Comparative Study of Cell Nuclei Segmentation Based on Computational and Handcrafted Features Using Machine Learning Algorithms
Abstract
1. Introduction
2. Literature Review
3. Materials and Methods
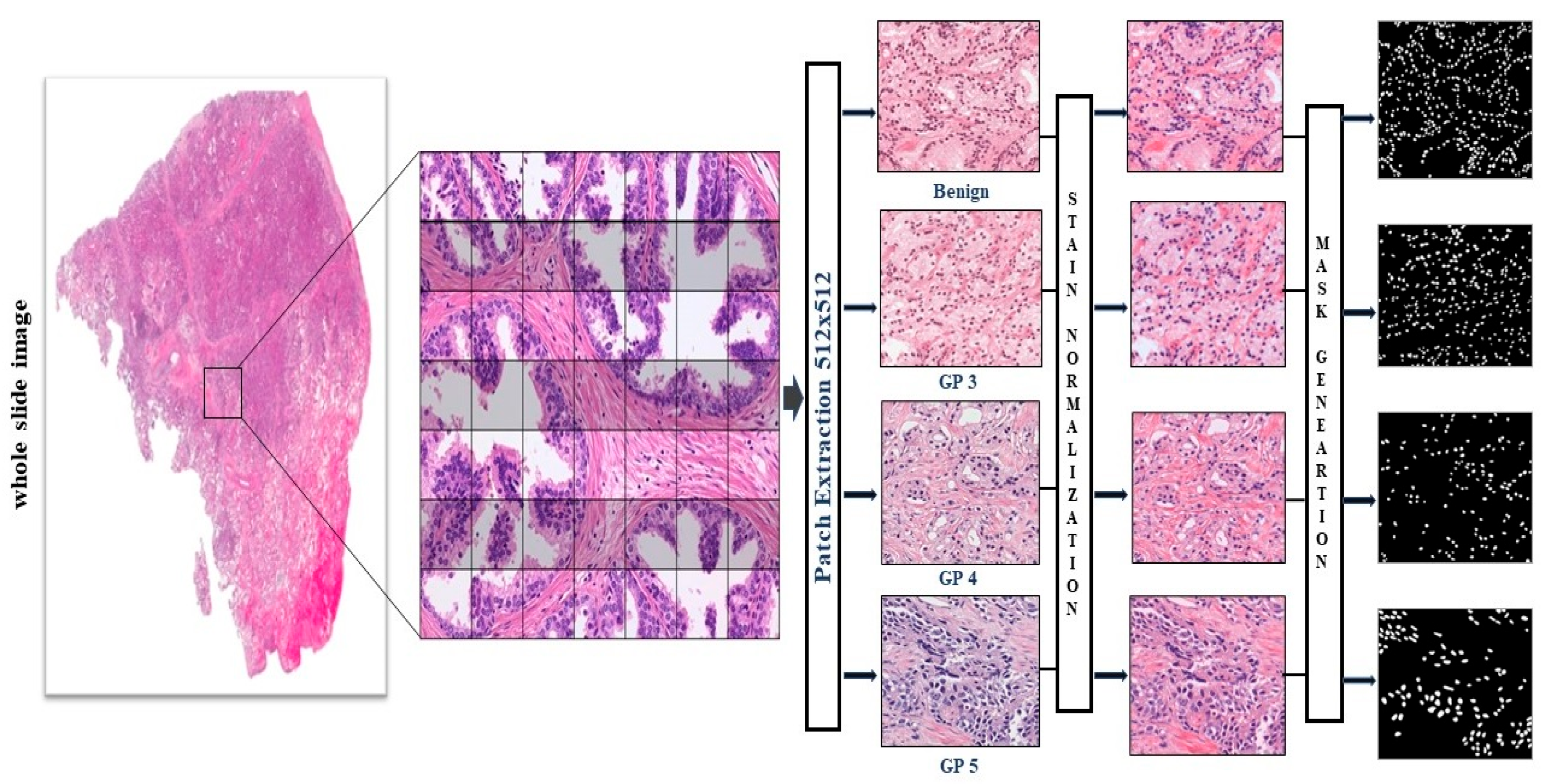
3.1. Image Acquisitions
3.2. Patch Generation from Whole Slide Image
3.3. Color Normalization
3.4. Feature Extraction-Based Segmentation
3.4.1. Handcrafted Feature
3.4.2. CNN Feature
3.5. Machine Learning Segmentation Algorithm
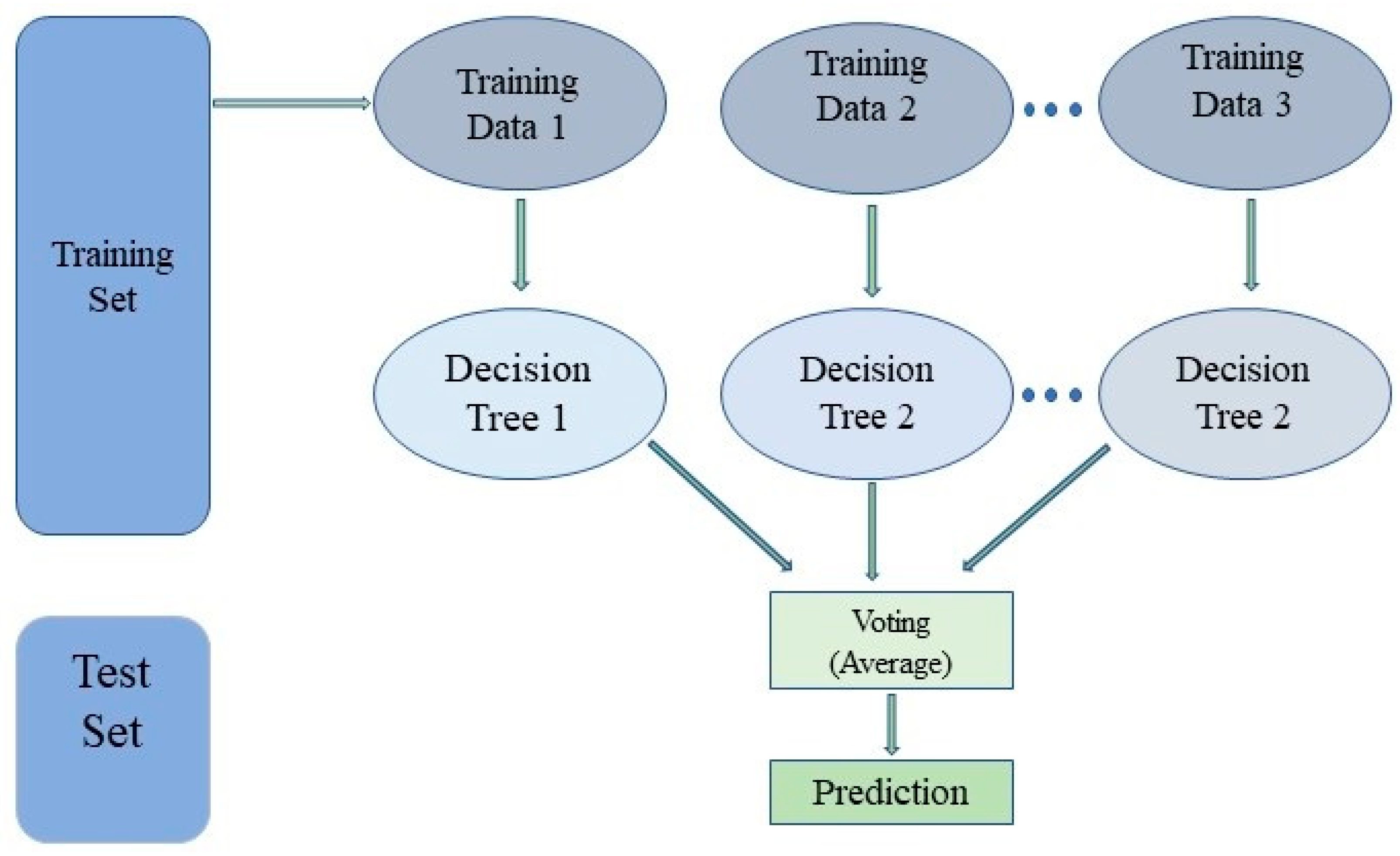
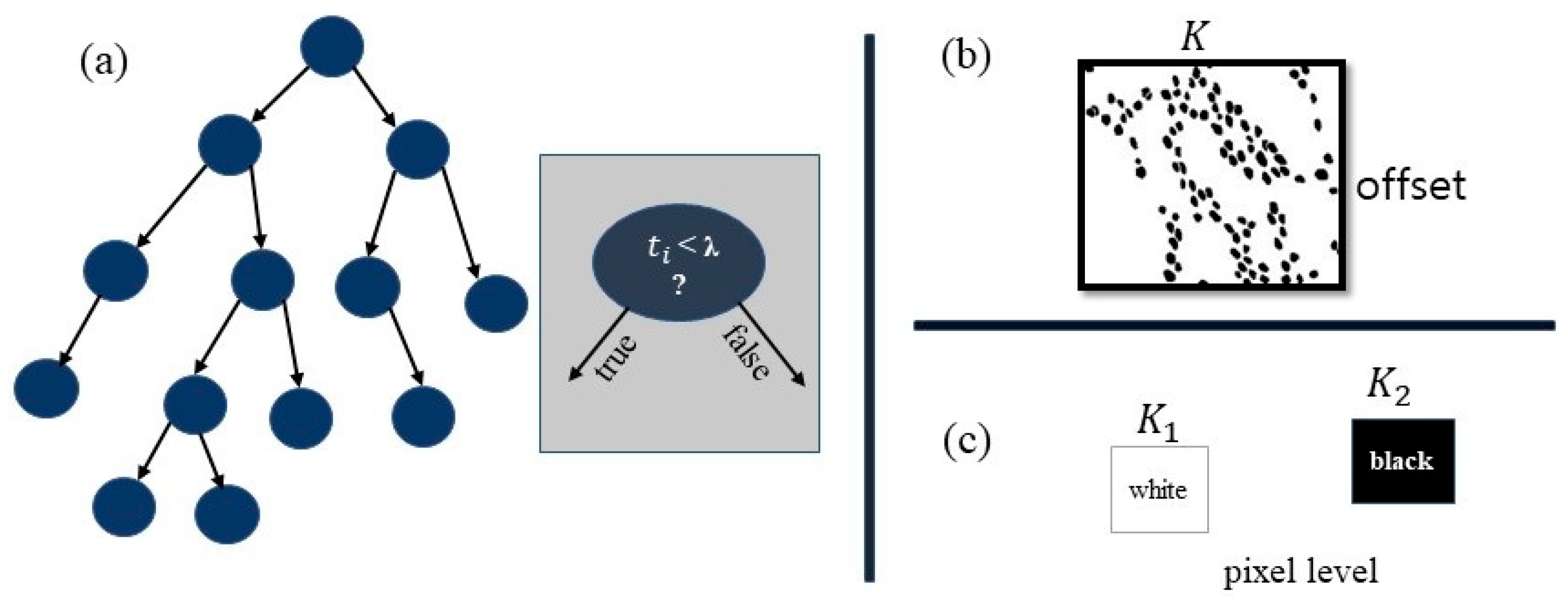
3.5.1. Random Forest
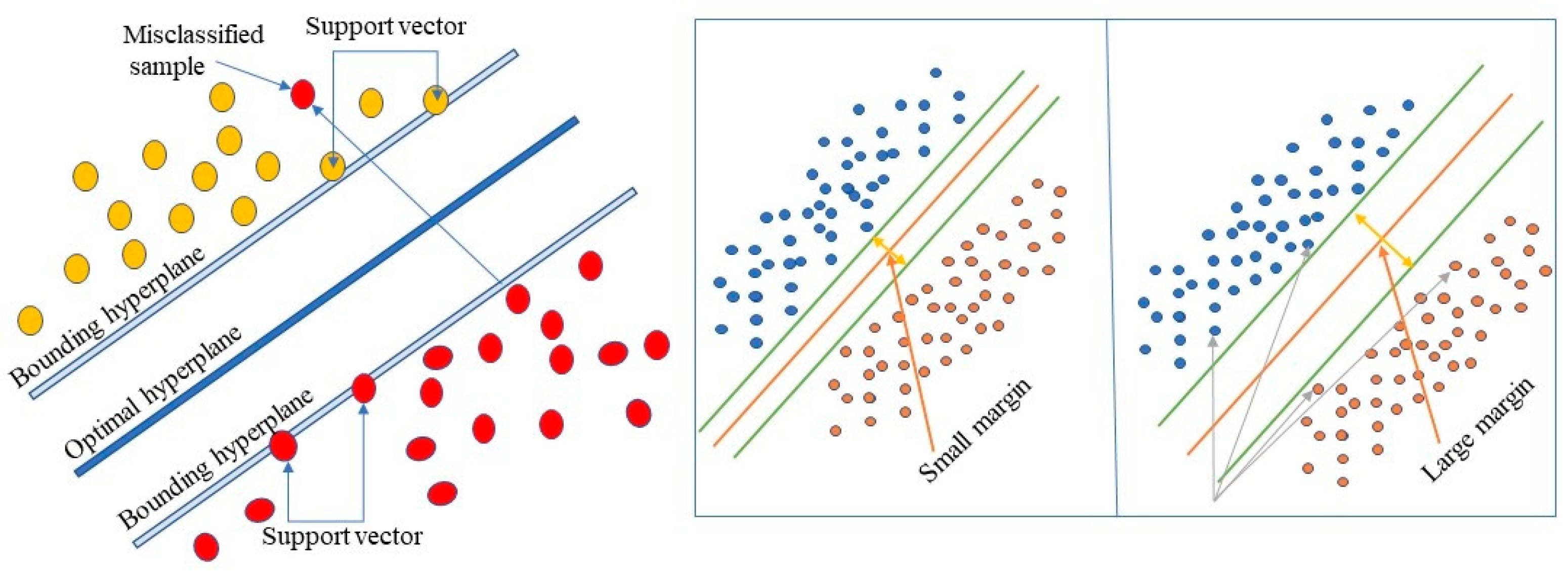
3.5.2. Support Vector Machine (SVM)
3.5.3. Logistic Regression
3.5.4. K-Means Clustering
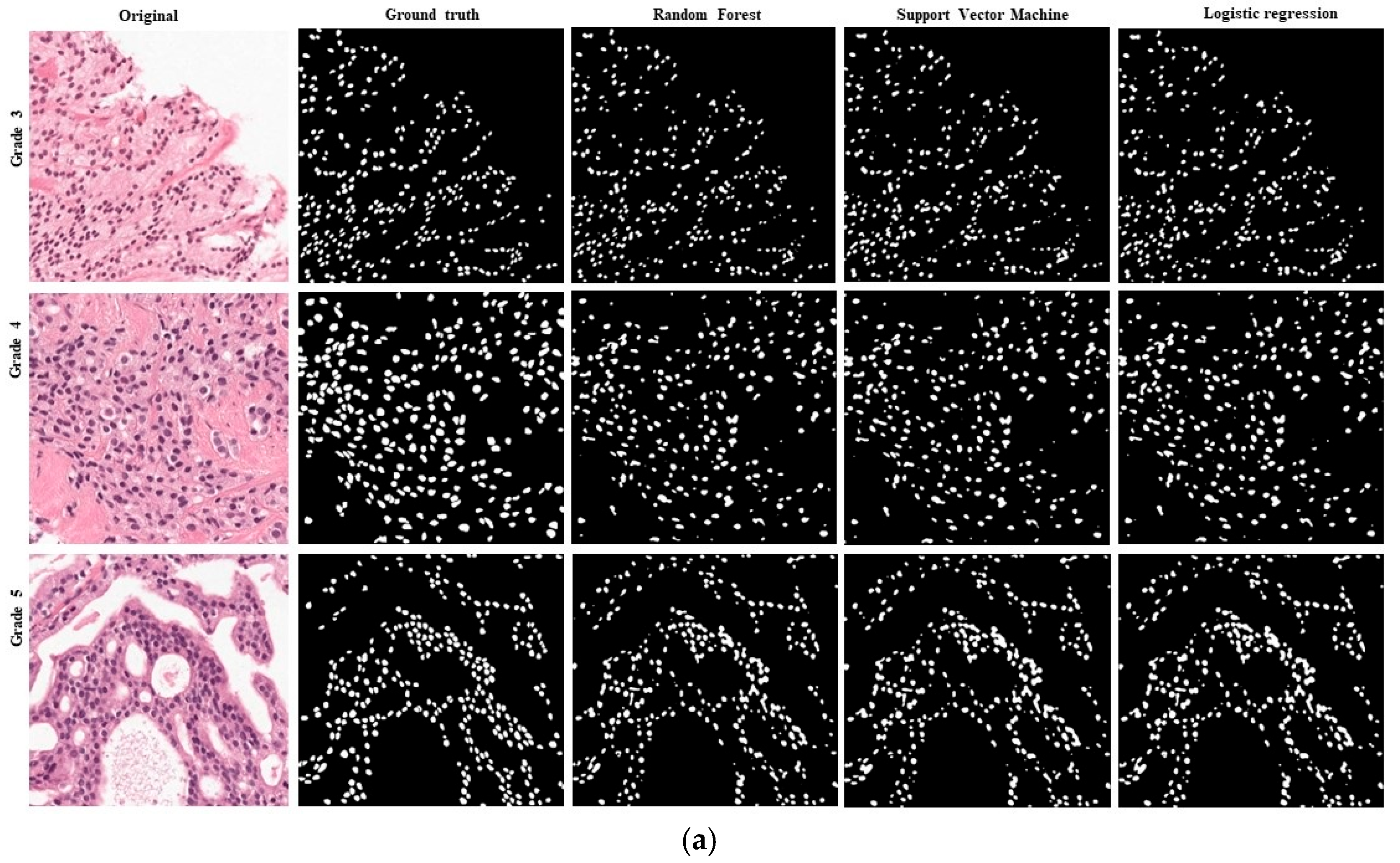
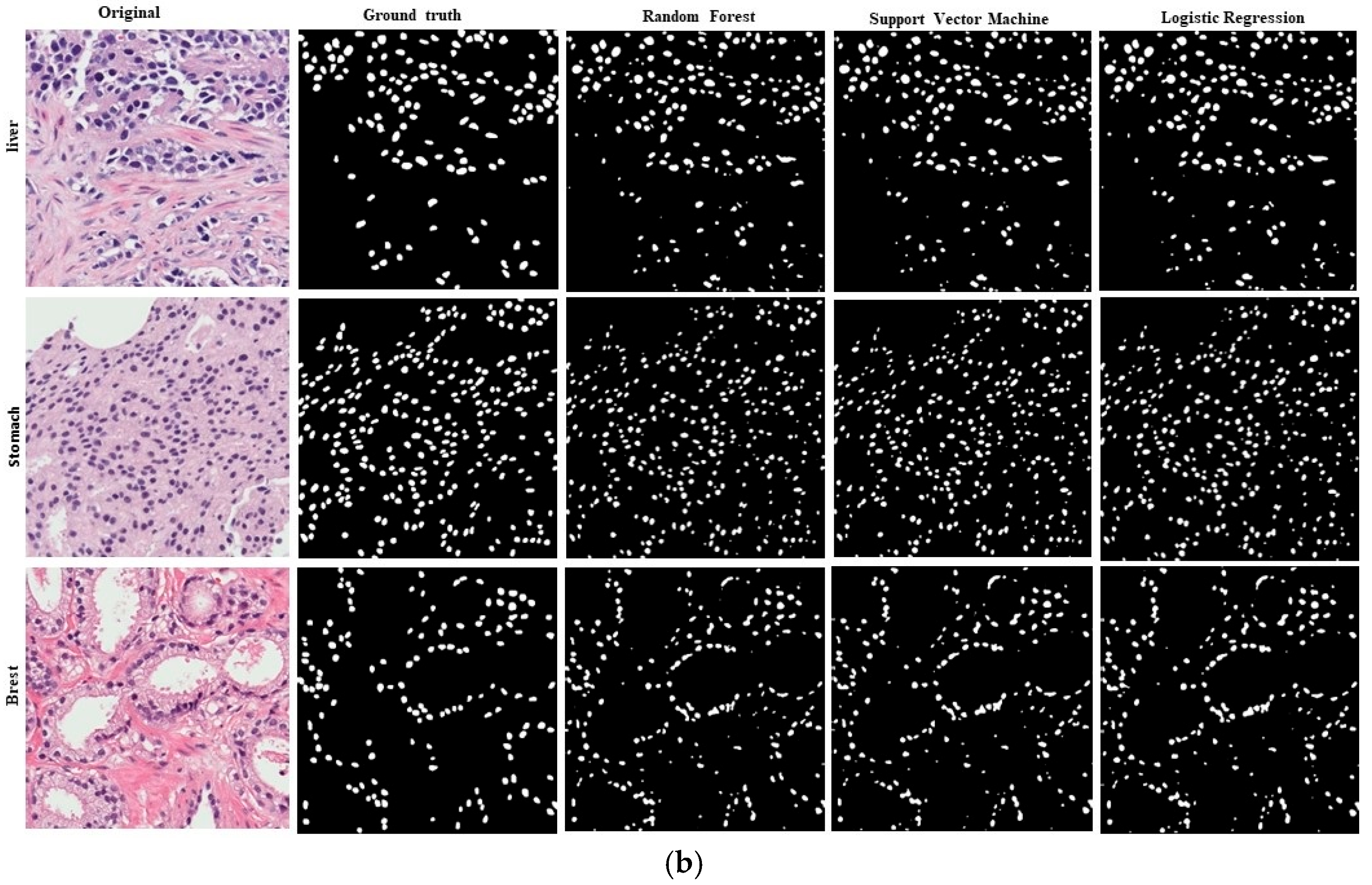
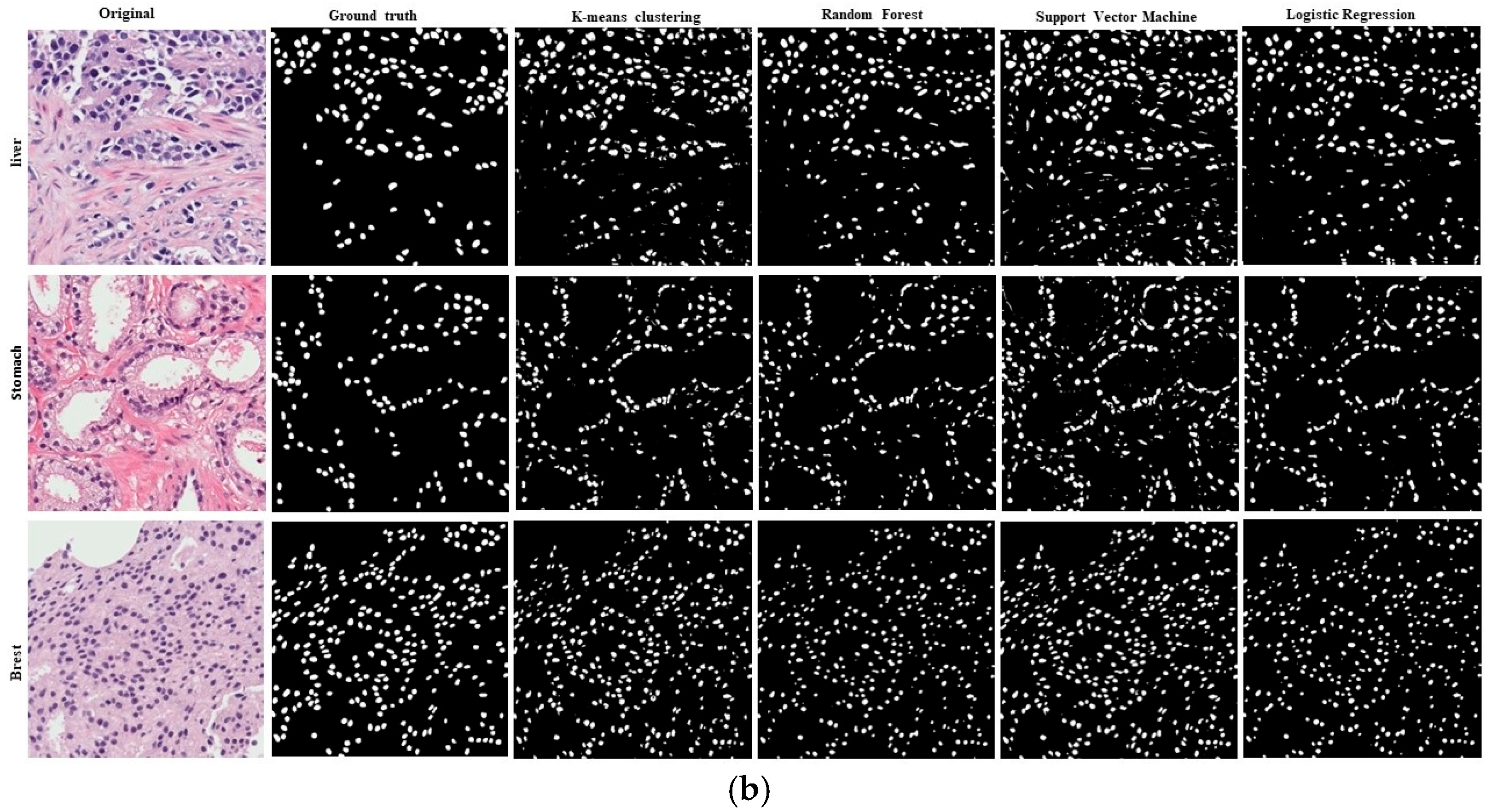
4. Results
5. Discussion
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Nasir, E.S.; Parvaiz, A.; Fraz, M.M. Nuclei and glands instance segmentation in histology images: A narrative review. Artif. Intell. Rev. 2023, 56, 7909–7964. [Google Scholar] [CrossRef]
- Albahra, S.; Gorbett, T.; Robertson, S.; D’Aleo, G.; Kumar, S.V.S.; Ockunzzi, S.; Lallo, D.; Hu, B.; Rashidi, H.H. Artificial intelligence and machine learning overview in pathology & laboratory medicine: A general review of data preprocessing and basic supervised concepts. Semin. Diagn. Pathol. 2023, 40, 71–87. [Google Scholar] [PubMed]
- Gill, A.Y.; Saeed, A.; Rasool, S.; Husnain, A.; Hussain, H. Revolutionizing healthcare: How machine learning is transforming patient diagnoses-a com-prehensive review of AI’s impact on medical diagnosis. J. World Sci. 2023, 2, 1638–1652. [Google Scholar] [CrossRef]
- Harry, A. Revolutionizing healthcare: How machine learning is transforming patient diagnoses-a comprehensive review of ai’s impact on medical diagnosis. BULLET J. Multidisiplin Ilmu 2023, 2, 1259–1266. [Google Scholar]
- Xie, S.; Hamid, N.; Zhang, T.; Zhang, Z.; Peng, L. Unraveling the Nexus: Microplastics, Antibiotics, and ARGs interactions, threats and control in Aquacul-ture-A Review. J. Hazard. Mater. 2024, 1, 134324. [Google Scholar] [CrossRef] [PubMed]
- Ikromjanov, K.; Bhattacharjee, S.; Sumon, R.I.; Hwang, Y.-B.; Rahman, H.; Lee, M.-J.; Kim, H.-C.; Park, E.; Cho, N.-H.; Choi, H.-K. Region Segmentation of Whole-Slide Images for Analyzing Histological Differentiation of Prostate Adenocarcinoma Using Ensemble EfficientNetB2 U-Net with Transfer Learning Mechanism. Cancers 2023, 15, 762. [Google Scholar] [CrossRef]
- Carleton, N.M.; Lee, G.; Madabhushi, A.; Veltri, R.W. Advances in the computational and molecular understanding of the prostate cancer cell nucleus. J. Cell. Biochem. 2018, 119, 7127–7142. [Google Scholar] [CrossRef]
- Alom, M.Z.; Taha, T.M.; Yakopcic, C.; Westberg, S.; Sidike, P.; Nasrin, M.S.; Van Esesn, B.C.; Awwal, A.A.S.; Asari, V.K. The history began from alexnet: A comprehensive survey on deep learning approaches. arXiv 2018, arXiv:1803.01164. [Google Scholar]
- Naik, S.; Doyle, S.; Agner, S.; Madabhushi, A.; Feldman, M.; Tomaszewski, J. Automated gland and nuclei segmentation for grading of prostate and breast cancer histopathology. In Proceedings of the 2008 5th IEEE International Symposium on Biomedical Imaging (ISBI 2008), Paris, France, 14–17 May 2008; pp. 284–287. [Google Scholar]
- Islam Sumon, R.; Bhattacharjee, S.; Hwang, Y.B.; Rahman, H.; Kim, H.C.; Ryu, W.S.; Kim, D.M.; Cho, N.H.; Choi, H.K. Densely Convolutional Spatial Attention Network for nuclei segmentation of histological images for computational pathology. Front. Oncol. 2023, 13, 1009681. [Google Scholar] [CrossRef]
- Wang, H.; Zheng, C.; Li, Y.; Zhu, H.; Yan, X. Application of support vector machines to classification of blood cells. J. Biomed. Eng. 2003, 20, 484–487. [Google Scholar]
- Glotsos, D.; Spyridonos, P.; Petalas, P.; Cavouras, D.; Ravazoula, P.; Dadioti, P.A.; Lekka, I.; Nikiforidis, G. Computer-based malignancy grading of astrocytomas employing a support vector machine classifier, the WHO grading system and the regular hematoxylin-eosin diagnostic staining procedure. Anal. Quant. Cytol. Histol. 2004, 26, 77–83. [Google Scholar] [PubMed]
- Wei, N.; You, J.; Friehs, K.; Flaschel, E.; Nattkemper, T.W. An in situ probe for on-line monitoring of cell density and viability on the basis of dark field microscopy in conjunction with image processing and supervised machine learning. Biotechnol. Bioeng. 2007, 97, 1489–1500. [Google Scholar] [CrossRef]
- Ikromjanov, K.; Bhattacharjee, S.; Hwang, Y.B.; Sumon, R.I.; Kim, H.C.; Choi, H.K. Whole slide image analysis and detection of prostate cancer using vision transformers. In Proceedings of the 2022 International Conference on Artificial Intelligence in Information and Communication (ICAIIC), Jeju, Republic of Korea, 21–24 February 2022. [Google Scholar]
- Haq, I.; Mazhar, T.; Asif, R.N.; Ghadi, Y.Y.; Saleem, R.; Mallek, F.; Hamam, H. A deep learning approach for the detection and counting of colon cancer cells (HT-29 cells) bunches and impurities. PeerJ Comput. Sci. 2023, 9, e1651. [Google Scholar] [CrossRef] [PubMed]
- Arsa DM, S.; Susila AA, N.H. VGG16 in batik classification based on random forest. In Proceedings of the 2019 International Conference on Information Management and Technology (ICIMTech), Jakart, Indonesia, 19–20 August 2019; Volume 1. [Google Scholar]
- Sumon, R.I.; Ali, H.; Akter, S.; Uddin, S.M.I.; Mozumder, A.I.; Kim, H.-C. A Deep Learning-Based Approach for Precise Emotion Recognition in Domestic Animals Using EfficientNetB5 Architecture. Eng 2025, 6, 9. [Google Scholar] [CrossRef]
- Karimi Jafarbigloo, S.; Danyali, H. Nuclear atypia grading in breast cancer histopathological images based on CNN feature extraction and LSTM classification. CAAI Trans. Intell. Technol. 2021, 6, 426–439. [Google Scholar] [CrossRef]
- Sumon, R.I.; Mazumdar, A.I.; Uddin, S.M.I.; Joo, M.-I.; Kim, H.-C. Enhanced Nuclei Segmentation in Histopathology Image Leveraging RGB Channels through Triple-Encoder and Single-Decoder Architectures. In Proceedings of the 2023 IEEE 14th Annual Ubiquitous Computing, Electronics & Mobile Communication Conference (UEMCON), New York, NY, USA, 12–14 October 2023; pp. 0830–0835. [Google Scholar]
- Jung, H.; Lodhi, B.; Kang, J. An automatic nuclei segmentation method based on deep convolutional neural networks for histopathology images. BMC Biomed. Eng. 2019, 1, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Sumon, R.I.; Mazumdar, A.I.; Uddin, S.M.I.; Kim, H.-C. Exploring Deep Learning and Machine Learning Techniques for Histopathological Image Classification in Lung Cancer Diagnosis. In Proceedings of the 2024 International Conference on Electrical, Computer and Energy Technologies (ICECET), Sydney, Australia, 25–27 July 2024; pp. 1–6. [Google Scholar]
- Naylor, P.; Laé, M.; Reyal, F.; Walter, T. Nuclei segmentation in histopathology images using deep neural networks. In Proceedings of the 2017 IEEE 14th International Symposium on Biomedical Imaging (ISBI 2017), Melbourne, Australia, 18–21 April 2017. [Google Scholar]
- Caicedo, J.C.; Goodman, A.; Karhohs, K.W.; Cimini, B.A.; Ackerman, J.; Haghighi, M.; Heng, C.; Becker, T.; Doan, M.; McQuin, C.; et al. Nucleus segmentation across imaging experiments: The 2018 Data Science Bowl. Nat. Methods 2019, 16, 1247–1253. [Google Scholar] [CrossRef]
- Kumar, N.; Verma, R.; Anand, D.; Zhou, Y.; Onder, O.F.; Tsougenis, E.; Chen, H.; Heng, P.-A.; Li, J.; Hu, Z.; et al. A Multi-organ nucleus segmentation challenge. IEEE Trans. Med. Imaging 2019, 39, 1380–1391. [Google Scholar] [CrossRef]
- Ker, D.F.E.; Eom, S.; Sanami, S.; Bise, R.; Pascale, C.; Yin, Z.; Huh, S.-I.; Osuna-Highley, E.; Junkers, S.N.; Helfrich, C.J.; et al. Phase contrast time-lapse microscopy datasets with automated and manual cell tracking annotations. Sci. Data 2018, 5, 180237. [Google Scholar] [CrossRef]
- Herráez-Aguilar, D.; Madrazo, E.; López-Menéndez, H.; Ramírez, M.; Monroy, F.; Redondo-Muñoz, J. Multiple particle tracking analysis in isolated nuclei reveals the mechanical phenotype of leukemia cells. Sci. Rep. 2020, 10, 1–12. [Google Scholar] [CrossRef]
- Grøvik, E.; Yi, D.; Iv, M.; Tong, E.; Rubin, D.; Zaharchuk, G. Deep learning enables automatic detection and segmentation of brain metastases on multisequence MRI. J. Magn. Reson. Imaging 2020, 51, 175–182. [Google Scholar] [CrossRef] [PubMed]
- Sumon, R.I.; Mazumder, A.I.; Akter, S.; Uddin, S.M.I.; Kim, H.-C. Innovative Deep Learning Strategies for Early Detection of Brain Tumours in MRI Scans with a Modified ResNet50V2 Approach. In Proceedings of the 2025 27th International Conference on Advanced Communications Technology (ICACT), Pyeongchang-gun, Republic of Korea, 16–19 February 2025; pp. 323–328. [Google Scholar]
- Mualla, F.; Scholl, S.; Sommerfeldt, B.; Maier, A.; Hornegger, J. Automatic cell detection in bright-field microscope images using SIFT, random forests, and hierarchical clustering. IEEE Trans. Med. Imaging 2013, 32, 2274–2286. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Khan, A.M.; ElDaly, H.; Rajpoot, N.M.; Am, K. A gamma-gaussian mixture model for detection of mitotic cells in breast cancer histopathology images. J. Pathol. Inform. 2013, 4, 11. [Google Scholar] [CrossRef]
- Akter, S.; Sumon, R.I.; Ali, H.; Kim, H.-C. Utilizing Convolutional Neural Networks for the Effective Classification of Rice Leaf Diseases Through a Deep Learning Approach. Electronics 2024, 13, 4095. [Google Scholar] [CrossRef]
- Jaganathan, P.; Rajkumar, N.; Kuppuchamy, R. A Comparative study of improved F-score with support vector machine and RBF network for breast cancer classification. Int. J. Mach. Learn. Comput. 2012, 2, 741–745. [Google Scholar] [CrossRef]
- Wang, H.; Li, Y.; Luo, Z. An improved breast cancer nuclei segmentation method based on unet++. In Proceedings of the 2020 6th International Conference on Computing and Artificial Intelligence, Tianjin, China, 23–26 April 2020. [Google Scholar]
- Kollem, S.; Reddy, K.R.L.; Rao, D.S. A Review of Image Denoising and Segmentation Methods Based on Medical Images. Int. J. Mach. Learn. Comput. 2019, 9, 288–295. [Google Scholar] [CrossRef]
- Gayathri, S.; Gopi, V.P.; Palanisamy, P. Diabetic retinopathy classification based on multipath CNN and machine learning classifiers. Phys. Eng. Sci. Med. 2021, 44, 639–653. [Google Scholar] [CrossRef]
- Imran Razzak, M.; Naz, S. Microscopic blood smear segmentation and classification using deep contour aware CNN and extreme machine learning. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition Workshops, Honolulu, HI, USA, 21–26 July 2017; pp. 49–55. [Google Scholar]
- Yang, H.-Y.; Wang, X.-Y.; Wang, Q.-Y.; Zhang, X.-J. LS-SVM based image segmentation using color and texture information. J. Vis. Commun. Image Represent. 2012, 23, 1095–1112. [Google Scholar] [CrossRef]
- Al-Doorı, S.K.S.; Taspınar, Y.S.; Koklu, M. Distracted Driving Detection with Machine Learning Methods by CNN Based Feature Extraction. Int. J. Appl. Math. Electron. Comput. 2021, 9, 116–121. [Google Scholar] [CrossRef]
- Shin, H.C. Hybrid clustering and logistic regression for multi-modal brain tumor segmentation. In Proceedings of the Workshops and Challanges in Medical Image Computing and Computer-Assisted Intervention (MICCAI’12), Nice, France, 1–5 October 2012. [Google Scholar]
- Ruusuvuori, P.; Manninen, T.; Huttunen, H. Image segmentation using sparse logistic regression with spatial prior. In Proceedings of the 2012 Proceedings of the 20th European Signal Processing Conference (EUSIPCO), Bucharest, Romania, 27–31 August 2012; pp. 2253–2257. [Google Scholar]
- Mahapatra, D. Automatic Cardiac Segmentation Using Semantic Information from Random Forests. J. Digit. Imaging 2014, 27, 794–804. [Google Scholar] [CrossRef]
- Ahmad, A.Y.A.B.; Alzubi, J.A.; Vasanthan, M.; Kondaveeti, S.B.; Shreyas, J.; Priyanka, T.P. Efficient hybrid heuristic adopted deep learning framework for diagnosing breast cancer using thermography images. Sci. Rep. 2025, 15, 1–30. [Google Scholar]
- Khan, A.M.; Rajpoot, N.; Treanor, D.; Magee, D. A nonlinear mapping approach to stain normalization in digital histopathology images using image-specific color deconvolution. IEEE Trans. Biomed. Eng. 2014, 61, 1729–1738. [Google Scholar] [CrossRef] [PubMed]
- Reinhard, E.; Adhikhmin, M.; Gooch, B.; Shirley, P. Color transfer between images. IEEE Comput. Graph. Appl. 2001, 21, 34–41. [Google Scholar] [CrossRef]
- Bejnordi, B.E.; Litjens, G.; Timofeeva, N.; Otte-Holler, I.; Homeyer, A.; Karssemeijer, N.; van der Laak, J.A. Stain Specific Standardization of Whole-Slide Histopathological Images. IEEE Trans. Med. Imaging 2015, 35, 404–415. [Google Scholar] [CrossRef] [PubMed]
- Janowczyk, A.; Basavanhally, A.; Madabhushi, A. Stain Normalization using Sparse AutoEncoders (StaNoSA): Application to digital pathology. Comput. Med. Imaging Graph. 2017, 57, 50–61. [Google Scholar] [CrossRef]
- Macenko, M.; Niethammer, M.; Marron, J.S.; Borland, D.; Woosley, J.T.; Guan, X.; Schmitt, C.; Thomas, N.E. A method for normalizing histology slides for quantitative analysis. In Proceedings of the 2009 IEEE International Sympo-Sium on Biomedical Imaging: From Nano to MACRO, Boston, MA, USA, 28 June–1 July 2009; pp. 1107–1110. [Google Scholar]
- Vahadane, A.; Peng, T.; Sethi, A.; Albarqouni, S.; Wang, L.; Baust, M.; Steiger, K.; Schlitter, A.M.; Esposito, I.; Navab, N. Structure-preserving color normalization and sparse stain separation for histological images. IEEE Trans. Med. Imaging 2016, 35, 1962–1971. [Google Scholar] [CrossRef]
- Haralick, R.M. Statistical and structural approaches to texture. Proc. IEEE 1979, 67, 786–804. [Google Scholar] [CrossRef]
- Wu, Z.; Matsui, O.; Kitao, A.; Kozaka, K.; Koda, W.; Kobayashi, S.; Ryu, Y.; Minami, T.; Sanada, J.; Gabata, T. Hepatitis C related chronic liver cirrhosis: Feasibility of texture analysis of MR images for classification of fibrosis stage and necroinflammatory activity grade. PLoS ONE 2015, 10, e0118297. [Google Scholar] [CrossRef]







| Value (Confidence Interval) | Feature Extraction | Average Dice Coefficient | Average Jaccard Index | Accuracy |
|---|---|---|---|---|
| Random Forest | CNN | 69.22 (58.1–80.3) | 53.46 (43.6–69.1) | 93.7 (90.9–95.1) |
| SVM | CNN | 65.72 (55.8–75.4) | 49.38 (38.7–60.5) | 97.8 (95.1–98.9) |
| Logistic Regression | CNN | 74.24 (59.6–79.6) | 55.61 (43.1–66.1) | 96.9 (95.9–97.1) |
| Value (Confidence Interval) | Feature Extraction | Average Dice Coefficient | Average Jaccard Index | Accuracy |
|---|---|---|---|---|
| Random Forest | Handcraft | 67.19 (52.2–78.7) | 51.34 (35.4–64.9) | 92.7 (92.6–96.4) |
| SVM | Handcraft | 68.90 (44.5–80.0) | 52.04 (28.7–66.5) | 98.0 (94.1–98.9) |
| K-Means | Handcraft | 61.23 (38.5–81.5) | 46.66 (29.9–68.8) | …… |
| Logistic Regression | Handcraft | 68.32 (42.9–81.1) | 53.81 (28.1–68.1) | 96.1 (91.9–96.1) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sumon, R.I.; Mozumdar, M.A.I.; Akter, S.; Uddin, S.M.I.; Al-Onaizan, M.H.A.; Alkanhel, R.I.; Muthanna, M.S.A. Comparative Study of Cell Nuclei Segmentation Based on Computational and Handcrafted Features Using Machine Learning Algorithms. Diagnostics 2025, 15, 1271. https://doi.org/10.3390/diagnostics15101271
Sumon RI, Mozumdar MAI, Akter S, Uddin SMI, Al-Onaizan MHA, Alkanhel RI, Muthanna MSA. Comparative Study of Cell Nuclei Segmentation Based on Computational and Handcrafted Features Using Machine Learning Algorithms. Diagnostics. 2025; 15(10):1271. https://doi.org/10.3390/diagnostics15101271
Chicago/Turabian StyleSumon, Rashadul Islam, Md Ariful Islam Mozumdar, Salma Akter, Shah Muhammad Imtiyaj Uddin, Mohammad Hassan Ali Al-Onaizan, Reem Ibrahim Alkanhel, and Mohammed Saleh Ali Muthanna. 2025. "Comparative Study of Cell Nuclei Segmentation Based on Computational and Handcrafted Features Using Machine Learning Algorithms" Diagnostics 15, no. 10: 1271. https://doi.org/10.3390/diagnostics15101271
APA StyleSumon, R. I., Mozumdar, M. A. I., Akter, S., Uddin, S. M. I., Al-Onaizan, M. H. A., Alkanhel, R. I., & Muthanna, M. S. A. (2025). Comparative Study of Cell Nuclei Segmentation Based on Computational and Handcrafted Features Using Machine Learning Algorithms. Diagnostics, 15(10), 1271. https://doi.org/10.3390/diagnostics15101271









