Identifying Key Hematological and Biochemical Indicators of Disease Severity in COVID-19 and Non-COVID-19 Patients
Abstract
1. Introduction
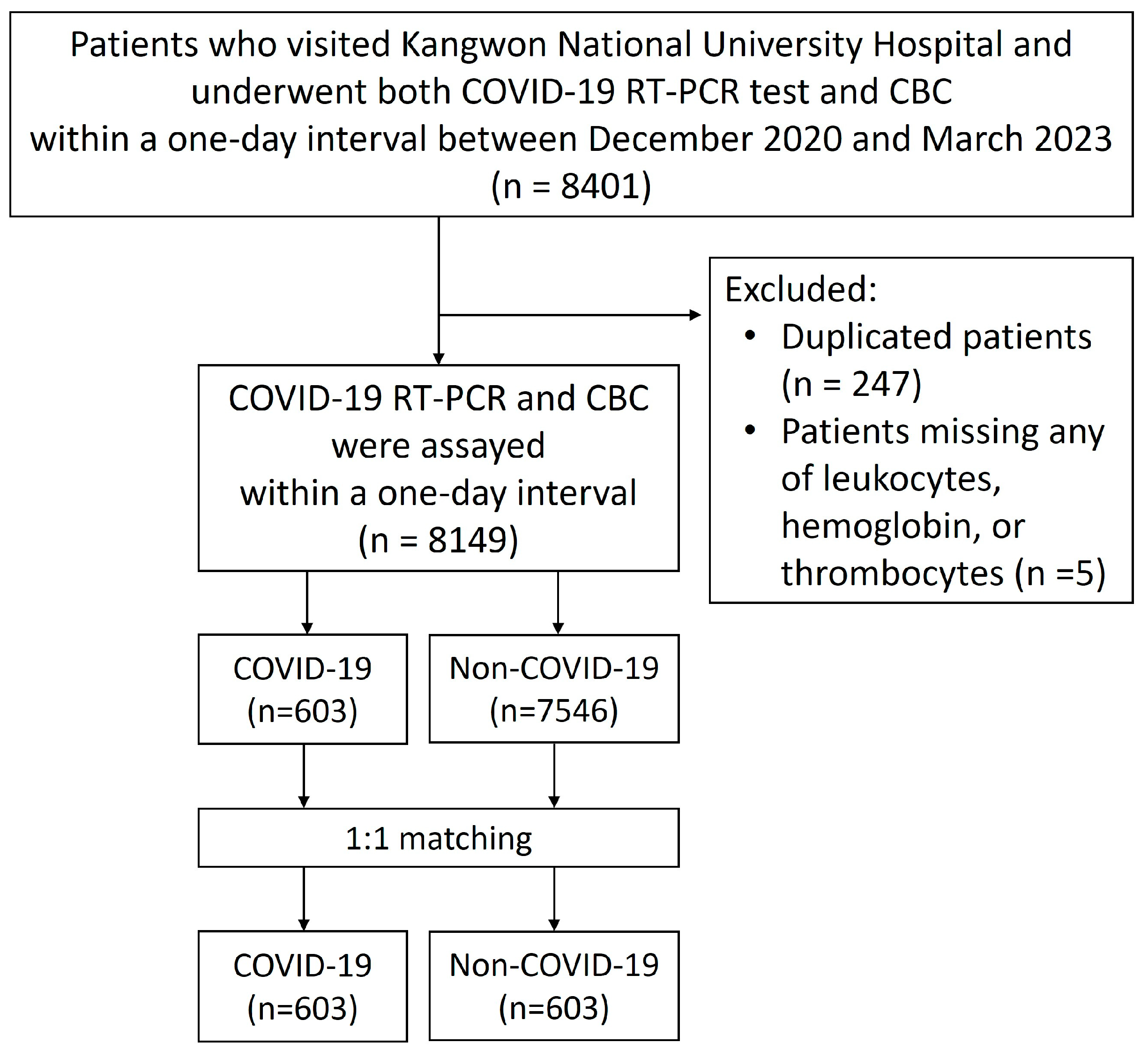
2. Materials and Methods
2.1. Patient Population and Data Collection
2.2. Hematologic and Biochemical Assays
2.3. Statistical Analysis
3. Results
3.1. Differences in Patient Characteristics Between COVID-19 and Non-COVID-19 Groups
3.2. Effect of COVID-19 on Severity Level
3.3. Laboratory Factors Associated with COVID-19 Severity
3.4. Laboratory Factors Associated with Non-COVID-19 Severity
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Ochani, R.; Asad, A.; Yasmin, F.; Shaikh, S.; Khalid, H.; Batra, S.; Sohail, M.R.; Mahmood, S.F.; Ochani, R.; Hussham Arshad, M.; et al. COVID-19 pandemic: From origins to outcomes. A comprehensive review of viral pathogenesis, clinical manifestations, diagnostic evaluation, and management. Infez. Med. 2021, 29, 20–36. [Google Scholar] [PubMed]
- Martens, R.J.H.; van Adrichem, A.J.; Mattheij, N.J.A.; Brouwer, C.G.; van Twist, D.J.L.; Broerse, J.; Magro-Checa, C.; van Dongen, C.M.P.; Mostard, R.L.M.; Ramiro, S.; et al. Hemocytometric characteristics of COVID-19 patients with and without cytokine storm syndrome on the sysmex XN-10 hematology analyzer. Clin. Chem. Lab. Med. 2021, 59, 783–793. [Google Scholar] [CrossRef] [PubMed]
- Urrechaga, E. Reviewing the value of leukocytes cell population data (CPD) in the management of sepsis. Ann. Transl. Med. 2020, 8, 953. [Google Scholar] [CrossRef]
- Wang, Z.; He, Y.; Shu, H.; Wang, P.; Xing, H.; Zeng, X.; Tang, Z. High-fluorescent lymphocytes are increased in patients with COVID-19. Br. J. Haematol. 2020, 190, e76–e78. [Google Scholar] [CrossRef]
- Urrechaga, E.; Aguirre, U.; España, P.P.; García de Guadiana, L. Complete blood counts and cell population data from Sysmex XN analyser in the detection of SARS-CoV-2 infection. Clin. Chem. Lab. Med. 2020, 59, e57–e60. [Google Scholar] [CrossRef]
- Dennison, D.; Al Khabori, M.; Al Mamari, S.; Aurelio, A.; Al Hinai, H.; Al Maamari, K.; Alshekaili, J.; Al Khadouri, G. Circulating activated neutrophils in COVID-19: An independent predictor for mechanical ventilation and death. Int. J. Infect. Dis. 2021, 106, 155–159. [Google Scholar] [CrossRef]
- Buoro, S.; Seghezzi, M.; Vavassori, M.; Dominoni, P.; Apassiti Esposito, S.; Manenti, B.; Mecca, T.; Marchesi, G.; Castellucci, E.; Azzarà, G.; et al. Clinical significance of cell population data (CPD) on Sysmex XN-9000 in septic patients with our without liver impairment. Ann. Transl. Med. 2016, 4, 418. [Google Scholar] [CrossRef]
- Sharafi, F.; Jafarzadeh Esfehani, R.; Moodi Ghalibaf, A.; Jarahi, L.; Shamshirian, A.; Mozdourian, M. Leukopenia and leukocytosis as strong predictors of COVID-19 severity: A cross-sectional study of the hematologic abnormalities and COVID-19 severity in hospitalized patients. Health Sci. Rep. 2023, 6, e1574. [Google Scholar] [CrossRef]
- Ghahramani, S.; Tabrizi, R.; Lankarani, K.B.; Kashani, S.M.A.; Rezaei, S.; Zeidi, N.; Akbari, M.; Heydari, S.T.; Akbari, H.; Nowrouzi-Sohrabi, P.; et al. Laboratory features of severe vs. non-severe COVID-19 patients in Asian populations: A systematic review and meta-analysis. Eur. J. Med. Res. 2020, 25, 30. [Google Scholar] [CrossRef]
- Sun, S.; Cai, X.; Wang, H.; He, G.; Lin, Y.; Lu, B.; Chen, C.; Pan, Y.; Hu, X. Abnormalities of peripheral blood system in patients with COVID-19 in Wenzhou, China. Clin. Chim. Acta 2020, 507, 174–180. [Google Scholar] [CrossRef]
- Schulte-Schrepping, J.; Reusch, N.; Paclik, D.; Baßler, K.; Schlickeiser, S.; Zhang, B.; Krämer, B.; Krammer, T.; Brumhard, S.; Bonaguro, L.; et al. Severe COVID-19 Is Marked by a Dysregulated Myeloid Cell Compartment. Cell 2020, 182, 1419–1440.e1423. [Google Scholar] [CrossRef] [PubMed]
- Huang, I.; Pranata, R. Lymphopenia in severe coronavirus disease-2019 (COVID-19): Systematic review and meta-analysis. J. Intensive Care 2020, 8, 36. [Google Scholar] [CrossRef] [PubMed]
- Lu, C.; Liu, Y.; Chen, B.; Yang, H.; Hu, H.; Liu, Y.; Zhao, Y. Prognostic value of lymphocyte count in severe COVID-19 patients with corticosteroid treatment. Signal Transduct. Target. Ther. 2021, 6, 106. [Google Scholar] [CrossRef] [PubMed]
- Iannetta, M.; Buccisano, F.; Fraboni, D.; Malagnino, V.; Campogiani, L.; Teti, E.; Spalliera, I.; Rossi, B.; Di Lorenzo, A.; Palmieri, R.; et al. Baseline T-lymphocyte subset absolute counts can predict both outcome and severity in SARS-CoV-2 infected patients: A single center study. Sci. Rep. 2021, 11, 12762. [Google Scholar] [CrossRef]
- Li, T.; Qiu, Z.; Zhang, L.; Han, Y.; He, W.; Liu, Z.; Ma, X.; Fan, H.; Lu, W.; Xie, J.; et al. Significant changes of peripheral T lymphocyte subsets in patients with severe acute respiratory syndrome. J. Infect. Dis. 2004, 189, 648–651. [Google Scholar] [CrossRef]
- Xu, H.; Zhong, L.; Deng, J.; Peng, J.; Dan, H.; Zeng, X.; Li, T.; Chen, Q. High expression of ACE2 receptor of 2019-nCoV on the epithelial cells of oral mucosa. Int. J. Oral Sci. 2020, 12, 8. [Google Scholar] [CrossRef]
- Lin, L.; Lu, L.; Cao, W.; Li, T. Hypothesis for potential pathogenesis of SARS-CoV-2 infection-a review of immune changes in patients with viral pneumonia. Emerg. Microbes Infect. 2020, 9, 727–732. [Google Scholar] [CrossRef]
- Lukaszewicz, A.C.; Grienay, M.; Resche-Rigon, M.; Pirracchio, R.; Faivre, V.; Boval, B.; Payen, D. Monocytic HLA-DR expression in intensive care patients: Interest for prognosis and secondary infection prediction. Crit. Care Med. 2009, 37, 2746–2752. [Google Scholar] [CrossRef]
- Navarro Conde, P.; Alemany Monraval, P.; Medina Medina, C.; Jiménez Sánchez, A.; Andrés Teruel, J.C.; Ferrando Marco, J.; Puglia Santos, V.; Mayordomo Aranda, E. Autopsy findings from the first known death from Severe Acute Respiratory Syndrome SARS-CoV-2 in Spain. Rev. Esp. Patol. 2020, 53, 188–192. [Google Scholar] [CrossRef]
- Kim, O.Y.; Monsel, A.; Bertrand, M.; Coriat, P.; Cavaillon, J.M.; Adib-Conquy, M. Differential down-regulation of HLA-DR on monocyte subpopulations during systemic inflammation. Crit. Care 2010, 14, R61. [Google Scholar] [CrossRef]
- Payen, D.; Cravat, M.; Maadadi, H.; Didelot, C.; Prosic, L.; Dupuis, C.; Losser, M.R.; De Carvalho Bittencourt, M. A Longitudinal Study of Immune Cells in Severe COVID-19 Patients. Front. Immunol. 2020, 11, 580250. [Google Scholar] [CrossRef] [PubMed]
- Faivre, V.; Lukaszewicz, A.C.; Payen, D. Downregulation of Blood Monocyte HLA-DR in ICU Patients Is Also Present in Bone Marrow Cells. PLoS ONE 2016, 11, e0164489. [Google Scholar] [CrossRef] [PubMed]
- Rolfes, V.; Ribeiro, L.S.; Hawwari, I.; Böttcher, L.; Rosero, N.; Maasewerd, S.; Santos, M.L.S.; Próchnicki, T.; Silva, C.M.S.; Wanderley, C.W.S.; et al. Platelets Fuel the Inflammasome Activation of Innate Immune Cells. Cell Rep. 2020, 31, 107615. [Google Scholar] [CrossRef]
- Rayes, J.; Bourne, J.H.; Brill, A.; Watson, S.P. The dual role of platelet-innate immune cell interactions in thrombo-inflammation. Res. Pract. Thromb. Haemost. 2020, 4, 23–35. [Google Scholar] [CrossRef]
- Elahi, S. Hematopoietic responses to SARS-CoV-2 infection. Cell Mol. Life Sci. 2022, 79, 187. [Google Scholar] [CrossRef]
- Lefrançais, E.; Ortiz-Muñoz, G.; Caudrillier, A.; Mallavia, B.; Liu, F.; Sayah, D.M.; Thornton, E.E.; Headley, M.B.; David, T.; Coughlin, S.R.; et al. The lung is a site of platelet biogenesis and a reservoir for haematopoietic progenitors. Nature 2017, 544, 105–109. [Google Scholar] [CrossRef]
- Choudhury, S.; Dubey, S.; Zumu, M.; Mustafa, P.P.M.; Burma, A.; Kumar, S. The Role of Platelet Parameters in Predicting the Disease Severity of COVID-19 Patients: A Hospital-Based Study. Cureus 2024, 16, e51523. [Google Scholar] [CrossRef]
- Barrett, T.J.; Bilaloglu, S.; Cornwell, M.; Burgess, H.M.; Virginio, V.W.; Drenkova, K.; Ibrahim, H.; Yuriditsky, E.; Aphinyanaphongs, Y.; Lifshitz, M.; et al. Platelets contribute to disease severity in COVID-19. J. Thromb. Haemost. 2021, 19, 3139–3153. [Google Scholar] [CrossRef]
- Kilercik, M.; Demirelce, Ö.; Serdar, M.A.; Mikailova, P.; Serteser, M. A new haematocytometric index: Predicting severity and mortality risk value in COVID-19 patients. PLoS ONE 2021, 16, e0254073. [Google Scholar] [CrossRef]
- Incir, S.; Komesli, Z.; Baygul, A.; Tasdemir, Z.A.; Palaoglu, K.E.; Kant, H.; Kapmaz, M.; Tekin, S.; Kilic, A.; Dagel, T.; et al. Immature platelet fraction: Is a novel early predictive marker for disease severity in patients with Covid-19 pneumonia? Turk. J. Biochem. 2021, 46, 359–366. [Google Scholar] [CrossRef]
- Cohen, A.; Harari, E.; Yahud, E.; Cipok, M.; Bryk, G.; Lador, N.K.; Mann, T.; Mayo, A.; Lev, E.I. Immature platelets in patients with Covid-19: Association with disease severity. J. Thromb. Thrombolysis 2021, 52, 708–714. [Google Scholar] [CrossRef] [PubMed]
- Jafar, N.; Edriss, H.; Nugent, K. The Effect of Short-Term Hyperglycemia on the Innate Immune System. Am. J. Med. Sci. 2016, 351, 201–211. [Google Scholar] [CrossRef]
- Lazarus, G.; Audrey, J.; Wangsaputra, V.K.; Tamara, A.; Tahapary, D.L. High admission blood glucose independently predicts poor prognosis in COVID-19 patients: A systematic review and dose-response meta-analysis. Diabetes Res. Clin. Pract. 2021, 171, 108561. [Google Scholar] [CrossRef]
- Sachdeva, S.; Desai, R.; Gupta, U.; Prakash, A.; Jain, A.; Aggarwal, A. Admission Hyperglycemia in Non-diabetics Predicts Mortality and Disease Severity in COVID-19: A Pooled Analysis and Meta-summary of Literature. SN Compr. Clin. Med. 2020, 2, 2161–2166. [Google Scholar] [CrossRef]
- Sharma, J.; Rajput, R.; Bhatia, M.; Arora, P.; Sood, V. Clinical Predictors of COVID-19 Severity and Mortality: A Perspective. Front. Cell Infect. Microbiol. 2021, 11, 674277. [Google Scholar] [CrossRef]
- Krishnan, A.; Prichett, L.; Tao, X.; Alqahtani, S.A.; Hamilton, J.P.; Mezey, E.; Strauss, A.T.; Kim, A.; Potter, J.J.; Chen, P.H.; et al. Abnormal liver chemistries as a predictor of COVID-19 severity and clinical outcomes in hospitalized patients. World J. Gastroenterol. 2022, 28, 570–587. [Google Scholar] [CrossRef]
- Dobrijević, D.; Katanić, J.; Todorović, M.; Vučković, B. Baseline laboratory parameters for preliminary diagnosis of COVID-19 among children: A cross-sectional study. Sao Paulo Med. J. 2022, 140, 691–696. [Google Scholar] [CrossRef]
- Dobrijević, D.; Vilotijević-Dautović, G.; Katanić, J.; Horvat, M.; Horvat, Z.; Pastor, K. Rapid Triage of Children with Suspected COVID-19 Using Laboratory-Based Machine-Learning Algorithms. Viruses 2023, 15, 1522. [Google Scholar] [CrossRef]
- Cheshmehzangi, A.; Su, Z.; Jin, R.; Dawodu, A.; Sedrez, M.; Pourroostaei Ardakani, S.; Zou, T. Space and social distancing in managing and preventing COVID-19 community spread: An overview. Heliyon 2023, 9, e13879. [Google Scholar] [CrossRef]
- Adab, P.; Haroon, S.; O’Hara, M.E.; Jordan, R.E. Comorbidities and covid-19. BMJ 2022, 377, o1431. [Google Scholar] [CrossRef]
- Russell, C.D.; Lone, N.I.; Baillie, J.K. Comorbidities, multimorbidity and COVID-19. Nat. Med. 2023, 29, 334–343. [Google Scholar] [CrossRef] [PubMed]
- Chatterjee, S.; Nalla, L.V.; Sharma, M.; Sharma, N.; Singh, A.A.; Malim, F.M.; Ghatage, M.; Mukarram, M.; Pawar, A.; Parihar, N.; et al. Association of COVID-19 with Comorbidities: An Update. ACS Pharmacol. Transl. Sci. 2023, 6, 334–354. [Google Scholar] [CrossRef]
- Clinical Management of COVID-19: Living Guideline, 18 August 2023. World Health Organization: Geneva. Available online: https://www.who.int/publications/i/item/WHO-2019-nCoV-clinical-2023.2 (accessed on 5 November 2024).
- COVID-19 Treatment Guidelines Panel. Coronavirus Disease 2019 (COVID-19) Treatment Guidelines. National Institutes of Health. Available online: https://www.covid19treatmentguidelines.nih.gov/ (accessed on 5 November 2024).
- Mohammad Reza, A.; Leila, E.; Morteza, A.; Nahid, M.; Saeed, J.; Raoof, N.; Mostafa, S. Machine Learning-Based Clinical Decision Support System for automatic diagnosis of COVID-19 based on the routine blood test. J. Biostat. Epidemiol. 2022, 8, 77–89. [Google Scholar] [CrossRef]
| Parameters | Alternative Name | Description | Meaning |
|---|---|---|---|
| MicroR | Micro RBC ratio | Microcytic population of red blood cells | |
| MacroR | Macro RBC ratio | Macrocytic population of red blood cells | |
| NE-FSC | NE-Z | Neutrophils cell size | Changes may reflect abnormal neutrophil size |
| NE-SFL | NEUT-RI, NE-Y | Neutrophils reactivity index. Metabolic activity | Increases with higher nucleic acid content, indicating immature or reactive neutrophils |
| NE-SSC | NEUT-GI, NE-X | Neutrophils granularity index | Increases with greater amounts of granules, vacuoles and other cytoplasmic inclusions |
| NE-FSC width | NE-WZ | Neutrophils cell size and the width of dispersion | Reflects neutrophil size heterogeneity |
| NE-SFL width | NE-WY | Neutrophils fluorescence intensity and the width of dispersion | Indicates heterogeneity in nucleic acid content among neutrophils |
| NE-SSC width | NE-WX | Neutrophils complexity and width of dispersion of the events measured | Indicates heterogeneity in granularity among neutrophils |
| LY-FSC | LY-Z | Lymphocytes cell size | Its change reflects presence of abnormal sized cells (e.g., increase with activated lymphocyte or decrease with pyknotic lymphocytes, etc.) |
| LY-SFL | LY-Y | Lymphocytes metabolic activity and/or permeability of the cell membrane. Lymphocytes fluorescence intensity | Increase in proportion to the amount of nucleic acid, such as in activated or abnormal lymphocytes and blast cells, etc. |
| LY-SSC | LY-X | Lymphocytes cell complexity | Increase in the presence of greater amounts of granules or vacuoles (e.g., large granular lymphocyte) |
| LY-FSC width | LY-WZ | Lymphocytes cell size and the width of dispersion | Reflects lymphocyte size heterogeneity |
| LY-SFL width | LY-WY | Lymphocytes fluorescence intensity and the width of dispersion | Indicates heterogeneity in nucleic acid content among lymphocytes |
| LY-SSC width | LY-WX | Lymphocytes complexity and width of dispersion of the events measured | Indicates heterogeneity in granularity among lymphocytes |
| HFLC | High fluorescence lymphocyte count | It represents activated cells (antibody-secreting B lymphocytes and plasma cells) if systemic hematological diseases can be excluded | |
| MO-FSC | MO-Z | Monocytes cell size | Its change reflects presence of abnormal sized cells |
| MO-SFL | MO-Y | Monocytes metabolic activity and/or permeability of cell membrane. Monocytes fluorescence intensity | Increase in proportion to the amount of cellular nucleic acid (e.g., activated monocytes and monoblasts |
| MO-SSC | MO-X | Monocytes cells complexity | Increase in the presence of greater amounts of granules, vacuoles and other cytoplasmic inclusions. Decrease in the presence of a lower cell complexity |
| MO-FSC width | MO-WZ | Monocytes cell size and the width of dispersion | Reflects monocyte size heterogeneity |
| MO-SFL width | MO-WY | Monocytes fluorescence intensity and the width of dispersion | Indicates heterogeneity in nucleic acid content among monocytes |
| MO-SSC width | MO-WX | Monocytes complexity and width of dispersion of the events measured | Indicates heterogeneity in granularity among monocytes |
| P-LCR | Platelet large cell ratio | Its increase suggests possible risk of thrombosis | |
| PDW | Platelet distribution width | Its increase reflects size variation. Its increase may be associated with vascular disease or certain cancers |
| COVID-19 | Non-COVID-19 | p-Value | |
|---|---|---|---|
| Mean (SD) | Mean (SD) | ||
| Age | 66.8 (22.2) | 66.8 (22.2) | 0.775 |
| Sex, male * | 314 (52.1%) | 314 (52.1%) | 1.000 |
| CBC | |||
| Hemoglobin, g/dL | 11.2 (2.2) | 11.9 (2.2) | <0.001 |
| Erythrocytes, ×1012/L | 3.7 (0.8) | 4.0 (0.8) | <0.001 |
| Leukocytes, ×109/L | 8.6 (5.8) | 8.6 (9.8) | 0.970 |
| Platelets, ×109/L | 224.7 (104.1) | 243.9 (99.5) | 0.001 |
| Hematocrit | 34 (6.3) | 36 (6.2) | <0.001 |
| Erythrocytes | |||
| MCV | 91.6 (6.3) | 91.7 (6.6) | 0.561 |
| MCH | 30.1 (2.4) | 30.4 (2.5) | 0.042 |
| MCHC | 32.9 (1.4) | 33.1 (1.3) | 0.002 |
| nRBC (>0) * | 73 (12.1%) | 134 (22.2%) | <0.001 |
| MicroR | 2.5 (4.2) | 2.4 (4.4) | 0.030 |
| MacroR | 4.7 (2.8) | 4.6 (2.8) | 0.673 |
| RDW-CV | 14.3 (2.6) | 14.0 (2.3) | 0.004 |
| RDW-SD | 47.4 (8.3) | 46.6 (7.6) | 0.049 |
| Granulocytes | |||
| Neutrophils, ×109/L | 6.3 (4.8) | 5.8 (5.8) | 0.035 |
| NE-FSC, ch | 89.8 (5.0) | 90.6 (4.6) | 0.006 |
| NE-SFL, ch | 50.7 (6.3) | 49.8 (4.0) | 0.070 |
| NE-SSC, ch | 154.9 (5.2) | 154.9 (4.5) | 0.798 |
| NE-SSC width, ch | 313.1 (26.4) | 306.8 (26.0) | <0.001 |
| NE-SFL width, ch | 650.2 (97.1) | 632.5 (127.8) | <0.001 |
| NE-FSC width, ch | 750.1 (96.3) | 738.5 (96.9) | 0.060 |
| Immature granulocytes, ×109/L | 0.1 (0.3) | 0.1 (1.9) | <0.001 |
| Basophils, ×109/L | 0 (0) | 0 (0) | <0.001 |
| Eosinophils, ×109/L | 0.1 (0.2) | 0.2 (0.3) | <0.001 |
| Lymphocytes | |||
| Lymphocytes, ×109/L | 1.4 (2.7) | 1.8 (1.1) | <0.001 |
| LY-SSC, ch | 78.8 (3.7) | 79.0 (3.6) | 0.246 |
| LY-SFL, ch | 69.4 (5.8) | 70.6 (4.8) | <0.001 |
| LY-FSC, ch | 58.2 (2.8) | 58.3 (2.1) | 0.170 |
| LY-SSC width, ch | 582.4 (108.7) | 553.3 (79.2) | <0.001 |
| LY-SFL width, ch | 900.7 (188.3) | 880.5 (110.4) | 0.179 |
| LY-FSC width, ch | 601.3 (137.5) | 569.6 (90.9) | <0.001 |
| HFLC, ×109/L | 0 (0.1) | 0 (0) | <0.001 |
| Monocytes | |||
| Monocytes, ×109/L | 0.7 (0.4) | 0.7 (1.4) | 0.142 |
| MO-SSC, ch | 121.2 (3.3) | 120.0 (3.1) | <0.001 |
| MO-SFL, ch | 114.6 (11.1) | 116.1 (8.7) | 0.017 |
| MO-FSC, ch | 66.8 (4.0) | 67.4 (3.6) | 0.005 |
| MO-SSC width, ch | 261.4 (37.3) | 259.7 (27.6) | 0.324 |
| MO-SFL width, ch | 709.0 (131.3) | 685.9 (94.9) | <0.001 |
| MO-FSC width, ch | 672.3 (126) | 668.5 (120.1) | 0.376 |
| TNC | 8.6 (5.8) | 8.6 (9.9) | 0.988 |
| Thrombocytes | |||
| Plateletcrit | 0.2 (0.1) | 0.2 (0.1) | 0.015 |
| P-LCR | 23.6 (8.6) | 21.7 (7.3) | <0.001 |
| MPV | 9.9 (1.1) | 9.6 (0.9) | <0.001 |
| PDW | 10.6 (2.5) | 10.2 (1.9) | 0.008 |
| Ratios | |||
| NLR | 7.2 (9.1) | 4.5 (6.3) | <0.001 |
| PLR | 222.2 (159.7) | 174.3 (127.0) | <0.001 |
| MLR | 0.6 (0.6) | 0.5 (0.7) | <0.001 |
| Chemical assay | |||
| Sodium | 138.7 (4.4) | 139.6 (3.7) | <0.001 |
| Glucose | 125.5 (46.7) | 127.2 (54.6) | 0.914 |
| Albumin | 3.3 (0.7) | 3.6 (0.7) | <0.001 |
| Renal function test | |||
| BUN | 20.8 (17.9) | 18.7 (13.8) | 0.039 |
| Creatinine | 1.1 (1.3) | 1.0 (1.1) | 0.060 |
| Liver function tests | |||
| AST | 49.5 (165.7) | 54.5 (271.5) | 0.005 |
| ALT | 50.8 (315.2) | 44.4 (254.4) | 0.079 |
| ALP | 110.7 (91.6) | 112.0 (99.3) | 0.955 |
| Total bilirubin | 0.9 (1.1) | 0.9 (1.5) | 0.259 |
| Estimate | p-Value | |
|---|---|---|
| Age | −0.1 | 0.421 |
| Sex, male | −0.4 | 0.072 |
| CBC | ||
| Hemoglobin, g/dL | 0.458 | 0.001 |
| Leukocytes, ×109/L | −0.442 | 0.001 |
| Granulocytes | ||
| NE-SFL width, ch | −0.261 | 0.028 |
| Lymphocytes | ||
| Lymphocytes, ×109/L | 0.278 | 0.038 |
| LY-SSC, ch | 0.407 | 0.008 |
| LY-FSC, ch | −0.334 | 0.015 |
| LY-SFL width, ch | −0.437 | 0.004 |
| Monocytes | ||
| MO-SSC, ch | −0.319 | 0.007 |
| Thrombocytes | ||
| Plateletcrit | 0.286 | 0.021 |
| Ratio | ||
| PLR | −0.353 | 0.003 |
| Chemical assay | ||
| Glucose | −0.233 | 0.047 |
| Albumin | 0.832 | <0.001 |
| Liver function tests | ||
| AST | −0.786 | 0.003 |
| Estimate | p-Value | |
|---|---|---|
| Age | −0.214 | <0.001 |
| Sex, male | −0.5 | <0.001 |
| CBC | ||
| Hemoglobin, g/dL | 0.537 | <0.001 |
| Leukocytes, ×109/L | −0.113 | 0.029 |
| Erythrocytes | ||
| MCV | −3.164 | <0.001 |
| MCH | 3.653 | <0.001 |
| MCHC | −1.886 | <0.001 |
| RDW-CV | −0.292 | <0.001 |
| Granulocytes | ||
| NE-FSC, ch | −0.161 | <0.001 |
| NE-SSC width, ch | −0.242 | <0.001 |
| Eosinophils, ×109/L | 0.142 | <0.001 |
| Lymphocytes | ||
| Lymphocytes, ×109/L | ||
| LY-SSC, ch | 0.263 | <0.001 |
| LY-FSC, ch | −0.28 | <0.001 |
| LY-SFL width, ch | −0.175 | <0.001 |
| LY-FSC width, ch | −0.143 | 0.003 |
| Monocytes | ||
| MO-SSC, ch | −0.438 | <0.001 |
| MO-SFL, ch | 0.175 | <0.001 |
| MO-FSC width, ch | −0.099 | 0.029 |
| Thrombocytes | ||
| P-LCR | −0.095 | 0.003 |
| Ratio | ||
| NLR | −0.319 | <0.001 |
| Chemical assay | ||
| Sodium | 0.123 | <0.001 |
| Albumin | 0.215 | <0.001 |
| Renal function test | ||
| BUN | −0.156 | 0.001 |
| Creatinine | 0.166 | <0.001 |
| Liver function tests | ||
| AST | −0.107 | 0.004 |
| ALP | −0.235 | <0.001 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, S.-K.; Pak, D.; Lee, J.-H.; Ryu, S.W. Identifying Key Hematological and Biochemical Indicators of Disease Severity in COVID-19 and Non-COVID-19 Patients. Diagnostics 2025, 15, 1374. https://doi.org/10.3390/diagnostics15111374
Kim S-K, Pak D, Lee J-H, Ryu SW. Identifying Key Hematological and Biochemical Indicators of Disease Severity in COVID-19 and Non-COVID-19 Patients. Diagnostics. 2025; 15(11):1374. https://doi.org/10.3390/diagnostics15111374
Chicago/Turabian StyleKim, Soo-Kyung, Daewoo Pak, Jong-Han Lee, and Sook Won Ryu. 2025. "Identifying Key Hematological and Biochemical Indicators of Disease Severity in COVID-19 and Non-COVID-19 Patients" Diagnostics 15, no. 11: 1374. https://doi.org/10.3390/diagnostics15111374
APA StyleKim, S.-K., Pak, D., Lee, J.-H., & Ryu, S. W. (2025). Identifying Key Hematological and Biochemical Indicators of Disease Severity in COVID-19 and Non-COVID-19 Patients. Diagnostics, 15(11), 1374. https://doi.org/10.3390/diagnostics15111374








