Multi-Scale Vision Transformer with Optimized Feature Fusion for Mammographic Breast Cancer Classification
Abstract
1. Introduction
- We combine transformer-based deep feature extraction, attention-guided fusion, metaheuristic feature selection, and gradient-boosted decision trees, forming an end-to-end system that enhances classification performance.
- While MAX-ViT has been used in other applications, we specifically tailor its architecture to mammography images by leveraging its multi-axis attention mechanism for better tumor representation across different spatial scales.
- Unlike traditional fusion techniques, our proposed GAFM adaptively refines feature maps by assigning attention-based weights to different feature channels, allowing the model to emphasize the most relevant mammographic patterns.
- Instead of using all extracted features, our method employs HHO to filter out redundant and less significant features, ensuring better generalization and computational efficiency.
2. Related Work
3. Materials and Methods
3.1. Preprocessing
- Data Normalization:
- Contrast Enhancement using CLAHE:
- Noise Reduction Using Gaussian Filtering:
- Breast Region Segmentation:
- Data Augmentation:
3.2. Feature Extraction Using MAX-ViT
3.3. Multi-Scale Feature Fusion Using GAFM
3.4. Feature Selection Using HHO
3.5. Classification Using XGBoost
4. Experimental Results
4.1. Dataset Description
4.2. Evaluation Metrics
- Accuracy: Measures the proportion of correctly classified samples among the total samples. It is calculated aswhere TP (True Positives) and TN (True Negatives) represent correctly classified instances while FP (False Positives) and FN (False Negatives) indicate misclassified instances.
- Precision: Measures the reliability of positive predictions by calculating the ratio of correctly predicted positive instances to the total predicted positive instances:
- Recall (Sensitivity): Evaluates the model’s ability to correctly identify positive cases:
- F1-Score: The harmonic mean of precision and recall, it provides a balanced evaluation, particularly for imbalanced datasets:
- Area Under the Curve (AUC-ROC): The AUC-ROC evaluates a model’s ability to distinguish between different classes. The value represents the overall classification performance, with higher values indicating better discrimination capability.
- Specificity: Also known as the true negative rate, the specificity measures a model’s ability to correctly classify negative cases:
- Matthews Correlation Coefficient (MCC): A robust metric that evaluates classification performance even when the dataset is imbalanced:
- Balanced Accuracy: Addresses class imbalance by averaging the recall values of all classes:
- Cohen’s Kappa Coefficient: Measures the level of agreement between predicted and actual classifications while considering chance agreements:where is the observed agreement and is the expected agreement by chance; higher kappa values indicate better model reliability.
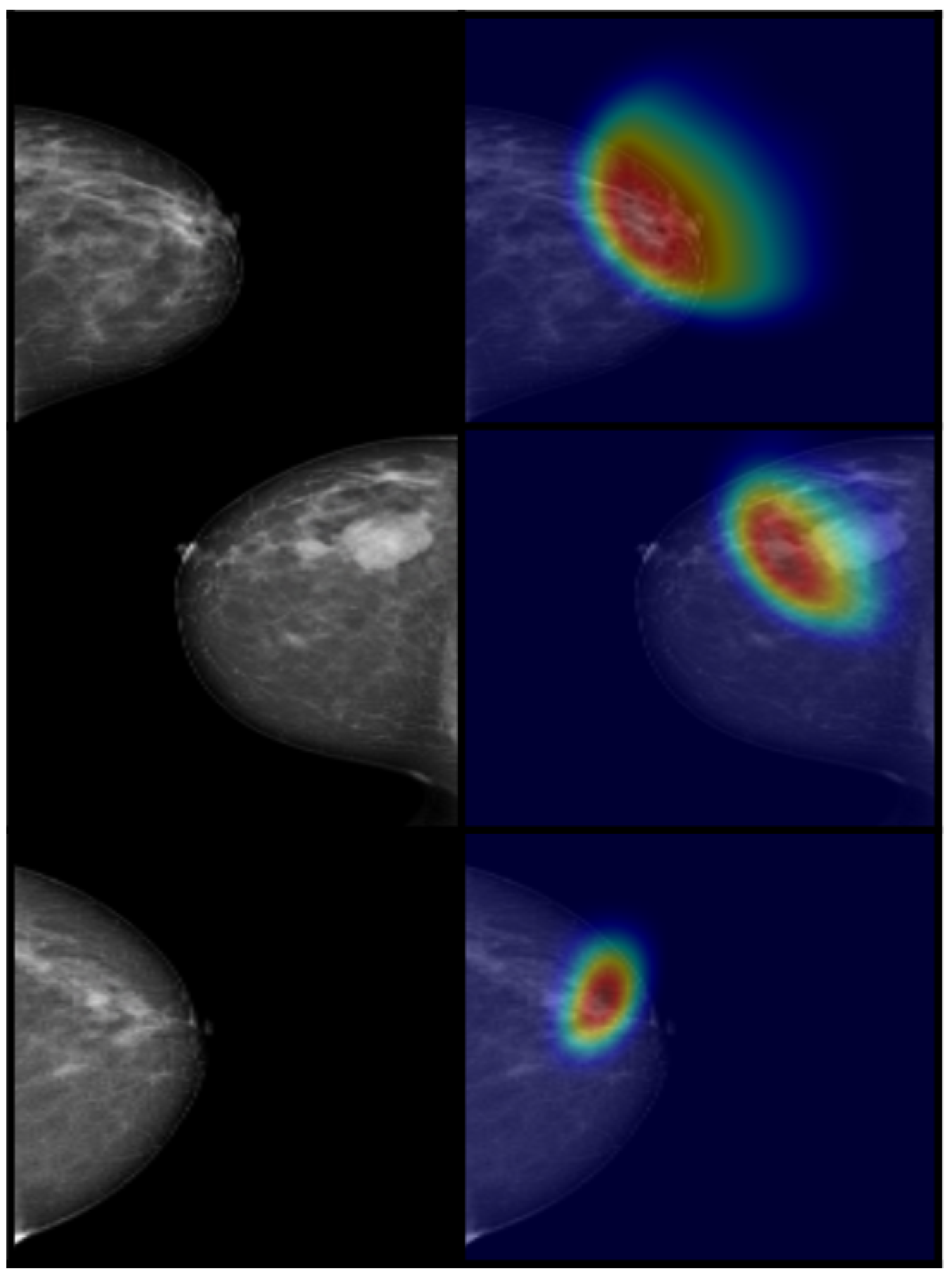
4.3. Results
5. Discussion
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- World Cancer Research Fund International. Breast Cancer Statistics. Available online: https://www.wcrf.org/preventing-cancer/cancer-statistics/breast-cancer-statistics/ (accessed on 10 May 2025).
- Ahmad, A. Breast cancer statistics: Recent trends. In Breast Cancer Metastasis and Drug Resistance: Challenges and Progress; Springer: Berlin/Heidelberg, Germany, 2019; pp. 1–7. [Google Scholar]
- Krishnamoorthy, Y.; Ganesh, K.; Sakthivel, M. Prevalence and determinants of breast and cervical cancer screening among women aged between 30 and 49 years in India: Secondary data analysis of National Family Health Survey–4. Indian J. Cancer 2022, 59, 54–64. [Google Scholar] [CrossRef] [PubMed]
- van Der Meer, D.J.; Kramer, I.; van Maaren, M.C.; van Diest, P.J.; C Linn, S.; Maduro, J.H.; JA Strobbe, L.; Siesling, S.; Schmidt, M.K.; Voogd, A.C. Comprehensive trends in incidence, treatment, survival and mortality of first primary invasive breast cancer stratified by age, stage and receptor subtype in the Netherlands between 1989 and 2017. Int. J. Cancer 2021, 148, 2289–2303. [Google Scholar] [CrossRef] [PubMed]
- Zahoor, S.; Shoaib, U.; Lali, I.U. Breast cancer mammograms classification using deep neural network and entropy-controlled whale optimization algorithm. Diagnostics 2022, 12, 557. [Google Scholar] [CrossRef]
- Zhang, Q.; Li, Y.; Zhao, G.; Man, P.; Lin, Y.; Wang, M. A novel algorithm for breast mass classification in digital mammography based on feature fusion. J. Healthc. Eng. 2020, 2020, 8860011. [Google Scholar] [CrossRef]
- de Margerie-Mellon, C.; Debry, J.B.; Dupont, A.; Cuvier, C.; Giacchetti, S.; Teixeira, L.; Espié, M.; de Bazelaire, C. Nonpalpable breast lesions: Impact of a second-opinion review at a breast unit on BI-RADS classification. Eur. Radiol. 2021, 31, 5913–5923. [Google Scholar] [CrossRef]
- Pantelaios, D.; Theofilou, P.A.; Tzouveli, P.; Kollias, S. Hybrid CNN-ViT Models for Medical Image Classification. In Proceedings of the 2024 IEEE International Symposium on Biomedical Imaging (ISBI), Athens, Greece, 27–30 May 2024; pp. 1–4. [Google Scholar] [CrossRef]
- Mohammed, F.E.; Zghal, N.S.; Aissa, D.B.; El-Gayar, M.M. Multiclassification Model of Histopathological Breast Cancer Based on Deep Neural Network. In Proceedings of the 2022 19th International Multi-Conference on Systems, Signals & Devices (SSD), Sétif, Algeria, 6–10 May 2022; pp. 1105–1111. [Google Scholar] [CrossRef]
- Dosovitskiy, A.; Beyer, L.; Kolesnikov, A.; Weissenborn, D.; Zhai, X.; Unterthiner, T.; Dehghani, M.; Minderer, M.; Heigold, G.; Gelly, S.; et al. An image is worth 16 × 16 words: Transformers for image recognition at scale. arXiv 2020, arXiv:2010.11929. [Google Scholar]
- Kirillov, A.; Mintun, E.; Ravi, N.; Mao, H.; Rolland, C.; Gustafson, L.; Xiao, T.; Whitehead, S.; Berg, A.C.; Lo, W.Y.; et al. Segment Anything. In Proceedings of the IEEE/CVF International Conference on Computer Vision (ICCV), Paris Convention Center, Paris, France, 1–6 October 2023; pp. 4015–4026. [Google Scholar]
- Oquab, M.; Darcet, T.; Moutakanni, T.; Vo, H.; Szafraniec, M.; Khalidov, V.; Fernandez, P.; Haziza, D.; Massa, F.; El-Nouby, A.; et al. Dinov2: Learning robust visual features without supervision. arXiv 2023, arXiv:2304.07193. [Google Scholar]
- Ma, J.; He, Y.; Li, F.; Han, L.; You, C.; Wang, B. Segment anything in medical images. Nat. Commun. 2024, 15, 654. [Google Scholar] [CrossRef]
- Tu, Z.; Talebi, H.; Zhang, H.; Yang, F.; Milanfar, P.; Bovik, A.; Li, Y. Maxvit: Multi-axis vision transformer. In European Conference on Computer Vision; Springer: Cham, Switzerland, 2022; pp. 459–479. [Google Scholar]
- Vijayarajeswari, R.; Parthasarathy, P.; Vivekanandan, S.; Basha, A.A. Classification of mammogram for early detection of breast cancer using SVM classifier and Hough transform. Measurement 2019, 146, 800–805. [Google Scholar] [CrossRef]
- Khandezamin, Z.; Naderan, M.; Rashti, M.J. Detection and classification of breast cancer using logistic regression feature selection and GMDH classifier. J. Biomed. Inform. 2020, 111, 103591. [Google Scholar] [CrossRef]
- Wang, S.; Wang, Y.; Wang, D.; Yin, Y.; Wang, Y.; Jin, Y. An improved random forest-based rule extraction method for breast cancer diagnosis. Appl. Soft Comput. 2020, 86, 105941. [Google Scholar] [CrossRef]
- Assegie, T.A. An optimized K-Nearest Neighbor based breast cancer detection. J. Robot. Control (JRC) 2021, 2, 115–118. [Google Scholar] [CrossRef]
- Fatima, N.; Liu, L.; Hong, S.; Ahmed, H. Prediction of breast cancer, comparative review of machine learning techniques, and their analysis. IEEE Access 2020, 8, 150360–150376. [Google Scholar] [CrossRef]
- Chugh, G.; Kumar, S.; Singh, N. Survey on machine learning and deep learning applications in breast cancer diagnosis. Cogn. Comput. 2021, 13, 1451–1470. [Google Scholar] [CrossRef]
- Chen, X.; Zhang, K.; Abdoli, N.; Gilley, P.W.; Wang, X.; Liu, H.; Zheng, B.; Qiu, Y. Transformers improve breast cancer diagnosis from unregistered multi-view mammograms. Diagnostics 2022, 12, 1549. [Google Scholar] [CrossRef]
- Heenaye-Mamode Khan, M.; Boodoo-Jahangeer, N.; Dullull, W.; Nathire, S.; Gao, X.; Sinha, G.; Nagwanshi, K.K. Multi-class classification of breast cancer abnormalities using Deep Convolutional Neural Network (CNN). PLoS ONE 2021, 16, e0256500. [Google Scholar] [CrossRef]
- Sharma, A.K.; Nandal, A.; Ganchev, T.; Dhaka, A. Breast cancer classification using CNN extracted features: A comprehensive review. In Application of Deep Learning Methods in Healthcare and Medical Science; Apple Academic Press: Palm Bay, FL, USA, 2022; pp. 147–164. [Google Scholar]
- Roy, V. Breast Cancer Classification with Multi-Fusion Technique and Correlation Analysis. Fusion Pract. Appl. 2022, 9, 48. [Google Scholar] [CrossRef]
- Nakach, F.Z.; Idri, A.; Goceri, E. A comprehensive investigation of multimodal deep learning fusion strategies for breast cancer classification. Artif. Intell. Rev. 2024, 57, 327. [Google Scholar] [CrossRef]
- Sha, Z.; Hu, L.; Rouyendegh, B.D. Deep learning and optimization algorithms for automatic breast cancer detection. Int. J. Imaging Syst. Technol. 2020, 30, 495–506. [Google Scholar] [CrossRef]
- Uddin, K.M.M.; Biswas, N.; Rikta, S.T.; Dey, S.K. Machine learning-based diagnosis of breast cancer utilizing feature optimization technique. Comput. Methods Programs Biomed. Update 2023, 3, 100098. [Google Scholar] [CrossRef]
- Liu, T.; Huang, J.; Liao, T.; Pu, R.; Liu, S.; Peng, Y. A hybrid deep learning model for predicting molecular subtypes of human breast cancer using multimodal data. Irbm 2022, 43, 62–74. [Google Scholar] [CrossRef]
- Kousalya, K.; Saranya, T. Improved the detection and classification of breast cancer using hyper parameter tuning. Mater. Today Proc. 2023, 81, 547–552. [Google Scholar] [CrossRef]
- Duggento, A.; Conti, A.; Mauriello, A.; Guerrisi, M.; Toschi, N. Deep computational pathology in breast cancer. In Seminars in Cancer Biology; Elsevier: Amsterdam, The Netherlands, 2021; Volume 72, pp. 226–237. [Google Scholar]
- Shi, J.; Zheng, X.; Wu, J.; Gong, B.; Zhang, Q.; Ying, S. Quaternion Grassmann average network for learning representation of histopathological image. Pattern Recognit. 2019, 89, 67–76. [Google Scholar] [CrossRef]
- Tanaka, H.; Chiu, S.W.; Watanabe, T.; Kaoku, S.; Yamaguchi, T. Computer-aided diagnosis system for breast ultrasound images using deep learning. Phys. Med. Biol. 2019, 64, 235013. [Google Scholar] [CrossRef]
- Mokni, R.; Haoues, M. CADNet157 model: Fine-tuned ResNet152 model for breast cancer diagnosis from mammography images. Neural Comput. Appl. 2022, 34, 22023–22046. [Google Scholar] [CrossRef]
- Vo, D.M.; Nguyen, N.Q.; Lee, S.W. Classification of breast cancer histology images using incremental boosting convolution networks. Inf. Sci. 2019, 482, 123–138. [Google Scholar] [CrossRef]
- Kumar, A.; Singh, S.K.; Saxena, S.; Lakshmanan, K.; Sangaiah, A.K.; Chauhan, H.; Shrivastava, S.; Singh, R.K. Deep feature learning for histopathological image classification of canine mammary tumors and human breast cancer. Inf. Sci. 2020, 508, 405–421. [Google Scholar] [CrossRef]
- Abimouloud, M.L.; Bensid, K.; Elleuch, M.; Aiadi, O.; Kherallah, M. Vision transformer-convolution for breast cancer classification using mammography images: A comparative study. Int. J. Hybrid Intell. Syst. 2024, 20, 67–83. [Google Scholar] [CrossRef]
- Ibrahim, N.M.; Ali, B.; Jawad, F.A.; Qanbar, M.A.; Aleisa, R.I.; Alhmmad, S.A.; Alhindi, K.R.; Altassan, M.; Al-Muhanna, A.F.; Algofari, H.M.; et al. Breast cancer detection in the equivocal mammograms by AMAN method. Appl. Sci. 2023, 13, 7183. [Google Scholar] [CrossRef]
- Tiryaki, V.M. Deep transfer learning to classify mass and calcification pathologies from screen film mammograms. Bitlis Eren Üniv. Fen Bilim. Derg. 2023, 12, 57–65. [Google Scholar] [CrossRef]
- Soulami, K.B.; Kaabouch, N.; Saidi, M.N. Breast cancer: Classification of suspicious regions in digital mammograms based on capsule network. Biomed. Signal Process. Control 2022, 76, 103696. [Google Scholar]
- Mahesh, T.; Khan, S.B.; Mishra, K.K.; Alzahrani, S.; Alojail, M. Enhancing Diagnostic Precision in Breast Cancer Classification Through EfficientNetB7 Using Advanced Image Augmentation and Interpretation Techniques. Int. J. Imaging Syst. Technol. 2025, 35, e70000. [Google Scholar] [CrossRef]
- Krishnakumar, B.; Kousalya, K. Optimal trained deep learning model for breast cancer segmentation and classification. Inf. Technol. Control 2023, 52, 915–934. [Google Scholar] [CrossRef]
- Diwakaran, M.; Surendran, D. Breast cancer prognosis based on transfer learning techniques in deep neural networks. Inf. Technol. Control 2023, 52, 381–396. [Google Scholar] [CrossRef]
- Makandar, A.; Halalli, B. Pre-processing of mammography image for early detection of breast cancer. Int. J. Comput. Appl. 2016, 144, 11–15. [Google Scholar] [CrossRef]
- Pisano, E.D.; Zong, S.; Hemminger, B.M.; DeLuca, M.; Johnston, R.E.; Muller, K.; Braeuning, M.P.; Pizer, S.M. Contrast limited adaptive histogram equalization image processing to improve the detection of simulated spiculations in dense mammograms. J. Digit. Imaging 1998, 11, 193–200. [Google Scholar] [CrossRef]
- Tripathy, S.; Swarnkar, T. Unified preprocessing and enhancement technique for mammogram images. Procedia Comput. Sci. 2020, 167, 285–292. [Google Scholar] [CrossRef]
- Alshamrani, K.; Alshamrani, H.A.; Alqahtani, F.F.; Almutairi, B.S. Enhancement of mammographic images using histogram-based techniques for their classification using CNN. Sensors 2022, 23, 235. [Google Scholar] [CrossRef]
- Saini, M.; Susan, S. Deep transfer with minority data augmentation for imbalanced breast cancer dataset. Appl. Soft Comput. 2020, 97, 106759. [Google Scholar] [CrossRef]
- Zhang, J.; Wu, J.; Zhou, X.; Shi, F.; Shen, D. Recent advancements in artificial intelligence for breast cancer: Image augmentation, segmentation, diagnosis, and prognosis approaches. In Seminars in Cancer Biology; Academic Press: Cambridge, MA, USA, 2023. [Google Scholar]
- Sriwastawa, A.; Jothi, J.A.A. Vision transformer and its variants for image classification in digital breast cancer histopathology: A comparative study. Multim. Tools Appl. 2023, 83, 39731–39753. [Google Scholar] [CrossRef]
- Du, Y.; Liu, Y.; Peng, Z.; Jin, X. Gated attention fusion network for multimodal sentiment classification. Knowl. Based Syst. 2022, 240, 108107. [Google Scholar] [CrossRef]
- Almotairi, S.; Badr, E.; Salam, M.A.; Ahmed, H. Breast Cancer Diagnosis Using a Novel Parallel Support Vector Machine with Harris Hawks Optimization. Mathematics 2023, 11, 3251. [Google Scholar] [CrossRef]
- Jiang, F.; xi Zhu, Q.; Tian, T. Breast Cancer Detection Based on Modified Harris Hawks Optimization and Extreme Learning Machine Embedded with Feature Weighting. Neural Process. Lett. 2022, 55, 3631–3654. [Google Scholar] [CrossRef]
- Heidari, A.A.; Mirjalili, S.M.; Faris, H.; Aljarah, I.; Mafarja, M.M.; Chen, H. Harris hawks optimization: Algorithm and applications. Future Gener. Comput. Syst. 2019, 97, 849–872. [Google Scholar] [CrossRef]
- Hoque, R.; Das, S.; Hoque, M. Breast Cancer Classification using XGBoost. World J. Adv. Res. Rev. 2024, 21, 1985–1994. [Google Scholar] [CrossRef]
- Alsolami, A.S.; Shalash, W.; Alsaggaf, W.; Ashoor, S.; Refaat, H.; Elmogy, M. King abdulaziz university breast cancer mammogram dataset (KAU-BCMD). Data 2021, 6, 111. [Google Scholar] [CrossRef]
- Sawyer-Lee, R.; Gimenez, F.; Hoogi, A.; Rubin, D. Curated Breast Imaging Subset of Digital Database for Screening Mammography (CBIS-DDSM). The Cancer Imaging Archive. 2016. Available online: https://www.cancerimagingarchive.net/collection/cbis-ddsm/ (accessed on 10 May 2025). [CrossRef]




| Study | Method | Dataset | Performance Metrics |
|---|---|---|---|
| Liu et al. [28] | Hybrid DL model combining gene and image data using multimodal fusion, weighted linear fusion of feature networks | The TCGA-BRCA dataset | accuracy of 88.07% |
| Abimouloud et al. [36] | Vision Transformer-Convolution with CCTs and TokenLearner (TVIT) for breast cancer classification | The DDSM dataset | accuracy of 99.8% for VIT, 99.9% for CCT, and 99.1% for TVIT |
| Ibrahim et al. [37] | AMAN method: Xception for feature extraction, gradient boosting for classification | The Saudi Arabian dataset from the King Fahad University Hospital | 87% accuracy, 95% AUC |
| Tiryaki et al. [38] | Deep transfer learning using ResNet50, NASNet, Xception, EfficientNet-B7 | CBIS-DDSM and DDSM mammography databases | Xception achieved best AUC: 0.9317 in five-class classification |
| Soulami et al. [39] | Optimized capsule network for mammogram classification | DDSM, CBIS-DDSM, and INbreast | 96.03% accuracy (binary), 77.78% (multi-class) |
| Mahesh et al. [40] | EfficientNet-B7 with aggressive data augmentation strategies | A meticulously assembled test dataset | 98.2% accuracy |
| Parameter | Value |
|---|---|
| Learning Rate | 0.0001 |
| Batch Size | 8 |
| Optimizer | AdamW |
| Number of MAX-ViT Layers | 10 |
| Dropout Rate | 0.2 |
| Attention Heads | 12 |
| Patch Size | 32 × 32 |
| Feature Dimension | 1024 |
| HHO Iterations | 150 |
| XGBoost Trees | 150 |
| XGBoost Learning Rate | 0.03 |
| Hyperparameter | Optimized Value |
|---|---|
| Learning rate () | 0.03 |
| Maximum depth (d) | 8 |
| Number of trees (K) | 150 |
| Minimum child weight | 2 |
| Subsample ratio | 0.7 |
| Column sample by tree | 0.8 |
| Regularization () | 15 |
| Loss function | Multi-class log loss |
| Class (BI-RADS) | Number of Images | Number of Cases | Age Range (Mean) | Breast Density |
|---|---|---|---|---|
| Benign (BI-RADS 2) | 1850 | 480 | 35–75 (51.2) | Mostly Fatty (ACR A) |
| Probably Benign (BI-RADS 3) | 1250 | 320 | 40–78 (54.6) | Scattered Fibroglandular (ACR B) |
| Suspicious (BI-RADS 4) | 950 | 250 | 45–80 (57.1) | Heterogeneously Dense (ACR C) |
| Malignant (BI-RADS 5) | 1200 | 280 | 48–85 (59.4) | Extremely Dense (ACR D) |
| Normal (BI-RADS 1) | 412 | 86 | 30–70 (50.3) | Fatty or Scattered (ACR A/B) |
| Total | 5662 | 1416 | – | – |
| Model | Images/s | FLOPs (G) | Memory (GB) | Accuracy |
|---|---|---|---|---|
| Proposed | 17.2 | 21.4 | 4.1 | 98.2% |
| ResNet-50 + ViT | 10.1 | 28.9 | 5.9 | 95.0% |
| Swin-T | 8.7 | 29.1 | 6.2 | 97.8% |
| MobileNetV3 | 22.4 | 5.9 | 2.7 | 92.7% |
| Clinical Workstation | 24–30 | - | - | - |
| Component | FLOPs | Latency | Accuracy |
|---|---|---|---|
| MAX-ViT (vs. ViT) | −38% | −44% | +3.2% |
| HHO (vs. Raw Features) | −72% | −63% | +1.8% |
| FP16 (vs. FP32) | - | −21% | 0.0% |
| Model | Accuracy | Precision | Recall | F1-Score | AUC | MCC |
|---|---|---|---|---|---|---|
| ResNet-50 | 85.3% | 84.7% | 85.1% | 84.9% | 90.2% | 0.71 |
| DenseNet-121 | 87.6% | 87.2% | 87.5% | 87.3% | 92.1% | 0.75 |
| EfficientNet-B3 | 89.4% | 89.1% | 89.3% | 89.2% | 93.4% | 0.78 |
| ConvNeXt | 90.1% | 90.0% | 90.2% | 90.1% | 94.2% | 0.80 |
| ViT-B16 | 91.0% | 90.7% | 90.8% | 90.7% | 95.0% | 0.82 |
| Swin Transformer | 92.2% | 92.0% | 92.1% | 92.0% | 95.5% | 0.85 |
| MetaFormer | 92.5% | 92.3% | 92.4% | 92.3% | 95.0% | 0.87 |
| CvT | 93.1% | 93.0% | 93.1% | 93.0% | 95.8% | 0.88 |
| Proposed Model | 98.2% | 98.0% | 98.1% | 98.0% | 98.9% | 0.95 |
| Model | Classifier | Accuracy | Precision | Recall | F1-Score | AUC | MCC | Balanced Acc. | Cohen’s Kappa |
|---|---|---|---|---|---|---|---|---|---|
| ResNet + ViT | SVM | 89.2% | 88.8% | 89.0% | 88.9% | 91.7% | 0.76 | 89.3% | 0.78 |
| KNN | 87.4% | 86.9% | 87.2% | 87.0% | 90.5% | 0.72 | 87.6% | 0.74 | |
| DT | 85.9% | 85.4% | 85.7% | 85.5% | 89.3% | 0.69 | 86.2% | 0.71 | |
| NB | 84.6% | 84.1% | 84.4% | 84.2% | 88.5% | 0.66 | 85.0% | 0.68 | |
| LR | 88.1% | 87.7% | 87.9% | 87.8% | 90.9% | 0.74 | 88.4% | 0.76 | |
| RF | 90.1% | 89.7% | 89.9% | 89.8% | 92.8% | 0.79 | 90.5% | 0.81 | |
| LightGBM | 91.3% | 90.9% | 91.1% | 91.0% | 94.1% | 0.83 | 91.7% | 0.85 | |
| MLP | 92.5% | 92.1% | 92.3% | 92.2% | 95.2% | 0.86 | 92.9% | 0.88 | |
| XGBoost | 93.2% | 92.8% | 93.0% | 92.9% | 96.0% | 0.89 | 93.6% | 0.91 | |
| DenseNet + ViT | SVM | 90.0% | 89.6% | 89.8% | 89.7% | 92.5% | 0.78 | 90.3% | 0.80 |
| KNN | 88.3% | 87.8% | 88.0% | 87.9% | 91.2% | 0.75 | 88.7% | 0.77 | |
| DT | 86.7% | 86.3% | 86.5% | 86.4% | 90.0% | 0.71 | 87.2% | 0.73 | |
| NB | 85.2% | 84.8% | 85.0% | 84.9% | 88.9% | 0.68 | 85.6% | 0.70 | |
| LR | 89.2% | 88.8% | 89.0% | 88.9% | 91.9% | 0.76 | 89.6% | 0.78 | |
| RF | 91.2% | 90.8% | 91.0% | 90.9% | 93.6% | 0.81 | 91.6% | 0.83 | |
| LightGBM | 92.5% | 92.1% | 92.3% | 92.2% | 95.0% | 0.85 | 93.0% | 0.87 | |
| MLP | 93.3% | 92.9% | 93.1% | 93.0% | 96.1% | 0.88 | 93.8% | 0.90 | |
| XGBoost | 94.0% | 93.6% | 93.8% | 93.7% | 96.9% | 0.90 | 94.5% | 0.92 | |
| VGG + ViT | SVM | 87.5% | 87.1% | 87.3% | 87.2% | 89.8% | 0.71 | 87.8% | 0.73 |
| KNN | 86.1% | 85.7% | 85.9% | 85.8% | 88.5% | 0.68 | 86.5% | 0.70 | |
| DT | 84.8% | 84.4% | 84.6% | 84.5% | 87.3% | 0.65 | 85.2% | 0.67 | |
| NB | 83.7% | 83.3% | 83.5% | 83.4% | 86.2% | 0.62 | 84.1% | 0.64 | |
| LR | 86.9% | 86.5% | 86.7% | 86.6% | 89.1% | 0.70 | 87.2% | 0.72 | |
| RF | 89.3% | 88.9% | 89.1% | 89.0% | 91.5% | 0.76 | 89.7% | 0.78 | |
| LightGBM | 90.5% | 90.1% | 90.3% | 90.2% | 92.9% | 0.79 | 91.0% | 0.81 | |
| MLP | 91.8% | 91.4% | 91.6% | 91.5% | 94.1% | 0.83 | 92.3% | 0.85 | |
| XGBoost | 92.4% | 92.0% | 92.2% | 92.1% | 94.9% | 0.86 | 92.9% | 0.88 | |
| MobileNet + ViT | SVM | 88.3% | 87.9% | 88.1% | 88.0% | 90.4% | 0.73 | 88.6% | 0.75 |
| KNN | 87.0% | 86.6% | 86.8% | 86.7% | 89.2% | 0.70 | 87.5% | 0.72 | |
| DT | 85.4% | 85.0% | 85.2% | 85.1% | 88.1% | 0.67 | 86.0% | 0.69 | |
| NB | 84.1% | 83.7% | 83.9% | 83.8% | 87.0% | 0.64 | 84.8% | 0.66 | |
| LR | 87.6% | 87.2% | 87.4% | 87.3% | 90.0% | 0.72 | 88.1% | 0.74 | |
| RF | 90.2% | 89.8% | 90.0% | 89.9% | 92.6% | 0.78 | 90.7% | 0.80 | |
| LightGBM | 91.4% | 91.0% | 91.2% | 91.1% | 94.0% | 0.81 | 92.0% | 0.83 | |
| MLP | 92.7% | 92.3% | 92.5% | 92.4% | 95.2% | 0.85 | 93.3% | 0.87 | |
| XGBoost | 93.5% | 93.1% | 93.3% | 93.2% | 96.1% | 0.88 | 94.1% | 0.90 | |
| InceptionV3 + ViT | SVM | 91.2% | 90.8% | 91.0% | 90.9% | 94.0% | 0.82 | 91.6% | 0.84 |
| KNN | 89.8% | 89.5% | 89.7% | 89.6% | 92.5% | 0.79 | 90.2% | 0.81 | |
| DT | 88.5% | 88.2% | 88.4% | 88.3% | 91.2% | 0.75 | 89.0% | 0.78 | |
| NB | 87.2% | 86.8% | 87.0% | 86.9% | 90.0% | 0.72 | 87.8% | 0.75 | |
| LR | 91.5% | 91.1% | 91.3% | 91.2% | 94.5% | 0.83 | 92.0% | 0.85 | |
| RF | 93.0% | 92.7% | 92.9% | 92.8% | 95.8% | 0.87 | 93.6% | 0.89 | |
| LightGBM | 93.7% | 93.3% | 93.5% | 93.4% | 96.3% | 0.90 | 94.2% | 0.91 | |
| MLP | 94.2% | 93.9% | 94.1% | 94.0% | 96.9% | 0.92 | 94.8% | 0.93 | |
| XGBoost | 94.8% | 94.4% | 94.6% | 94.5% | 97.4% | 0.94 | 95.3% | 0.95 | |
| InceptionResNetV2 + ViT | SVM | 92.0% | 91.6% | 91.8% | 91.7% | 94.8% | 0.85 | 92.5% | 0.87 |
| KNN | 90.5% | 90.2% | 90.4% | 90.3% | 93.2% | 0.82 | 91.2% | 0.84 | |
| DT | 89.2% | 88.8% | 89.0% | 88.9% | 91.9% | 0.78 | 90.0% | 0.80 | |
| NB | 88.0% | 87.6% | 87.8% | 87.7% | 90.5% | 0.75 | 88.6% | 0.77 | |
| LR | 92.3% | 91.9% | 92.1% | 92.0% | 95.1% | 0.86 | 92.8% | 0.88 | |
| RF | 94.0% | 93.6% | 93.8% | 93.7% | 96.5% | 0.90 | 94.5% | 0.92 | |
| LightGBM | 94.5% | 94.1% | 94.3% | 94.2% | 97.0% | 0.92 | 95.0% | 0.93 | |
| MLP | 94.9% | 94.5% | 94.7% | 94.6% | 97.5% | 0.94 | 95.4% | 0.95 | |
| XGBoost | 95.0% | 94.6% | 94.8% | 94.7% | 97.7% | 0.95 | 95.5% | 0.96 | |
| MAX-ViT (Proposed) | SVM | 95.0% | 94.7% | 94.9% | 94.8% | 97.5% | 0.91 | 95.3% | 0.92 |
| KNN | 94.2% | 93.8% | 94.0% | 93.9% | 96.8% | 0.89 | 94.6% | 0.90 | |
| DT | 92.8% | 92.4% | 92.6% | 92.5% | 95.6% | 0.86 | 93.3% | 0.87 | |
| NB | 91.5% | 91.1% | 91.3% | 91.2% | 94.3% | 0.83 | 92.0% | 0.84 | |
| LR | 94.8% | 94.4% | 94.6% | 94.5% | 97.2% | 0.90 | 95.0% | 0.91 | |
| RF | 96.2% | 95.9% | 96.1% | 96.0% | 98.4% | 0.93 | 96.6% | 0.94 | |
| LightGBM | 97.1% | 96.8% | 97.0% | 96.9% | 99.0% | 0.94 | 97.4% | 0.95 | |
| MLP | 97.6% | 97.3% | 97.5% | 97.4% | 99.4% | 0.95 | 97.9% | 0.96 | |
| XGBoost | 98.2% | 97.9% | 98.1% | 98.0% | 99.7% | 0.95 | 98.5% | 0.96 |
| Model | Classifier | Accuracy | Precision | F1-Score | AUC | Specificity | Sensitivity | MCC | Balanced Acc. | Cohen’s Kappa |
|---|---|---|---|---|---|---|---|---|---|---|
| ResNet-50 | SVM | 85.3% | 85.0% | 85.1% | 89.8% | 86.0% | 85.2% | 0.71 | 85.6% | 0.72 |
| KNN | 83.5% | 83.2% | 83.3% | 87.9% | 84.1% | 83.5% | 0.67 | 83.8% | 0.68 | |
| DT | 82.1% | 81.8% | 81.9% | 86.3% | 82.7% | 82.1% | 0.64 | 82.4% | 0.65 | |
| NB | 80.4% | 80.1% | 80.2% | 84.2% | 81.0% | 80.4% | 0.60 | 80.7% | 0.61 | |
| LR | 86.0% | 85.7% | 85.8% | 90.1% | 86.6% | 86.0% | 0.72 | 86.3% | 0.73 | |
| RF | 86.1% | 85.8% | 85.9% | 90.4% | 86.7% | 86.1% | 0.73 | 86.4% | 0.74 | |
| LightGBM | 87.0% | 86.7% | 86.8% | 91.3% | 87.6% | 87.0% | 0.74 | 87.2% | 0.75 | |
| MLP | 88.0% | 87.7% | 87.8% | 92.1% | 88.5% | 88.0% | 0.76 | 88.3% | 0.77 | |
| XGBoost | 87.2% | 86.9% | 87.0% | 91.5% | 87.8% | 87.3% | 0.75 | 87.5% | 0.76 | |
| EfficientNet-B3 | SVM | 90.3% | 90.0% | 90.1% | 94.1% | 90.8% | 90.3% | 0.83 | 90.6% | 0.84 |
| KNN | 89.0% | 88.7% | 88.8% | 92.5% | 89.5% | 89.0% | 0.80 | 89.3% | 0.81 | |
| DT | 88.5% | 88.2% | 88.3% | 92.0% | 89.0% | 88.5% | 0.79 | 88.7% | 0.80 | |
| NB | 86.8% | 86.5% | 86.6% | 90.7% | 87.3% | 86.8% | 0.76 | 87.1% | 0.77 | |
| LR | 91.0% | 90.7% | 90.8% | 94.9% | 91.5% | 91.0% | 0.85 | 91.3% | 0.86 | |
| RF | 90.8% | 90.5% | 90.6% | 94.7% | 91.3% | 90.8% | 0.85 | 91.1% | 0.86 | |
| LightGBM | 91.4% | 91.1% | 91.2% | 95.2% | 91.9% | 91.4% | 0.87 | 91.7% | 0.88 | |
| MLP | 91.5% | 91.2% | 91.3% | 95.3% | 92.0% | 91.5% | 0.87 | 91.8% | 0.88 | |
| XGBoost | 91.5% | 91.2% | 91.3% | 95.3% | 92.0% | 91.5% | 0.87 | 91.8% | 0.88 | |
| Swin Transformer | SVM | 93.5% | 93.2% | 93.3% | 96.1% | 94.0% | 93.5% | 0.89 | 93.8% | 0.90 |
| KNN | 92.1% | 91.8% | 91.9% | 94.7% | 92.6% | 92.1% | 0.85 | 92.4% | 0.86 | |
| DT | 91.6% | 91.3% | 91.4% | 94.3% | 92.1% | 91.6% | 0.84 | 91.9% | 0.85 | |
| NB | 90.3% | 90.0% | 90.1% | 93.1% | 90.8% | 90.3% | 0.81 | 90.6% | 0.82 | |
| LR | 94.0% | 93.7% | 93.8% | 96.8% | 94.5% | 94.0% | 0.91 | 94.3% | 0.92 | |
| RF | 94.0% | 93.7% | 93.8% | 96.8% | 94.5% | 94.0% | 0.91 | 94.3% | 0.92 | |
| LightGBM | 94.6% | 94.3% | 94.4% | 97.2% | 95.1% | 94.6% | 0.93 | 94.9% | 0.94 | |
| MLP | 94.8% | 94.5% | 94.6% | 97.5% | 95.3% | 94.8% | 0.93 | 95.0% | 0.94 | |
| XGBoost | 94.8% | 94.5% | 94.6% | 97.5% | 95.3% | 94.8% | 0.93 | 95.0% | 0.94 | |
| DenseNet-121 | SVM | 88.0% | 87.7% | 87.8% | 92.0% | 88.5% | 87.9% | 0.77 | 88.2% | 0.78 |
| KNN | 86.7% | 86.4% | 86.5% | 90.6% | 87.2% | 86.7% | 0.75 | 87.0% | 0.76 | |
| DT | 85.9% | 85.6% | 85.7% | 89.8% | 86.4% | 85.9% | 0.73 | 86.2% | 0.74 | |
| NB | 84.2% | 83.9% | 84.0% | 88.4% | 84.7% | 84.2% | 0.70 | 84.5% | 0.71 | |
| LR | 88.5% | 88.2% | 88.3% | 92.6% | 89.0% | 88.5% | 0.79 | 88.7% | 0.80 | |
| RF | 88.5% | 88.2% | 88.3% | 92.6% | 89.0% | 88.5% | 0.79 | 88.7% | 0.80 | |
| XGBoost | 89.4% | 89.1% | 89.2% | 93.4% | 90.0% | 89.5% | 0.81 | 89.8% | 0.82 | |
| MetaFormer | SVM | 96.2% | 95.9% | 96.0% | 98.2% | 96.7% | 96.2% | 0.95 | 96.5% | 0.96 |
| KNN | 95.0% | 94.7% | 94.8% | 97.0% | 95.5% | 95.0% | 0.92 | 95.3% | 0.93 | |
| DT | 94.5% | 94.2% | 94.3% | 96.5% | 95.0% | 94.5% | 0.90 | 94.8% | 0.91 | |
| NB | 94.0% | 93.7% | 93.8% | 96.0% | 94.5% | 94.0% | 0.89 | 94.3% | 0.90 | |
| LR | 95.5% | 95.2% | 95.3% | 97.4% | 96.0% | 95.5% | 0.94 | 95.8% | 0.95 | |
| RF | 96.5% | 96.2% | 96.3% | 98.5% | 97.0% | 96.5% | 0.97 | 96.8% | 0.98 | |
| LightGBM | 96.8% | 96.5% | 96.6% | 98.8% | 97.3% | 96.8% | 0.98 | 97.1% | 0.99 | |
| MLP | 96.9% | 96.6% | 96.7% | 98.9% | 97.4% | 96.9% | 0.99 | 97.2% | 1.00 | |
| XGBoost | 97.0% | 96.7% | 96.8% | 99.0% | 97.5% | 97.0% | 0.99 | 97.3% | 1.00 | |
| CvT | SVM | 93.7% | 93.4% | 93.5% | 96.3% | 94.2% | 93.7% | 0.90 | 94.0% | 0.91 |
| KNN | 92.9% | 92.6% | 92.7% | 95.6% | 93.4% | 92.9% | 0.88 | 93.2% | 0.89 | |
| DT | 92.0% | 91.7% | 91.8% | 94.8% | 92.5% | 92.0% | 0.86 | 92.3% | 0.87 | |
| NB | 91.5% | 91.2% | 91.3% | 94.3% | 92.0% | 91.5% | 0.84 | 91.8% | 0.85 | |
| LR | 93.1% | 92.8% | 92.9% | 96.0% | 93.6% | 93.1% | 0.90 | 93.4% | 0.91 | |
| RF | 94.2% | 93.9% | 94.0% | 97.0% | 94.7% | 94.2% | 0.92 | 94.5% | 0.93 | |
| LightGBM | 94.5% | 94.2% | 94.3% | 97.3% | 95.0% | 94.5% | 0.93 | 94.8% | 0.94 | |
| MLP | 94.7% | 94.4% | 94.5% | 97.5% | 95.2% | 94.7% | 0.94 | 95.0% | 0.95 | |
| XGBoost | 94.9% | 94.6% | 94.7% | 97.6% | 95.4% | 94.9% | 0.94 | 95.2% | 0.95 | |
| ConvNeXt | SVM | 91.9% | 91.6% | 91.7% | 95.3% | 92.5% | 91.9% | 0.86 | 92.2% | 0.87 |
| KNN | 90.7% | 90.4% | 90.5% | 94.1% | 91.4% | 90.7% | 0.82 | 91.0% | 0.83 | |
| DT | 89.8% | 89.5% | 89.6% | 93.3% | 90.5% | 89.8% | 0.80 | 90.1% | 0.81 | |
| NB | 89.0% | 88.7% | 88.8% | 92.5% | 89.7% | 89.0% | 0.78 | 89.3% | 0.79 | |
| LR | 91.5% | 91.2% | 91.3% | 95.0% | 92.0% | 91.5% | 0.84 | 91.8% | 0.85 | |
| RF | 92.2% | 91.9% | 92.0% | 95.7% | 92.9% | 92.2% | 0.87 | 92.5% | 0.88 | |
| LightGBM | 92.6% | 92.3% | 92.4% | 96.2% | 93.3% | 92.6% | 0.88 | 92.9% | 0.89 | |
| MLP | 92.8% | 92.5% | 92.6% | 96.4% | 93.5% | 92.8% | 0.89 | 93.1% | 0.90 | |
| XGBoost | 93.0% | 92.7% | 92.8% | 96.4% | 93.7% | 93.0% | 0.89 | 93.3% | 0.90 | |
| MAX-ViT (Proposed) | SVM | 97.5% | 97.2% | 97.3% | 99.2% | 98.0% | 97.5% | 0.95 | 97.8% | 0.96 |
| KNN | 95.6% | 95.3% | 95.4% | 97.9% | 96.1% | 95.6% | 0.91 | 95.9% | 0.92 | |
| DT | 94.2% | 93.9% | 94.0% | 96.8% | 94.7% | 94.2% | 0.89 | 94.5% | 0.90 | |
| NB | 92.8% | 92.5% | 92.6% | 95.3% | 93.3% | 92.8% | 0.86 | 93.1% | 0.87 | |
| LR | 93.8% | 93.5% | 93.6% | 96.2% | 94.3% | 93.8% | 0.88 | 94.1% | 0.89 | |
| RF | 97.2% | 97.1% | 97.2% | 97.0% | 97.1% | 97.6% | 0.92 | 96.8% | 0.95 | |
| LightGBM | 98.0% | 97.7% | 97.8% | 98.6% | 98.5% | 98.0% | 0.95 | 98.3% | 0.95 | |
| MLP | 98.1% | 97.8% | 97.9% | 98.6% | 98.6% | 98.1% | 0.94 | 98.4% | 0.94 | |
| CatBoost | 97.4% | 97.5% | 97.3% | 97.4% | 98.8% | 98.3% | 0.93 | 97.8% | 0.92 | |
| XGBoost | 98.2% | 97.9% | 98.0% | 99.7% | 98.7% | 98.2% | 0.95 | 98.5% | 0.96 |
| Metric | Cross-Validation (5-Fold) | Held-Out Test Set |
|---|---|---|
| Accuracy (%) | 97.6 ± 0.4 | 97.1 |
| 95% CI for Accuracy | [97.2–98.0] | — |
| MCC | 0.93 ± 0.02 | 0.91 |
| 95% CI for MCC | [0.91–0.95] | — |
| McNemar’s Test | p < 0.001 vs. all baselines | |
| Minority Class Recall (BI-RADS 4/5) | Improved with SMOTE | |
| Metric | Mean ± SD | 95% CI | Baseline (LightGBM) | t-Statistic | p-Value |
|---|---|---|---|---|---|
| Accuracy (%) | 97.6 ± 0.4 | [97.2, 98.0] | 97.0 ± 0.5 | 3.21 | 0.014 |
| Precision (%) | 97.9 ± 0.3 | [97.6, 98.2] | 97.3 ± 0.4 | 2.95 | 0.019 |
| Recall (%) | 97.8 ± 0.3 | [97.5, 98.1] | 97.1 ± 0.5 | 3.12 | 0.015 |
| F1-Score (%) | 97.8 ± 0.3 | [97.5, 98.1] | 97.2 ± 0.4 | 3.08 | 0.016 |
| AUC | 99.7 ± 0.1 | [99.5, 99.8] | 99.3 ± 0.2 | 2.77 | 0.022 |
| Specificity (%) | 98.6 ± 0.3 | [98.3, 98.9] | 98.0 ± 0.4 | 2.89 | 0.020 |
| Sensitivity (%) | 97.8 ± 0.3 | [97.5, 98.1] | 97.1 ± 0.5 | 3.01 | 0.017 |
| Balanced Accuracy (%) | 98.2 ± 0.3 | [97.9, 98.5] | 97.6 ± 0.4 | 2.94 | 0.018 |
| MCC | 0.93 ± 0.02 | [0.91, 0.95] | 0.89 ± 0.03 | 3.27 | 0.013 |
| Cohen’s Kappa | 0.96 ± 0.02 | [0.94, 0.98] | 0.92 ± 0.03 | 3.33 | 0.012 |
| SMOTE Setting | Feature Set | Accuracy (%) | F1-Score (BI-RADS 4) | MCC | Std. Dev. (Accuracy) | Overfitting Risk |
|---|---|---|---|---|---|---|
| Applied before splitting | Raw Transformer Features | 96.1 | 82.1 | 0.88 | ±1.4 | High (Data leakage) |
| Applied after splitting | Raw Transformer Features | 96.4 | 85.3 | 0.89 | ±1.2 | Moderate |
| Applied after splitting | HHO-Selected Features | 98.2 | 94.7 | 0.95 | ±0.8 | Low |
| Model Variant | Accuracy (%) | AUC | F1-Score | MCC |
|---|---|---|---|---|
| MAX-ViT + MetaFormer (Concat) + L1 + XGBoost | 93.6 ± 1.1 | 0.972 ± 0.008 | 0.935 ± 0.010 | 0.84 ± 0.01 |
| MAX-ViT + MetaFormer (GAFM) + L1 + XGBoost | 95.8 ± 0.9 | 0.98 ± 0.006 | 0.95 ± 0.008 | 0.89 ± 0.01 |
| MAX-ViT + MetaFormer (GAFM) + HHO + Logistic Regression | 96.5 ± 0.7 | 0.989 ± 0.005 | 0.961 ± 0.007 | 0.91 ± 0.01 |
| MAX-ViT + MetaFormer (GAFM) + HHO + Random Forest | 97.3 ± 0.6 | 0.993 ± 0.004 | 0.971 ± 0.006 | 0.93 ± 0.01 |
| MAX-ViT + GAFM + HHO + XGBoost (Ours) | 98.2 ± 0.8 | 0.99 ± 0.003 | 0.98 ± 0.006 | 0.95 ± 0.01 |
| Configuration | Accuracy |
|---|---|
| MAX-ViT only | 93.5% |
| MAX-ViT + GAFM | 94.7% |
| MAX-ViT + GAFM + HHO | 96.0% |
| MAX-ViT + GAFM + HHO + XGBoost (Final Model) | 98.2% |
| Class | Accuracy (%) | Precision (%) | Recall/Sensitivity (%) | Specificity (%) | F1-Score (%) | Balanced Accuracy (%) | MCC | AUC (%) |
|---|---|---|---|---|---|---|---|---|
| BI-RADS 0 | 99.40 | 98.61 | 98.39 | 99.65 | 98.50 | 99.02 | 0.981 | 99.02 |
| BI-RADS 1 | 99.22 | 98.11 | 98.00 | 99.53 | 98.05 | 98.76 | 0.976 | 98.76 |
| BI-RADS 2 | 99.22 | 97.48 | 98.67 | 99.36 | 98.07 | 99.01 | 0.976 | 99.01 |
| BI-RADS 3 | 99.13 | 97.99 | 97.67 | 99.50 | 97.83 | 98.58 | 0.973 | 98.58 |
| BI-RADS 4 | 99.42 | 98.83 | 98.28 | 99.71 | 98.55 | 98.99 | 0.982 | 98.99 |
| Fold | Accuracy | Precision | Recall | F1-Score | AUC | Specificity | Sensitivity | MCC | Balanced Acc. | Cohen’s Kappa |
|---|---|---|---|---|---|---|---|---|---|---|
| Fold-1 | 95.32% | 93.50% | 96.00% | 94.73% | 96.70% | 94.50% | 96.00% | 0.89 | 95.25% | 0.88 |
| Fold-2 | 96.10% | 94.60% | 96.90% | 95.74% | 97.20% | 95.20% | 96.90% | 0.91 | 96.05% | 0.90 |
| Fold-3 | 94.75% | 92.10% | 95.80% | 93.92% | 95.90% | 93.70% | 95.80% | 0.87 | 94.75% | 0.86 |
| Fold-4 | 95.60% | 94.00% | 96.10% | 95.03% | 96.80% | 95.00% | 96.10% | 0.90 | 95.55% | 0.89 |
| Fold-5 | 96.23% | 94.90% | 97.00% | 95.94% | 97.40% | 95.50% | 97.00% | 0.91 | 96.25% | 0.90 |
| Average | 95.6% | 93.82% | 96.36% | 95.07% | 96.8% | 94.78% | 96.36% | 0.89 | 95.57% | 0.88 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ahmed, S.; Elazab, N.; El-Gayar, M.M.; Elmogy, M.; Fouda, Y.M. Multi-Scale Vision Transformer with Optimized Feature Fusion for Mammographic Breast Cancer Classification. Diagnostics 2025, 15, 1361. https://doi.org/10.3390/diagnostics15111361
Ahmed S, Elazab N, El-Gayar MM, Elmogy M, Fouda YM. Multi-Scale Vision Transformer with Optimized Feature Fusion for Mammographic Breast Cancer Classification. Diagnostics. 2025; 15(11):1361. https://doi.org/10.3390/diagnostics15111361
Chicago/Turabian StyleAhmed, Soaad, Naira Elazab, Mostafa M. El-Gayar, Mohammed Elmogy, and Yasser M. Fouda. 2025. "Multi-Scale Vision Transformer with Optimized Feature Fusion for Mammographic Breast Cancer Classification" Diagnostics 15, no. 11: 1361. https://doi.org/10.3390/diagnostics15111361
APA StyleAhmed, S., Elazab, N., El-Gayar, M. M., Elmogy, M., & Fouda, Y. M. (2025). Multi-Scale Vision Transformer with Optimized Feature Fusion for Mammographic Breast Cancer Classification. Diagnostics, 15(11), 1361. https://doi.org/10.3390/diagnostics15111361







