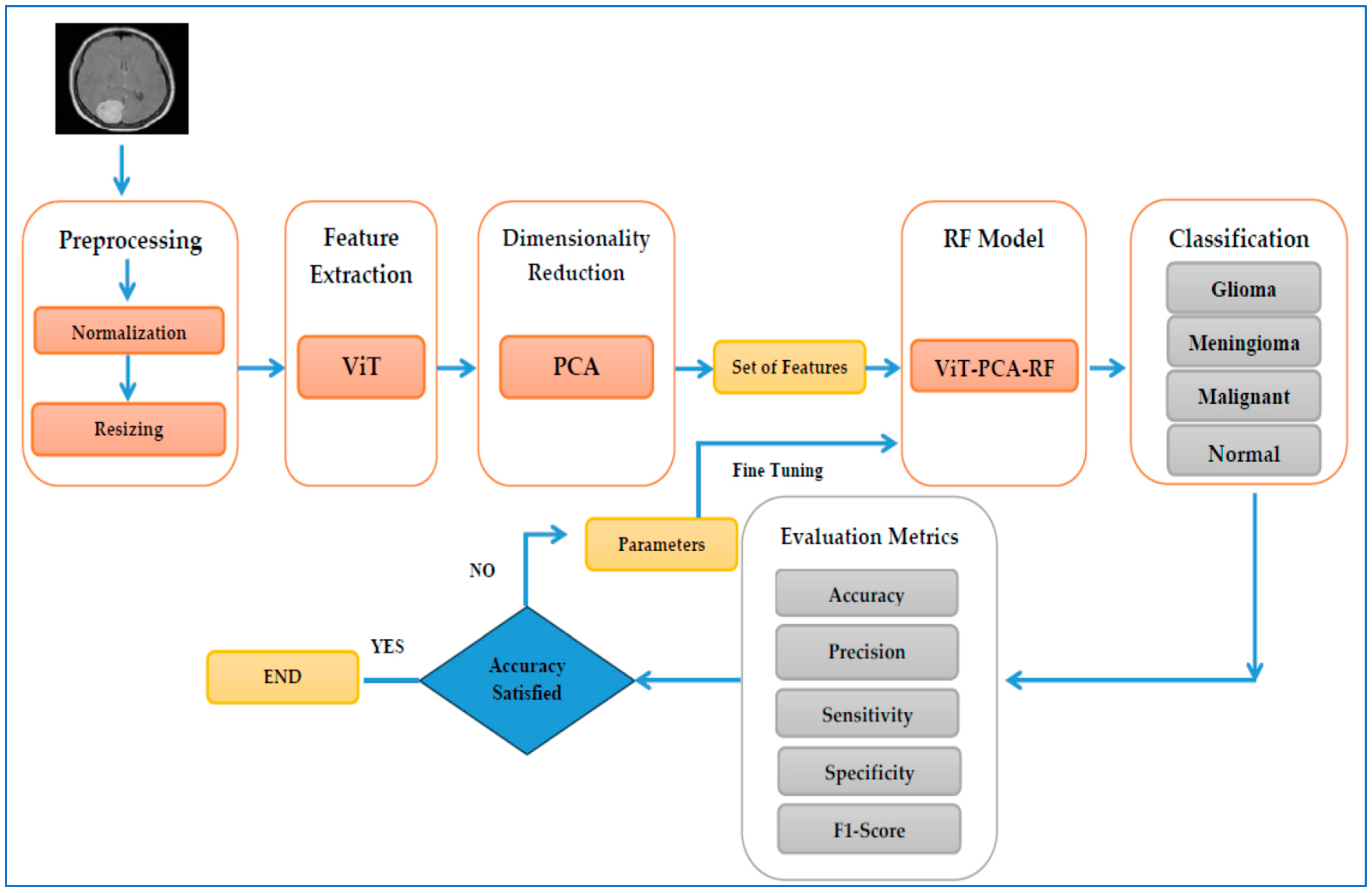
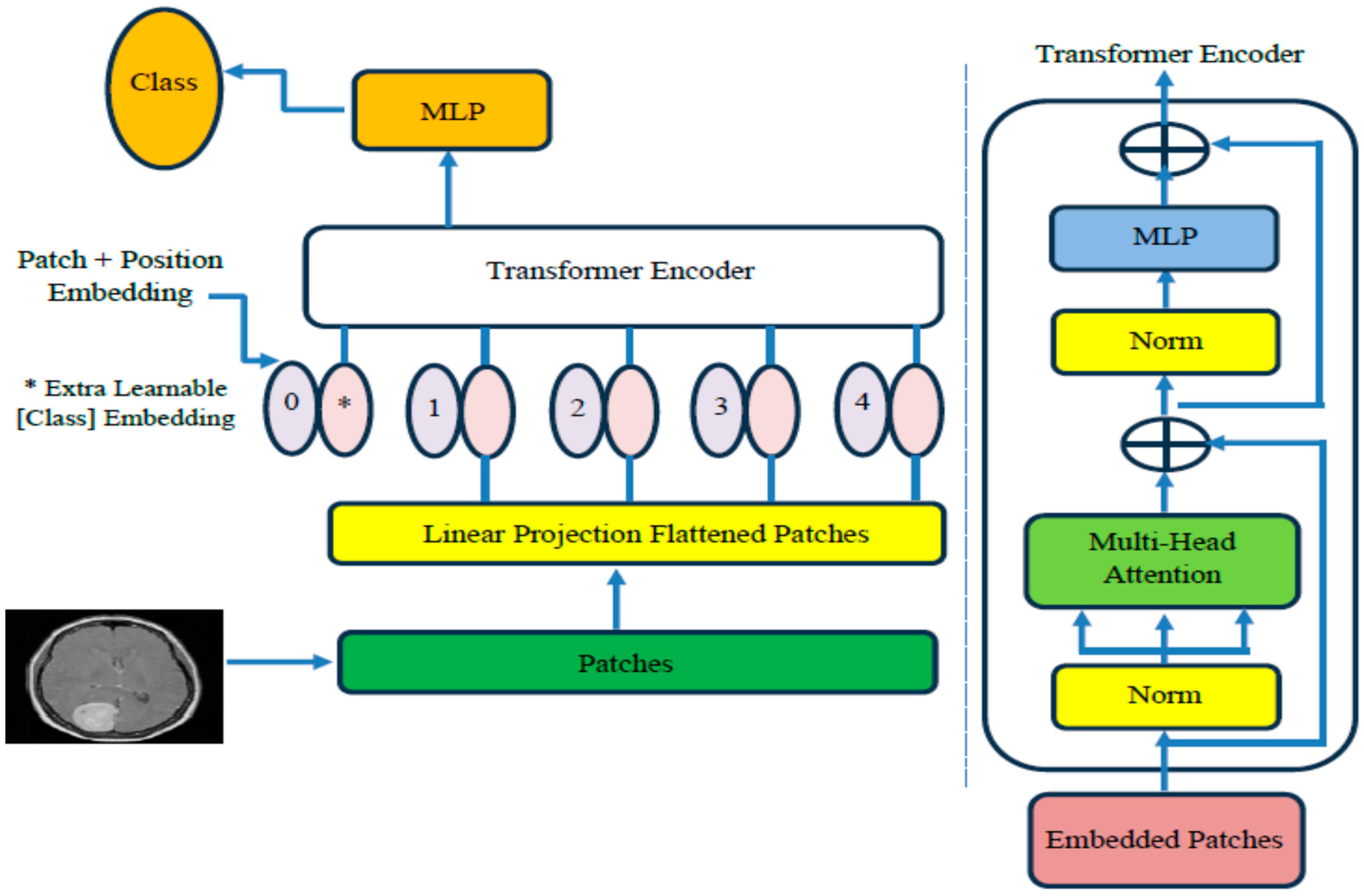
4.3. The ViT-PCA-RF Model Assessment
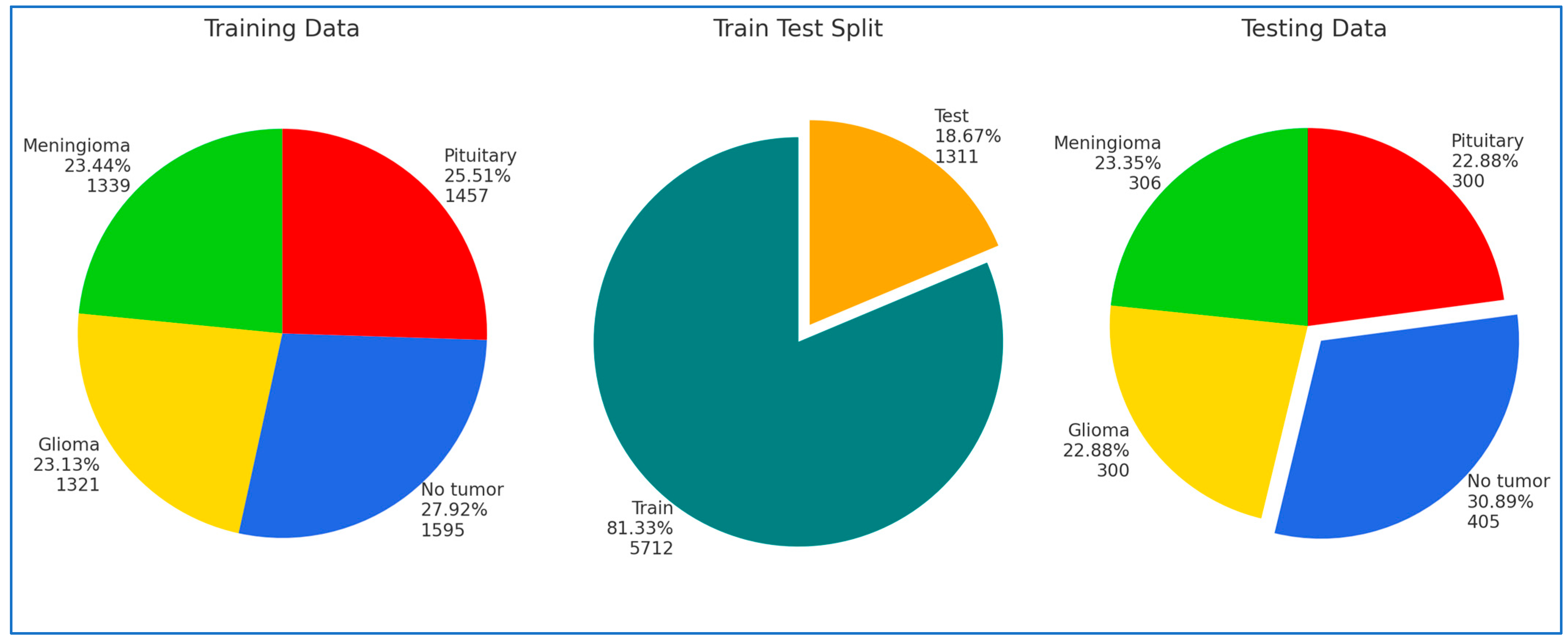
In our research, we conducted four experiments on the Kaggle platform utilizing BTM and Figshare datasets. For the training phase, we partitioned the BTM dataset into 81.33% of the total images, amounting to 5712 MRI images. The remaining 18.67%, consisting of 1311 MRI images, was designated as the test set. The Figshare dataset was divided into 70% for training, 15% for testing, and 15% for validation.
The first experiment compared the ViT-PCA-RF model against three other hybrid machine learning models: ViT-PCA-DT, ViT-PCA-XGB, and ViT-PCA-SVM. In the second experiment, we evaluated ViT’s feature extraction capabilities in comparison to VGG16, ResNet50, DenseNet201, and Xception. The third and fourth experiments involved external validation of the ViT-PCA-RF model using an external dataset from Figshare. Following each experiment, the identified metrics (Equations (9)–(15)) were employed to assess the performance of the hybrid models.
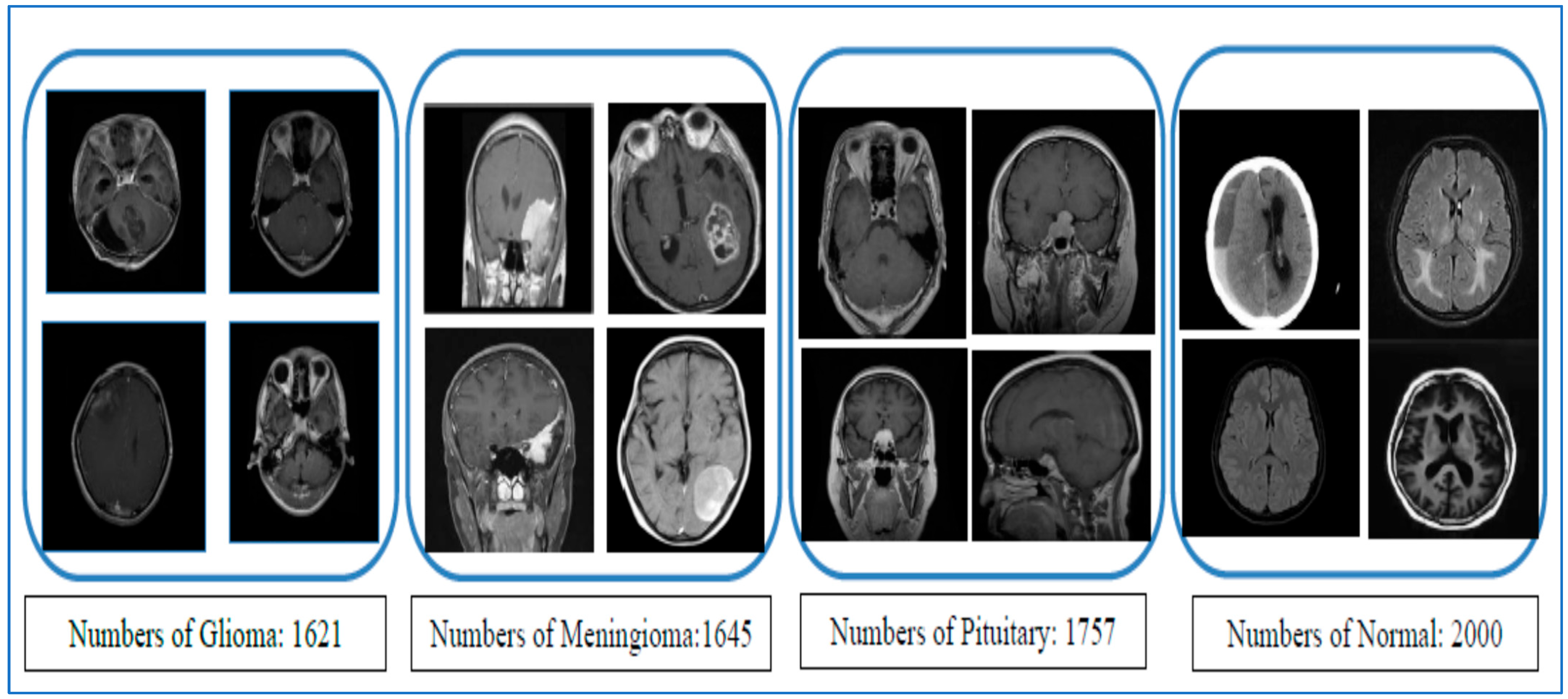
Our first experiment categorized brain tumors into multiple classes using the test set of the BTM dataset. This involved utilizing ViT for feature extraction, PCA to adjust feature dimensions, and RF, DT, XGB, and SVM for classification purposes. The primary goal of this experiment was to distinguish between different types of brain tumors, leading to improved patient outcomes and a more efficient diagnostic process, ultimately reducing both time and costs for patients. The BTM dataset comprises four main classes: Normal, Meningioma, Pituitary, and Glioma. The outcomes of the ViT-PCA-RF, ViT-PCA-DT, ViT-PCA-XGB, and ViT-PCA-SVM hybrid models are detailed in
Table 8, respectively. These tables present the average evaluation metrics for the four hybrid models used in the multi-classification task over the BTM dataset’s test set. The average accuracies achieved were 99%, 98.2%, 98.9%, and 98.8% for ViT-PCA-RF, ViT-PCA-DT, ViT-PCA-XGB, and ViT-PCA-SVM, respectively. Thus, the model ViT-PCA-RF demonstrated the highest level of accuracy.
ViT-PCA-DT achieved an average of 98.9% specificity, 3.8% FNR, 98.9% NPV, 96.5% precision, 96.5% recall, and 96.5% F1 score. ViT-PCA-XGB achieved the following average scores: 99.2% for specificity, 2.5% for FNR, 99.3% for NPV, 97.7% for precision, 97.7% for recall, and 97.7% for F1 score. ViT-PCA-SVM achieved average scores of 99.2% for specificity, 2.5% for FNR, 99.2% for NPV, 97.6% for precision, 97.6% for recall, and 97.6% for F1 score.
Therefore, ViT-PCA-RF demonstrated outstanding performance across various metrics. It achieved the highest averages for accuracy, specificity, NPV, precision, recall, and F1 score, at 99.4%, 99.4%, 98.1%, 98.1%, and 98.1%, respectively. Additionally, it boasted the lowest average FNR at 2.1%. This signifies that ViT-PCA-RF excels in multi-classification tasks, showcasing strong performance across various evaluation criteria.
The test set of the BTM dataset was divided into four main groups: Glioma, Meningioma, Normal, and Pituitary. We evaluated the effectiveness of four new hybrid models—ViT-PCA-RF, ViT-PCA-DT, ViT-PCA-XGB, and ViT-PCA—using a range of assessment measures such as accuracy, specificity, FNR, NPV, precision, recall, and F1 score for each category as shown in
Table 9,
Table 10,
Table 11 and
Table 12.
Among the different models assessed, ViT_PCA_RF stood out in the Glioma category, achieving exceptional results with the highest accuracy, specificity, precision, and F1 score, at 98.47%, 99.60%, 98.61%, and 96.60%, respectively. On the other hand, ViT-PCA-SVM demonstrated the highest NPV and recall, at 98.7% and 95.7%, while also exhibiting the lowest FNR of 4.3%.
In class Meningioma, ViT_PCA_RF achieved the highest performance metrics with the following scores: accuracy: 98.55%, specificity: 98.81%, NPV: 99.30%, precision: 96.14%, recall: 97.71%, F1 score: 96.92%. Furthermore, it demonstrated the lowest FNR at 2.29%.
In the Normal class, the ViT-PCA-RF model showed the most substantial performance metrics, boasting the highest accuracy, specificity, precision, and F1 score rates of 99.85%, 99.78%, 99.51%, and 99.75%, respectively. Furthermore, within the ViT-PCA-RF, ViT-PCA-DT, and ViT-PCA-XGB models, the NPV and recall rates reached 100%, while also showcasing the lowest false negative rate of 0%.
In the Pituitary class, ViT-PCA-SVM achieved the highest F1 score accuracy, specificity, precision, and F1 score scores, with 99.5%, 99.5%, 98.3%, and 98.8%, respectively. ViT-PCA-SVM and ViT-PCA-RF both reached the greatest NPV at 99.8%. Additionally, ViT-PCA-RF obtained the top overall score at 99.33%. Moreover, ViT-PCA-RF showcased the best recall rate at 99.33% and achieved the lowest FNR at 0.67%.
Figure 6 illustrates the performance results of the hybrid models ViT-PCA-RF, ViT-PCA-DT, ViT-PCA-XGB, and ViT-PCA-SVM when evaluated using the test set of the BTM dataset across four distinct classes: Glioma, Meningioma, Normal, and Pituitary. Within the BTM test set, there are 300 MRI scans for Glioma, 306 for Meningioma, 405 for Normal, and 300 for Pituitary.
In classifying MRI images related to Glioma, the hybrid ViT-PCA models yielded the following results: ViT-PCA-RF successfully classified 284 out of 300 MRI images, achieving an accuracy rate of 94.6% for the Glioma class. ViT-PCA-DT accurately classified 272 MRI images, with an accuracy rate of 90.6% for the Glioma class. ViT-PCA-XGB correctly classified 283 MRI scans, attaining an accuracy of 94.3%, specifically for the Glioma class. ViT-PCA-SVM demonstrated the strongest performance by correctly classifying 287 MRI scans, resulting in an accuracy rate of 95.6% for the Glioma class. Consequently, the models ranked based on their performance in the Glioma class are as follows: ViT-PCA-SVM, ViT-PCA-RF, ViT-PCA-XGB, and ViT-PCA-DT.
ViT-PCA-RF effectively classified 299 out of 306 MRI images, resulting in a 97.7% accuracy for the Meningioma category. ViT-PCA-DT and ViT-PCA-SVM accurately identified 295 MRI images, achieving a 96.4% accuracy rate for the Meningioma class. ViT-PCA-XGB accurately categorized 296 MRI scans, with a specific accuracy of 96.7% for the Meningioma class. The models ranked by their performance in the Meningioma class are as follows, from best to least performing: ViT-PCA-RF, ViT-PCA-XGB, ViT-PCA-SVM, and ViT-PCA-DT.
The hybrid models ViT-PCA-RF, ViT-PCA-XGB, and ViT-PCA-DT successfully categorized all 405 MRI images, resulting in a 100% accuracy rate for the Normal category. Among these models, ViT-PCA-SVM accurately identified 400 MRI images, achieving a 98.7% accuracy rate specifically for the Normal category. Therefore, when considering performance within the Normal class, the models can be ranked as ViT-PCA-RF, ViT-PCA-XGB, ViT-PCA-DT, and ViT-PCA-SVM.
ViT-PCA-RF and ViT-PCA-SVM effectively classified 298 out of 300 MRI images, resulting in an accuracy rate of 99.3% for the Pituitary category. ViT-PCA-DT correctly identified 293 MRI images, achieving a 97.6% accuracy for the Pituitary class. Additionally, ViT-PCA-XGB successfully categorized 297 MRI scans, with a notable accuracy of 99% for the Pituitary category. When considering the models’ performance in the Pituitary class, they can be ranked from best to least performing as follows: ViT-PCA-RF, ViT-PCA-SVM, ViT-PCA-XGB, and ViT-PCA-DT.
In our second experiment, we categorized brain tumors using the BTM dataset with the assistance of five DL models, ViT, VGG16, ResNet50, DenseNet201, and Xception, for feature extraction. Subsequently, we utilized PCA to reduce the features’ dimensions and applied the RF algorithm for classification. The primary aim of this experiment was to assess the feature extraction effectiveness of ViT compared to VGG16, ResNet50, DenseNet201, and Xception. The BTM dataset comprised four key classes: Normal, Meningioma, Pituitary, and Glioma. Results from our second experiment, including hybrid models like ViT-PCA-RF, VGG16-PCA-RF, ResNet50-PCA-RF, DenseNet201-PCA-RF, and Xception-PCA-RF, are detailed in
Table 13. These tables demonstrate the average performance metrics of the five hybrid models in the multi-classification task on the test set of the BTM dataset. The average accuracies obtained were 99% for ViT-PCA-RF, 97% for VGG16-PCA-RF, 95.7% for ResNet50-PCA-RF, 96.2% for DenseNet201-PCA-RF, and 95.3% for Xception-PCA-RF. Consequently, the ViT-PCA-RF model displayed the highest accuracy level among all the models.
VGG16-PCA-RF recorded an average of 98% specificity, 6.5% FNR, 98.1% NPV, 94% precision, 94% recall, and 93.9% F1 score. ResNet50-PCA-RF recorded the following average scores: 97.2% for specificity, 9.4% for FNR, 97.2% for NPV, 91.2% for precision, 91.3% for recall, and 91.2% for F1 score. DenseNet201-PCA-RF recorded average scores of 97.5% for specificity, 8.3% for FNR, 97.6% for NPV, 93.0% for precision, 92.4% for recall, and 92.3% for F1 score. Xception-PCA-RF recorded the following average scores: 96.9% for specificity, 10.2% for FNR, 97% for NPV, 90.6% for precision, 90.5% for recall, and 90.5% for F1 score.
Therefore, ViT-PCA-RF showed excellent performance in different areas. It had the highest average scores for accuracy, specificity, NPV, precision, recall, and F1 score, at 99%, 99.4%, 99.4%, 98.1%, 98.1%, and 98.1%, respectively. Additionally, it had the lowest average FNR at 2.1%. This indicates that ViT-PCA-RF is very good at handling tasks involving multiple categories and performs well across various evaluation standards.
The test set of the BTM dataset was categorized into four primary groups: Glioma, Meningioma, Normal, and Pituitary. We assessed the performance of four novel hybrid models—ViT-PCA-RF, VGG16-PCA-RF, ResNet50-PCA-RF, DenseNet201-PCA-RF, and Xception-PCA-RF—by measuring various evaluation metrics, including accuracy, specificity, FNR, NPV, precision, recall, and F1 score for every classification as shown in
Table 9 and
Table 14,
Table 15,
Table 16,
Table 17.
In the Glioma class, ViT_PCA_RF stood out among all the models assessed for its exceptional performance. It delivered impressive outcomes in accuracy, specificity, NPV, precision, recall, and F1 score, achieving scores of 98.47%, 99.60%, 98.44, 98.61%, 94.67, and 96.60%. Notably, ViT-PCA-RF exhibited the lowest FNR at 5.33%.
In the class Meningioma, the ViT_PCA_RF model showed outstanding performance with the following results: accuracy: 98.55%, specificity: 98.81%, NPV: 99.30%, precision: 96.14%, recall: 97.71%, F1 score: 96.92%. Moreover, it exhibited the smallest FNR at 2.29%.
In the Normal class, the DenseNet201-PCA-RF model displayed outstanding performance metrics. It achieved the highest accuracy, specificity, precision, and F1 score rates, all at 99.9% or 100%. Additionally, among the models ViT-PCA-RF, VGG16-PCA-RF, and ResNet50-PCA-RF, both the NPV and recall rates achieved 100%, along with maintaining the lowest FNR of 0%.
In the Pituitary class, ViT-PCA-RF achieved top scores in accuracy, specificity, NPV, precision, recall, and F1 score, with 99.31%, 99.31%, 99.80%, 97.70%, 99.33%, and 98.51%, respectively. Furthermore, ViT-PCA-RF attained the lowest FNR at 0.67%.
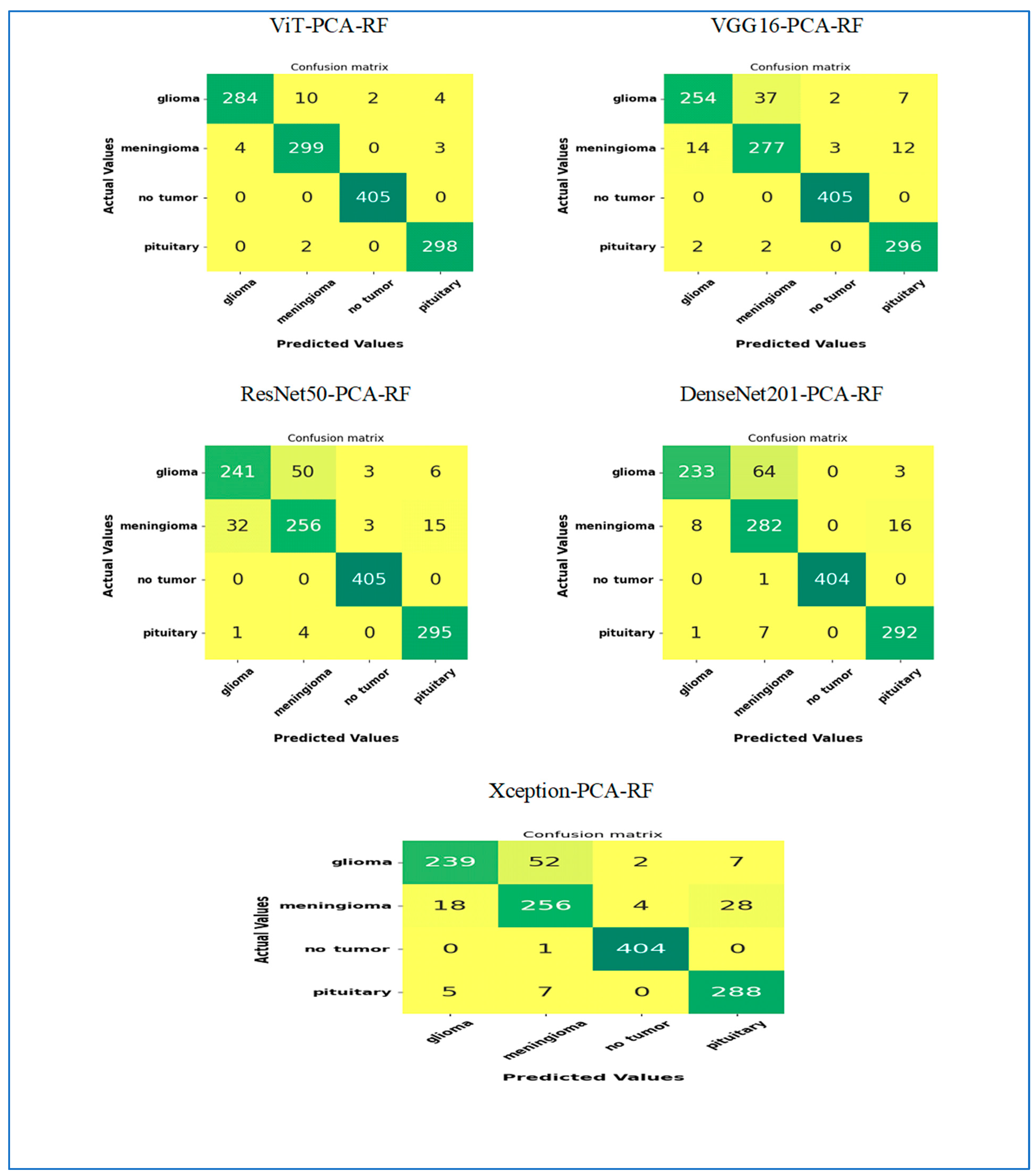
Figure 7 illustrates the performance results of the hybrid models ViT-PCA-RF, VGG16-PCA-DT, ResNet50-PCA-RF, DenseNet201-PCA-RF, and Xception-PCA-RF when evaluated using the BTM dataset across four distinct classes: Glioma, Meningioma, Normal, and Pituitary. Within the BTM test set, there are 300 MRI scans for Glioma, 306 for Meningioma, 405 for Normal, and 300 for Pituitary.
In classifying MRI images of the class Glioma, the hybrid ViT-PCA-RF accurately classified 284 out of 300 MRI images, achieving a 94.6% accuracy rate for Glioma. VGG16-PCA-RF achieved an 84.7% accuracy in classifying 254 MRI images related to Glioma. ResNet50-PCA-RF correctly classified 241 MRI scans with an accuracy rate of 80.3% for the Glioma category. DenseNet201-PCA-RF demonstrated the strongest performance by accurately classifying 233 MRI scans, resulting in a 77.6% accuracy for Glioma. Xception-PCA-RF achieved a 79.6% accuracy in classifying 239 MRI images specifically for Glioma. The models’ rank in terms of Glioma classification performance is as follows: ViT-PCA-RF, VGG16-PCA-RF, ResNet50-PCA-RF, Xception-PCA-RF, and DenseNet201-PCA-RF.
In classifying MRI images of the class Meningioma, the hybrid ViT-PCA-RF accurately classified 299 out of 306 MRI images, achieving a 97.7% accuracy rate for Meningioma. VGG16-PCA-RF achieved a 90.5% accuracy in classifying 277 MRI images related to Meningioma. ResNet50-PCA-RF and Xception-PCA-RF correctly classified 256 MRI scans with an accuracy rate of 83.6% for the Meningioma category. DenseNet201-PCA-RF demonstrated the strongest performance by accurately classifying 282 MRI scans, resulting in a 92.1% accuracy for Meningioma. The models’ rank in terms of Meningioma classification performance is ViT-PCA-RF, DenseNet201-PCA-RF, VGG16-PCA-RF, ResNet50-PCA-RF, and Xception-PCA-RF.
In the classification of MRI images of the class Normal, the hybrid models ViT-PCA-RF, VGG16-PCA-RF, and ResNet50-PCA-RF all achieved a perfect classification, accurately identifying all 405 MRI images in the Normal class, resulting in a 100% accuracy rate. DenseNet201-PCA-RF and Xception-PCA-RF also performed exceptionally well, correctly classifying 404 MRI scans, leading to a 99.75% accuracy rate for the Normal class. Ranking of models based on their performance in Normal classification: ViT-PCA-RF, VGG16-PCA-RF, ResNet50-PCA-RF, Xception-PCA-RF, and DenseNet201-PCA-RF.
In the classification of MRI images of the class Pituitary, the following models achieved the following accuracy rates: ViT-PCA-RF: 99.3% accuracy (298 out of 300 images), VGG16-PCA-RF: 98.6% accuracy (296 out of 300 images), ResNet50-PCA-RF: 98.3% accuracy (295 out of 300 images), DenseNet201-PCA-RF: 97.3% accuracy (292 out of 300 images), and Xception-PCA-RF: 96% accuracy (288 out of 300 images). Rank of models based on Pituitary classification performance: ViT-PCA-RF, VGG16-PCA-RF, ResNet50-PCA-RF, DenseNet201-PCA-RF, and Xception-PCA-RF.
4.4. External Validationof the ViT-PCA-RF Model
In the third experiment, we conducted external validation of the ViT-PCA-RF model using the external dataset from Figshare. This brain tumor dataset contains 3064 T1-weighted contrast-enhanced images from 233 patients, featuring three types of brain tumors: meningioma (708 slices), glioma (1426 slices), and pituitary tumor (930 slices). The dataset was divided into 70% for training, 15% for testing, and 15% for validation. The testing set has three brain tumors: meningioma, glioma, and pituitary tumor.
In the third experiment, we utilized ViT for feature extraction, applied PCA to modify feature dimensions, and employed RF, DT, XGB, and SVM for classification. The main objective of this experiment was to externally validate the combination of ViT, PCA, and RF.
The results of the hybrid models ViT-PCA-RF, ViT-PCA-DT, ViT-PCA-XGB, and ViT-PCA-SVM are summarized in
Table 18.
Table 18 presents the average evaluation metrics for the four hybrid models applied to the multi-classification task, using the test set from the Figshare dataset. The average accuracies obtained were 95.85% for ViT-PCA-RF, 95.36% for ViT-PCA-DT, 95.36% for ViT-PCA-XGB, and 95.36% for ViT-PCA-SVM. Therefore, the ViT-PCA-RF model recorded the highest accuracy.
The performance of five models—ViT-PCA-RF, ViT-PCA-DT, ViT-PCA-XGB, ViT-PCA-SVM, and A-SVM—was assessed using various metrics: accuracy, specificity, false FNR, NPV, precision, recall, and F1 score.
The ViT-PCA-RF model exhibited the highest specificity (96.54%) and NPV (97.05%), showcasing its strong capability to accurately identify negative cases while minimizing false positives. Its precision was 93.94%, and recall stood at 91.90%, leading to the highest F1 score of 92.74% among the models tested. Additionally, it recorded the lowest FNR at 8.10%, indicating fewer false negatives than the other models.
The ViT-PCA-DT model showed specificity at 96.44% and an NPV of 96.47%. Its precision (91.96%) and recall (91.81%) were slightly lower than those of ViT-PCA-RF, resulting in an F1 score of 91.88%. The FNR for this model was 8.19%, which was marginally higher than that of ViT-PCA-RF, suggesting a slightly greater number of false negatives.
Similarly, the ViT-PCA-XGB model had slightly lower specificity (96.17%) and a higher FNR (8.82%) compared to ViT-PCA-DT. Its NPV was 96.62%. While it achieved a precision of 92.91%, its recall of 91.18% was lower, leading to an F1 score of 91.89%.
In contrast, the ViT-PCA-SVM model had significantly lower specificity (64.24%) and NPV (64.99%) compared to the other models. It recorded a substantially lower FNR of 2.49%, indicating better identification of positive cases. However, its precision (62.57%) and recall (64.51%) were considerably lower, resulting in the lowest F1 score of 63.50% among the models.
In summary, the ViT-PCA-RF model exhibited superior performance across most metrics, particularly in specificity, precision, recall, and F1 score. On the other hand, the ViT-PCA-SVM model fell short in precision, specificity, and F1 score, despite its low false negative rate.
Table 19,
Table 20,
Table 21 and
Table 22 present the class-wise results of the four hybrid models.
The ViT-PCA-DT model demonstrated strong classification performance across three types of brain tumors: Glioma, Meningioma, and Pituitary tumors.
For the Glioma class, the model achieved an accuracy of 95.52%, with a specificity of 95.65%. The FNR was 4.62%, and the NPV was also 95.65%. Precision, recall, and F1 score were consistently high at 95.38%, indicating a balanced performance in identifying Glioma cases.
For the Meningioma class, the model reached an accuracy of 94.03%, but the specificity was notably lower at 96.51%, indicating a high false positive rate. Nevertheless, the FNR remained low at 14.94%, and the NPV was 95.90%. Precision was recorded at 87.06%, while recall was 85.06%, resulting in an F1 score of 86.05%. The low specificity highlighted the model’s difficulty in accurately identifying non-Meningioma cases.
For the Pituitary class, the model performed best in the Pituitary class, achieving an accuracy of 96.52% and a specificity of 97.16%. The FNR was 5.00%, and the NPV reached 97.86%. Precision was 93.44%, and recall was 95.00%, leading to an F1 score of 94.21%, indicating robust and balanced performance.
The
ViT-PCA-SVM demonstrated strong performance across most evaluation metrics, with some variation depending on the tumor type.
For Glioma, the model achieved an accuracy of 95.02%, a specificity of 95.65%, and an FNR of 5.64%. The NPV was recorded at 94.74%, while precision was 95.34%, recall was 94.36%, and the F1 score was 94.85%. These results indicate that the model was highly effective in distinguishing Glioma cases, maintaining both high precision and recall, and minimizing false positives and false negatives [
1].
In the case of Meningioma, the model reported a slightly lower accuracy of 94.28%. However, it showed a very low specificity of 97.46% and an FNR of 17.24%. The NPV was 95.34%, precision was 90%, recall was 83.76%, and the F1 score was 86.23%. The unusually low specificity and NPV suggest challenges in accurately identifying non-Meningioma cases, potentially due to class imbalance or overlapping features, despite having acceptable accuracy.
For Pituitary tumors, the model achieved the highest accuracy at 96.77%, with a specificity of 96.10% and an FNR of 1.67%. It demonstrated an outstanding NPV of 99.27%, along with a precision of 91.47%, a recall of 98.33%, and an F1 score of 94.78%. These metrics illustrate excellent sensitivity and reliability in detecting Pituitary tumors, with very few false negatives.
In the fourth experiment, we validated the ViT-PCA-RF model using an external dataset from Figshare. We employed five DL models—ViT, VGG16, ResNet50, DenseNet201, and Xception—for feature extraction. We then applied PCA to reduce the dimensionality of the features and utilized the RF algorithm for classification.
The primary aim was to evaluate the feature extraction effectiveness of the ViT model in comparison to VGG16, ResNet50, DenseNet201, and Xception. The Figshare dataset consisted of three main classes: Meningioma, Pituitary, and Glioma. Results from this experiment, including hybrid models like ViT-PCA-RF, VGG16-PCA-RF, ResNet50-PCA-RF, DenseNet201-PCA-RF, and Xception-PCA-RF, are displayed in
Table 23. This table illustrates the average performance metrics of the five hybrid models in the multi-classification task on the test set of the Figshare dataset. The average accuracies recorded were ViT-PCA-RF: 99%, VGG16-PCA-RF: 88.39%, ResNet50-PCA-RF: 90.71%, DenseNet201-PCA-RF: 92.37%, Xception-PCA-RF: 89.72%. As a result, the ViT-PCA-RF model demonstrated the highest accuracy among all models.
ViT-PCA-RF achieved the highest overall performance with specificity of 99.4% and recall of 98.1%. Its FNR was the lowest at 2.1%, while both precision and F1 score were at 98.1%, indicating a highly balanced and reliable classification. The high specificity and NPV of 99.4% suggested that this model effectively identified negative cases, minimizing false positives and false negatives.
DenseNet201-PCA-RF followed with specificity of 94.01% and recall of 85.77%. Its FNR was 14.23%, exhibiting a relatively lower rate of false negatives compared to most models, except for ViT-PCA-RF. Precision was at 86.88%, and the F1 score was 86.02%, reflecting a solid balance between precision and recall, though it underperformed relative to ViT-PCA-RF.
ResNet50-PCA-RF achieved specificity of 91.98%, recall of 80.98%, and an F1 score of 82.15%. Its FNR was 19.02%, indicating a greater tendency to miss positive cases. Precision was at 86.30%, slightly better than DenseNet201-PCA-RF, although the model’s balance leaned more towards higher precision at the expense of recall.
Xception-PCA-RF recorded specificity of 91.00%. Its recall was 78.57%, with an F1 score of 79.95% and an FNR of 21.43%. The model’s lower recall and higher FNR suggested more frequent misclassification of positive instances, despite a reasonable precision of 85.43%.
Lastly,
VGG16-PCA-RF exhibited the lowest performance among the five models, with specificity of 89.70%, and recall of 75.82%. It had the highest FNR at 24.18% and the lowest F1 score at 77.02%, indicating struggles in correctly identifying positive cases and maintaining a balance between precision (84.20%) and recall (75.82%) [
5].
In summary, ViT-PCA-RF outperformed the other models across all metrics, demonstrating superior accuracy, sensitivity, and specificity. DenseNet201-PCA-RF was the second-best model, while VGG16-PCA-RF lagged in all aspects. These results are consistent with prior findings suggesting that ViT can achieve state-of-the-art performance when combined with PCA and RF.