma |
| Djuričić et al. [21] | 2023 | Retrospective | MRI | Fractal Radiomics + LASSO | AUC 0.95 | Osteosarcoma |
| Zhang et al. [22] | 2024 | Retrospective | DWI-MRI | ML Radiomics Nomogram | AUC 0.848 | Osteosarcoma |
| Huang et al. [23] | 2020 | Retrospective | Multi-parametric MRI | ML Model | AUCs: 0.93–0.97 | Osteosarcoma |
| Zhang et al. [24] | 2021 | Retrospective | DCE-MRI | KNN, SVM, LR | AUCs: 0.86, 0.92, 0.93 | Osteosarcoma |
| Zhang et al. [25] | 2024 | Retrospective | MRI | Radiomics (pre/post NAC) | AUC 0.999 (post), 0.915 (pre) | Osteosarcoma |
| Mori et al. [26] | 2024 | Retrospective | MRI (T1, T2) | Texture Analysis | AUCs 0.99 (T1), 0.94 (T2) | Osteosarcoma |
| Chen et al. [27] | 2021 | Multicenter | MRI | LASSO-LR | Radiomics signature prediction (no specific AUC reported) | Osteosarcoma |
| Miedler et al. [28] | 2023 | Retrospective | MRI | Radiomics | Predictive potential (no numerical metrics) | Ewing Sarcoma |
| Chaber et al. [29] | 2019 | Retrospective | IR Spectroscopy | ML | Accuracy 92% | Ewing Sarcoma |
| Dufau et al. [30] | 2019 | Retrospective | PET | ML + Radiomics | AUC 0.98, sensitivity 100% | Osteosarcoma |
| Jeong et al. [31] | 2019 | Retrospective | PET | Linear SVM + PCA | Improved AUC (no number) | Osteosarcoma |
| Bouhamama et al. [32] | 2022 | Retrospective | MRI | Radiomics | AUC 0.97 | Osteosarcoma |
| Kim et al. [33] | 2021 | Retrospective | PET | CNN | Predictive (no numerical metrics) | Osteosarcoma |
| Helen et al. [34] | 2024 | Retrospective | PET | Binary CNN | Improved prediction | Osteosarcoma |
| Im et al. [35] | 2017 | Retrospective | PET | ML Using FDG-PET | Prognostic FDG-based features for NAC prediction | Osteosarcoma |
| Sheen et al. [36] | 2019 | Retrospective | PET | Logistic Model | SUVmax + GLZLM_SZLGE as predictors | Osteosarcoma |
| White et al. [37] | 2023 | Retrospective | T2 MRI | Radiomics | AUC 0.708 ± 0.046 | High-Grade Osteosarcoma |
| Author | Year | Study Type | Imaging Modality | AI Model | Performance Metrics | Tumor Type |
|---|---|---|---|---|---|---|
| Sampath et al. [38] | 2024 | Retrospective | CT | AlexNet | Accuracy 100% | Parosteal Osteosarcoma, Osteochondroma, Enchondroma |
| Sun et al. [39] | 2021 | Retrospective | CT | Radiomics + Clinical Model | AUC 0.823 | Bone Tumors |
| Sanmartín et al. [40] | 2024 | Retrospective | Histology | FP-Growth + Transfer Learning + Stacking | Noise reduction and variation minimization | Osteosarcoma |
| Gawade et al. [41] | 2023 | Retrospective | MRI | ResNet101 (best among VGG16, VGG19, DenseNet) | Accuracy 90.36%, precision 89.51%, AUC 0.9461 | Osteosarcoma |
| Bansal et al. [42] | 2022 | Retrospective | WSI | IF-FSM-C | Accuracy 96.08% | Osteosarcoma |
| Deng et al. [43] | 2024 | Retrospective | Histopathology | CNN | 99.8% (normal vs. tumor), 71.2% (benign vs. malignant), PPV 91.9% | Bone Tumors |
| Rao et al. [44] | 2024 | Retrospective | Histology | BCDNet | Accuracy: 96.29% (binary), 94.69% (multi-class) | Bone Cancer |
| Shao et al. [45] | 2024 | Multicenter | X-Ray | DL model | Accuracy 93.1% | Osteosarcoma vs. GCT |
| Wang et al. [46] | 2024 | Retrospective | X-Ray + Labs | DL + ALP + LDH | Accuracy 97.17% | Osteosarcoma |
| Yang et al. [47] | 2023 | Retrospective | Nuclear Medicine | CNN | Accuracy 96.17%, specificity 91.67% | Pediatric Bone Disease |
| Ren et al. [48] | 2024 | Retrospective | X-Ray | ChatGPT-4 | Specificity is 100%, but lower sensitivity | Osteosarcoma |
| Loraksa et al. [49] | 2022 | Retrospective | X-Ray | CNN | Accuracy 96.4% (internal), 92.0% (external) | Osteosarcoma |
| Hasei et al. [50] | 2024 | Retrospective | X-Ray | U-Net | Sensitivity 95.52%, specificity 96.21% | Pediatric Osteosarcoma |
| Ling et al. [51] | 2022 | Retrospective | MRI | DUconViT (Transformer + CNN) | Dice similarity coefficient 92.4% | Osteosarcoma |
| Xia et al. [52] | 2023 | Retrospective | X-Ray | Mask R-CNN | Precision 92% | Osteosarcoma, Osteochondroma |
| Author | Year | Study Type | Imaging Modality | AI Model | Performance Metrics | Tumor Type |
|---|---|---|---|---|---|---|
| Song et al. [53] | 2024 | Retrospective | X-ray, CT, MRI | Multimodal DL Model | Micro-average AUC 0.847 | Primary Bone Tumors |
| Xie et al. [54] | 2024 | Multicenter | Radiograph | DL + Radiologist | Macro-average AUC 0.904/0.873 | Primary Bone Tumors |
| He et al. [55] | 2020 | Preliminary | Radiograph | DL Model | AUC: benign/non-benign 0.894/0.877; malignant 0.907/0.916 | Primary Bone Tumors |
| Obaid et al. [56] | 2023 | Retrospective | CT | DL + Remora Optimization | High accuracy (not specified) | Osteosarcoma |
| He & Bi [57] | 2024 | Retrospective | MRI | Optimized DenseNet | Improved classification performance | Spinal Osteosarcoma vs. GCT |
| Malibari et al. [58] | 2022 | Retrospective | Image | Elephant Herd Optimization + DL | Effective classification | Osteosarcoma |
| Rahouma et al. [59] | 2023 | Retrospective | CT | XGBoost, SVM, KNN | Diagnostic model for osteosarcoma | Osteosarcoma |
| Wang et al. [60] | 2024 | Retrospective | CT | PCA-IPSO + SVM | Outperforms traditional feature selection | Osteosarcoma |
| Georgeanu et al. [61] | 2021 | Retrospective | MRI | CNN | Automated detection and classification | Bone Tumors |
| Sagar & Bhan [62] | 2024 | Retrospective | Not Specified | ML Model | Osteosarcoma grading classification | Osteosarcoma |
| Gitto et al. [63] | 2019 | Retrospective | MRI | Texture Analysis + ML | Low vs. high-grade chondrosarcoma classification | Chondrosarcoma |
| Gitto et al. [64] | 2022 | Retrospective | MRI | Radiomics + ML | ACT vs. grade II chondrosarcoma | Chondrosarcoma |
| Gitto et al. [65] | 2020 | Retrospective | MRI | Radiomics + ML | Bone chondrosarcoma classification | Chondrosarcoma |
| Vaiyapuri et al. [66] | 2022 | Retrospective | Image | Honey Badger Opt. + Transfer Learning | High diagnostic accuracy | Osteosarcoma |
| Jha et al. [67] | 2022 | Retrospective | MRI | Radiomic Signature | High vs. low-grade classification | Chondrosarcoma |
| Shen et al. [68] | 2018 | Retrospective | X-ray + Metabolomics | ML Model | Enhanced classification using combined features | Osteosarcoma |
| Li et al. [69] | 2023 | Retrospective | Full-field Radiograph | YOLO DL Model | Multi-class: normal, benign, intermediate, malignant | Primary Bone Tumors |
| Hadi et al. [70] | 2023 | Retrospective | Image | Bald Eagle Optimization + ANN | High accuracy | Osteosarcoma |
| Guo et al. [71] | 2024 | Retrospective | Radiograph | AlexNet and ResNet | Tumor malignancy classification | Spinal Bone Tumors |
| Li et al. [72] | 2023 | Meta-analysis | Multiple | ML Models | Diagnostic value confirmed | Malignant Bone Tumors |
| Gitto et al. [73] | 2021 | Retrospective | CT | Radiomics + ML | ACT vs. appendicular chondrosarcoma | Chondrosarcoma |
| Pan et al. [74] | 2021 | Retrospective | Radiograph | ML Model | Radiographic feature classification | Bone Tumors |
| Von Schacky et al. [75] | 2022 | Retrospective | X-Ray | ANN + RFC + GNB | AUC 0.79/0.90 | Primary Bone Tumors |
| Gitto et al. [76] | 2024 | Retrospective | X-Ray | Radiomics + ML | ACT vs. high-grade chondrosarcoma | Chondrosarcoma |
| von Schacky et al. [77] | 2021 | Retrospective | Radiograph | Multitask DL | Accuracy 80.2%, better than residents, comparable to radiologists | Primary Bone Tumors |
| Author | Year | Study Type | Imaging Modality | AI Model | Performance Metrics | Tumor Type |
|---|---|---|---|---|---|---|
| Zhong et al. [78] | 2023 | Systematic Review | MRI | Manual Segmentation | 0.90–0.94 (AUC) | Chondrosarcoma |
| Wu et al. [79] | 2022 | Retrospective | MRI | ETUNet + SBF + NLM + CRF | DSC > 90%, Accuracy 95.67% | Osteosarcoma |
| Zhan et al. [80] | 2023 | Retrospective | MRI | SEAGNET | DSC 0.967, Accuracy 0.996 | Bone Tumors |
| Zhong et al. [81] | 2024 | Retrospective | MRI | NSRDN with HRNet | DSC 96.4%, IoU 92.8%, Accuracy 95.5% | Osteosarcoma |
| Lv et al. [82] | 2023 | Retrospective | MRI | TBNet | DSC 0.949, Accuracy 0.997 | Osteosarcoma |
| Wang et al. [83] | 2022 | Retrospective | MRI | Eformer + DFANet | Accuracy 0.995 | Osteosarcoma |
| Liu et al. [84] | 2022 | Retrospective | MRI | OSTransNet | DSC 0.949, IoU 0.904 | Osteosarcoma |
| Wu et al. [85] | 2022 | Retrospective | MRI | BA-GCA Net | DSC 0.927, IoU 0.880 | Osteosarcoma |
| Lim et al. [86] | 2023 | Retrospective | MRI | 3D U-Net (MONAI) | DSC 83.75–87.62% | Osteosarcoma |
| Wu et al. [87] | 2024 | Retrospective | MRI | DECIDE | DSC 70.40%, IoU 54.50% | Osteosarcoma |
| Wu et al. [88] | 2022 | Retrospective | MRI | OSDCN (SepUNet + CRF) | DSC 0.914, IoU 0.883 | Osteosarcoma |
| Dionísio et al. [89] | 2020 | Retrospective | MRI | Manual and Semi-Automatic | DSC 0.71–0.97 | Bone Sarcomas |
| Zhang et al. [90] | 2018 | Retrospective | CT | MSRN | DSC 89.22%, F1 0.9305 | Osteosarcoma |
| Shen et al. [91] | 2022 | Retrospective | MRI | OSGABN (FaBiNet) | DSC 0.915, IoU 0.853 | Osteosarcoma |
| Ørum et al. [92] | 2019 | Retrospective | PET/CT | U-Net | Precision 0.71, sensitivity 0.39–0.54 | Pediatric Sarcoma |
| Kaur et al. [93] | 2024 | Retrospective | MRI | Modified DeepLabV3+ (ASPP) | DSC 70.40%, IoU 54.50% | Bone Cancer |
| Ouyang et al. [94] | 2022 | Retrospective | MRI | UATransNet | DSC 0.921, IoU 0.922 | Osteosarcoma |
| Zou et al. [95] | 2023 | Retrospective | MRI | RTUNet++ | DSC 0.82 | Osteosarcoma |
| Kayal et al. [96] | 2020 | Retrospective | DWI-MRI | SLIC-S and FCM | DSC ~82%, ~79% | Osteosarcoma |
| Zhou et al. [97] | 2024 | Retrospective | MRI | MPFNet | DSC 84.19%, HQSR 94.38% | Osteosarcoma |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Papageorgiou, P.S.; Christodoulou, R.; Korfiatis, P.; Papagelopoulos, D.P.; Papakonstantinou, O.; Pham, N.; Woodward, A.; Papagelopoulos, P.J. Artificial Intelligence in Primary Malignant Bone Tumor Imaging: A Narrative Review. Diagnostics 2025, 15, 1714. https://doi.org/10.3390/diagnostics15131714
Papageorgiou PS, Christodoulou R, Korfiatis P, Papagelopoulos DP, Papakonstantinou O, Pham N, Woodward A, Papagelopoulos PJ. Artificial Intelligence in Primary Malignant Bone Tumor Imaging: A Narrative Review. Diagnostics. 2025; 15(13):1714. https://doi.org/10.3390/diagnostics15131714
Chicago/Turabian StylePapageorgiou, Platon S., Rafail Christodoulou, Panagiotis Korfiatis, Dimitra P. Papagelopoulos, Olympia Papakonstantinou, Nancy Pham, Amanda Woodward, and Panayiotis J. Papagelopoulos. 2025. "Artificial Intelligence in Primary Malignant Bone Tumor Imaging: A Narrative Review" Diagnostics 15, no. 13: 1714. https://doi.org/10.3390/diagnostics15131714
APA StylePapageorgiou, P. S., Christodoulou, R., Korfiatis, P., Papagelopoulos, D. P., Papakonstantinou, O., Pham, N., Woodward, A., & Papagelopoulos, P. J. (2025). Artificial Intelligence in Primary Malignant Bone Tumor Imaging: A Narrative Review. Diagnostics, 15(13), 1714. https://doi.org/10.3390/diagnostics15131714








