Color-Transfer-Enhanced Data Construction and Validation for Deep Learning-Based Upper Gastrointestinal Landmark Classification in Wireless Capsule Endoscopy
Abstract
1. Introduction
2. Materials and Methods
2.1. Related Research and Model Selection
2.2. Original Dataset
2.3. Hypothesis Formulation
2.4. Validation of Alternative WCE Datasets
2.4.1. Selection of the Image Similarity Measurement Method
2.4.2. Comparative Analysis of the Kvasir and Kvasir-Capsule Datasets
2.4.3. WCE Dataset Construction Using Color Transfer
2.4.4. Validation of Image Similarity: Measurement of Euclidean Distance
2.4.5. Establishing Threshold and Final Acceptable Ranges
2.5. Experimental Methods
- Accuracy comparison across three datasets based on five image sizes;
- Accuracy evaluation for datasets with five image sizes and different image preprocessing techniques (sharpen and detail filters).
3. Results
3.1. Accuracy Comparison Experiment of L, R, and G Datasets
3.1.1. Accuracy Comparison among Three Datasets
3.1.2. Classification Accuracy for Different Input Image Sizes
3.1.3. Classification Accuracy among Different Classes
3.2. Classification Accuracy through Image Preprocessing Using Sharpen and Detail Filters
3.2.1. Accuracy Comparison between the Datasets
3.2.2. Classification Accuracy across Different Input Image Sizes
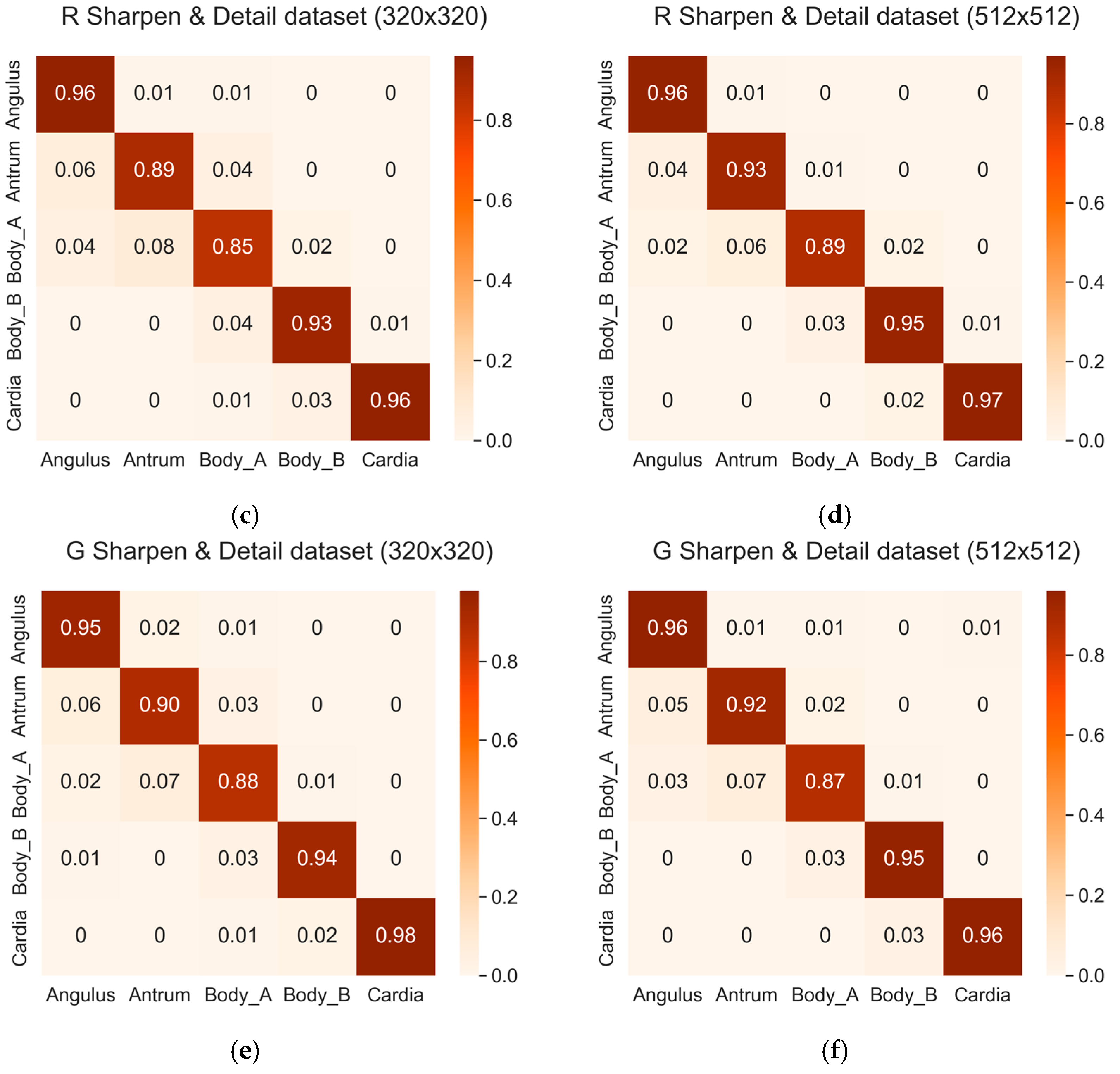
3.2.3. Classification Accuracy among Different Classes
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Siddhi, S.; Dhar, A.; Sebastian, S. Best practices in environmental advocacy and research in endoscopy. Tech. Innov. Gastrointest. Endosc. 2021, 23, 376–384. [Google Scholar] [CrossRef]
- Xiao, Z.; Lu, J.; Wang, X.; Li, N. WCE-DCGAN: A data augmentation method based on wireless capsule endoscopy images for gastrointestinal disease detection. IET Image Process. 2023, 17, 1170–1180. [Google Scholar] [CrossRef]
- Soffer, S.; Klang, E.; Shimon, O.; Nachmias, N.; Eliakim, R.; Benhorin, S.; Kopylov, U.; Barash, Y. Deep learning for wireless capsule endoscopy: A systematic review and meta-analysis. Gastrointest. Endosc. 2020, 92, 831–839. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Meng, M.Q.H. Tumor recognition in wireless capsule endoscopy images using textural features and SVM-based feature selection. IEEE Trans. Inf. Technol. Biomed. 2012, 16, 323–329. [Google Scholar] [PubMed]
- Lonescu, A.G.; Glodeanu, A.D.; Lonescu, M.; Zaharie, S.L.; Ciurea, A.M.; Golli, A.L.; Mavritsakis, N.; Popa, D.L.; Vere, C.C. Clinical impact of wireless capsule endoscopy for small bowel investigation. Exp. Ther. Med. 2022, 23, 262. [Google Scholar] [CrossRef] [PubMed]
- Saito, H.; Aoki, T.; Mmath, K.A.; Kato, Y.; Tsuboi, A.; Yamada, A.; Fujishiro, M.; Oka, S.; Ishihara, S.; Matsuda, T.; et al. Automatic detection and classification of protruding lesions in wireless capsule endoscopy images based on a deep convolutional neural network. Gastrointest. Endosc. 2020, 92, 144–151. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Zhang, Y.; Huang, X. Development and application of magnetically controlled capsule endoscopy in detecting gastric lesions. Gastroenterol. Res. Pract. 2021, 2021, 2716559. [Google Scholar] [CrossRef]
- Jiang, X.; Pan, J.; Li, Z.S.; Liao, Z. Standardized examination procedure of magnetically controlled capsule endoscopy. VideoGIE 2019, 4, 239–243. [Google Scholar] [CrossRef]
- Hoang, M.C.; Nguyen, K.T.; Le, V.H.; Kim, J.; Choi, E.; Kang, B.; Park, J.O.; Kim, C.S. Independent electromagnetic field control for practical approach to actively locomotive wireless capsule endoscope. IEEE Trans. Syst. Man Cybern. Syst. 2019, 51, 3040–3052. [Google Scholar] [CrossRef]
- Zhang, Y.; Qu, L.; Hao, J.; Pan, Y.; Huang, X. In vitro and in vivo evaluation of a novel wired transmission magnetically controlled capsule endoscopy system for upper gastrointestinal examination. Surg. Endosc. 2022, 36, 9454–9461. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.H.; Nam, S.J. Capsule endoscopy for gastric evaluation. Diagnostics 2021, 11, 1792. [Google Scholar] [CrossRef]
- Serrat, J.A.A.; Córdova, H.; Moreira, L.; Pocurull, A.; Ureña, R.; Delgadoguillena, P.G.; Garcésdurán, R.; Sendino, O.; Garcíarodríguez, A.; Gonzálezsuárez, B.; et al. Evaluation of long-term adherence to oesophagogastroduodenoscopy quality indicators. Gastroenterol. Hepatol. (Engl. Ed.) 2020, 43, 589–597. [Google Scholar]
- Tran, T.H.; Nguyen, P.T.; Tran, D.H.; Manh, X.H.; Vu, D.H.; Ho, N.K.; Do, K.L.; Nguyen, V.T.; Nguyen, L.T.; Dao, V.H.; et al. Classification of anatomical landmarks from upper gastrointestinal endoscopic images. In Proceedings of the 2021 8th NAFOSTED Conference on Information and Computer Science (NICS), Hanoi, Vietnam, 21–22 December 2021; IEEE: New York, NY, USA, 2021; pp. 278–283. [Google Scholar]
- Adewole, S.; Yeghyayan, M.; Hyatt, D.; Ehsan, L.; Jablonski, J.; Copland, A.; Syed, S.; Brown, D. Deep learning methods for anatomical landmark detection in video capsule endoscopy images. In Proceedings of the Future Technologies Conference (FTC) 2020, Vancouver, Canada, 5–6 November 2020; Springer International Publishing: Berlin/Heidelberg, Germany, 2021; Volume 1, pp. 426–434. [Google Scholar]
- Xu, Z.; Tao, Y.; Wenfang, Z.; Ne, L.; Zhengxing, H.; Jiquan, L.; Wiling, H.; Huilong, D.; Jianmin, S. Upper gastrointestinal anatomy detection with multi-task convolutional neural networks. Healthc. Technol. Lett. 2019, 6, 176–180. [Google Scholar] [CrossRef]
- Takiyama, H.; Ozawa, T.; Ishihara, S.; Fujishiro, M.; Shichijo, S.; Nomura, S.; Miura, M.; Tada, T. Automatic anatomical classification of esophagogastroduodenoscopy images using deep convolutional neural networks. Sci. Rep. 2018, 8, 7497. [Google Scholar] [CrossRef]
- Cogan, T.; Cogan, M.; Tamil, L. MAPGI: Accurate identification of anatomical landmarks and diseased tissue in gastrointestinal tract using deep learning. Comput. Biol. Med. 2019, 111, 103351. [Google Scholar] [CrossRef]
- Jha, D.; Ali, S.; Hicks, S.; Thambawita, V.; Borgli, H.; Smedsrud, P.H.; de Lange, T.; Pogorelov, K.; Wang, X.; Harzig, P.; et al. A comprehensive analysis of classification methods in gastrointestinal endoscopy imaging. Med. Image Anal. 2021, 70, 102007. [Google Scholar] [CrossRef] [PubMed]
- Smedsrud, P.H.; Thambawita, V.; Hicks, S.A.; Gjestang, H.; Nedrejord, O.O.; Næss, E.; Borgli, H.; Jha, D.; Berstad, T.J.B.; Eskeland, S.L.; et al. Kvasir-Capsule, a video capsule endoscopy dataset. Sci. Data 2021, 8, 142. [Google Scholar] [CrossRef] [PubMed]
- Jha, D.; Smedsrud, P.H.; Riegler, M.A.; Halvorsen, P.; Lange, T.; Johansen, D.; Johansen, H. Kvasir-seg: A segmented polyp dataset. In MultiMedia Modeling: 26th International Conference, MMM 2020, Daejeon, South Korea, January 5–8 2020, Proceedings, Part II 26; Springer International Publishing: Berlin/Heidelberg, Germany, 2020; pp. 451–462. [Google Scholar]
- Pogorelov, K.; Randel, K.R.; Griwodz, C.; Eskeland, S.L.; Lange, T.; Johansen, D.; Spampinato, C.; Dangnguyen, D.T.; Lux, M.; Schmidt, P.T.; et al. Kvasir: A multi-class image dataset for computer aided gastrointestinal disease detection. In Proceedings of the Proceedings of the 8th ACM on Multimedia Systems Conference, Taipei, Taiwan, 20–23 June 2017; pp. 164–169. [Google Scholar]
- Crum, W.R.; Camara, O.; Hill, D.L.G. Generalized overlap measures for evaluation and validation in medical image analysis. IEEE Trans. Med. Imaging 2006, 25, 1451–1461. [Google Scholar] [CrossRef]
- Tian, H.; Li, Y.; Pian, W.; Kaboré, A.K.; Liu, K.; Habib, A.; Klein, J.; Bissyandé, T.F. Predicting patch correctness based on the similarity of failing test cases. ACM Trans. Softw. Eng. Methodol. (TOSEM) 2022, 31, 1–30. [Google Scholar] [CrossRef]
- Kandel, I.; Castelli, M. Transfer learning with convolutional neural networks for diabetic retinopathy image classification: A review. Appl. Sci. 2021, 10, 2021. [Google Scholar] [CrossRef]
- Zhang, W.; Zhong, J.; Yang, S.; Gao, Z.; Hu, J.; Chen, Y.; Yi, Z. Automated identification and grading system of diabetic retinopathy using deep neural networks. Knowl.-Based Syst. 2019, 175, 12–25. [Google Scholar] [CrossRef]
- Krizhevsky, A.; Sutskever, I.; Hinton, G.E. Imagenet classification with deep convolutional neural networks. Adv. Neural Inf. Process. Syst. 2012, 25, 1097–1105. [Google Scholar] [CrossRef]
- Kaiming, H.; Zhang, X.; Ren, S.; Sun, J. Deep residual learning for image recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016; pp. 770–778. [Google Scholar]
- Szegedy, C.; Liu, W.; Jia, Y.; Sermanet, P.; Reed, S.; Anguelov, D.; Erhan, D.; Vanhoucke, V.; Rabinovich, A. Going deeper with convolutions. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Boston, MA, USA, 7–12 June 2015; pp. 1–9. [Google Scholar]
- Rahman, M.T.; Dola, A. Automated grading of diabetic retinopathy using densenet-169 architecture. In Proceedings of the 2021 5th International Conference on Electrical Information and Communication Technology (EICT), Khulna, Bangladesh, 17–19 December 2021; IEEE: New York, NY, USA, 2021; pp. 1–4. [Google Scholar]
- Chen, H.; Wu, X.; Tao, G.; Peng, Q. Automatic content understanding with cascaded spatial–temporal deep framework for capsule endoscopy videos. Neurocomputing 2017, 229, 77–87. [Google Scholar] [CrossRef]
- Jang, H.W.; Lim, C.N.; Park, Y.S.; Lee, G.J.; Lee, J.W. Estimating gastrointestinal transition location using CNN-based gastrointestinal landmark classifier. KIPS Trans. Softw. Data Eng. 2020, 9, 101–108. [Google Scholar]
- Wang, W.; Yang, X.; Li, X.; Tang, J. Convolutional-capsule network for gastrointestinal endoscopy image classification. Int. J. Intell. Syst. 2022, 37, 5796–5815. [Google Scholar] [CrossRef]
- Alam, J.; Rashid, R.B.; Fattah, S.A.; Saquib, M. RAt-CapsNet: A Deep Learning Network Utilizing Attention and Regional Information for Abnormality Detection in Wireless Capsule Endoscopy. IEEE J. Transl. Eng. Health Med. 2022, 10, 3300108. [Google Scholar] [CrossRef]
- Pascual, G.; Laiz, P.; García, A.; Wenzek, H.; Vitrià, J.; Seguí, S. Time-based self-supervised learning for Wireless Capsule Endoscopy. Comput. Biol. Med. 2022, 146, 105631. [Google Scholar] [CrossRef] [PubMed]
- Athanasiou, S.A.; Sergaki, E.S.; Polydorou, A.A.; Stavrakakis, G.S.; Afentakis, N.M.; Vardiambasis, I.O.; Zervakis, M.E. Revealing the Boundaries of Selected Gastro-Intestinal (GI) Organs by Implementing CNNs in Endoscopic Capsule Images. Diagnostics 2023, 13, 865. [Google Scholar] [CrossRef]
- Laiz, P.; Vitrià, J.; Gilabert, P.; Wenzek, H.; Malagelada, C.; Watson, A.J.M.; Seguí, S. Anatomical landmarks localization for capsule endoscopy studies. Comput. Med. Imaging Graph. 2023, 108, 102243. [Google Scholar] [CrossRef]
- Vaghela, H.; Sarvaiya, A.; Premlani, P.; Agarwal, A.; Upla, K.; Raja, K.; Pedersen, M. DCAN: DenseNet with Channel Attention Network for Super-resolution of Wireless Capsule Endoscopy. In Proceedings of the 2023 11th European Workshop on Visual Information Processing, Gjovik, Norway, 11–14 September 2023; IEEE: New York, NY, USA, 2023; Volume 10, p. 10323037. [Google Scholar]
- Cai, Q.; Lis, X.; Guo, Z. Identifying architectural distortion in mammogram images via a se-densenet model and twice transfer learning. In Proceedings of the 2018 11th International Congress on Image and Signal Processing, BioMedical Engineering and Informatics (CISP-BMEI), Beijing, China, 13–15 October 2018; IEEE: New York, NY, USA, 2018; pp. 1–6. [Google Scholar]
- Vulli, A.; Srinivasu, P.N.; Sashank, M.S.K.; Shafi, J.; Choi, J.; Ijaz, M.F. Fine-tuned DenseNet-169 for breast cancer metastasis prediction using FastAI and 1-cycle policy. Sensors 2022, 22, 2988. [Google Scholar] [CrossRef]
- Fekriershad, S.; Alimari, M.J.; Hamad, M.H.; Alsaffar, M.F.; Hassan, F.G.; Hadi, M.E.; Mahdi, K.S. Cell phenotype classification based on joint of texture information and multilayer feature extraction in DenseNet. Comput. Intell. Neurosci. 2022, 2022, 6895833. [Google Scholar] [PubMed]
- Abbas, Q.; Qureshi, I.; Ibrahim, M.E.A. An Automatic Detection and Classification System of Five Stages for Hypertensive Retinopathy Using Semantic and Instance Segmentation in DenseNet Architecture. Sensors 2021, 21, 6936. [Google Scholar] [CrossRef] [PubMed]
- Farag, M.M.; Fouad, M.; Abdelhamid, A.T. Automatic severity classification of diabetic retinopathy based on densenet and convolutional block attention module. IEEE Access 2022, 10, 38299–38308. [Google Scholar] [CrossRef]
- Varma, N.M.; Choudhary, A. Evaluation of Distance Measures in Content Based Image Retrieval. In Proceedings of the 2019 3rd International conference on Electronics, Communication and Aerospace Technology (ICECA), Coimbatore, India, 12–14 June 2019; IEEE: New York, NY, USA, 2019; pp. 696–701. [Google Scholar]
- Korenius, T.; Laurikkala, J.; Juhola, M. On principal component analysis, cosine and Euclidean measures in information retrieval. Inf. Sci. 2007, 177, 4893–4905. [Google Scholar] [CrossRef]
- Srikaewsiew, T.; Khianchainat, K.; Tharatipyakul, A.; Pongnumkul, S.; Kanjanawattana, S. A Comparison of the Instructor-Trainee Dance Dataset Using Cosine similarity, Euclidean distance, and Angular difference. In Proceedings of the 2022 26th International Computer Science and Engineering Conference (ICSEC), Sakon Nakhon, Thailand, 21–23 December 2022; IEEE: New York, NY, USA, 2022; pp. 235–240. [Google Scholar]
- Zenggang, X.; Zhiwen, T.; Xiaowen, C.; Xuemin, Z.; Kaibin, Z.; Conghuan, Y. Research on image retrieval algorithm based on combination of color and shape features. J. Signal Process. Syst. 2021, 93, 139–146. [Google Scholar] [CrossRef]
- Heidari, H.; Chalechale, A.; Mohammadabadi, A.A. Parallel implementation of color based image retrieval using CUDA on the GPU. Int. J. Inf. Technol. Comput. Sci. (IJITCS) 2013, 6, 33–40. [Google Scholar] [CrossRef][Green Version]
- Wang, L.; Zhang, Y.; Feng, J. On the Euclidean distance of images. IEEE Trans. Pattern Anal. Mach. Intell. 2005, 27, 1334–1339. [Google Scholar] [CrossRef]
- Xia, P.; Zhang, L.; Li, F. Learning similarity with cosine similarity ensemble. Inf. Sci. 2015, 307, 39–52. [Google Scholar] [CrossRef]
- Zhang, D.; Lu, G. Evaluation of similarity measurement for image retrieval. In Proceedings of the International Conference on Neural Networks and Signal Processing 2003. Proceedings of the 2003, Nanjing, China, 14–17 December 2003; IEEE: New York, NY, USA, 2003; pp. 928–931. [Google Scholar]
- Ferreira, J.R.; Oliveira, M.C.; Freitas, A.L. Performance evaluation of medical image similarity analysis in a heterogeneous architecture. In Proceedings of the 2014 IEEE 27th International Symposium on Computer-Based Medical Systems, New York, NY, USA, 27–29 May 2014; IEEE: New York, NY, USA, 2014; pp. 159–164. [Google Scholar]
- Garcia, N.; Vogiatzis, G. Learning non-metric visual similarity for image retrieval. Image Vis. Comput. 2019, 82, 18–25. [Google Scholar] [CrossRef]
- Reinhard, E.; Adhikhmin, M.; Gooch, B.; Shirley, P. Color transfer between images. IEEE Comput. Graph. Appl. 2001, 21, 34–41. [Google Scholar] [CrossRef]
- Yin, L.; Jia, J.; Morrissey, J. Towards race-related face identification: Research on skin color transfer. In Proceedings of the Sixth IEEE International Conference on Automatic Face and Gesture Recognition, 2004. Proceedings, Seoul, Republic of Korea, 19 May 2004; IEEE: New York, NY, USA, 2004; pp. 362–368. [Google Scholar]
- Mohapatra, S.; Pati, G.K.; Mishra, M.; Swarnkar, T. Gastrointestinal abnormality detection and classification using empirical wavelet transform and deep convolutional neural network from endoscopic images. Ain Shams Eng. J. 2023, 14, 101942. [Google Scholar] [CrossRef]
- Liao, Z.; Duan, X.D.; Xin, L.; Bo, L.M.; Wang, X.H.; Xiao, G.H.; Hu, L.H.; Zhuang, S.L.; Li, Z.S. Feasibility and safety of magnetic-controlled capsule endoscopy system in examination of human stomach: A pilot study in healthy volunteers. J. Interv. Gastroenterol. 2012, 2, 155–160. [Google Scholar] [CrossRef]
- Rahman, I.; Pioche, M.; Shim, C.S.; Lee, S.P.; Sung, I.K.; Saurin, J.C.S.; Patel, P. Magnetic-assisted capsule endoscopy in the upper GI tract by using a novel navigation system (with video). Gastrointest. Endosc. 2016, 83, 889–895. [Google Scholar] [CrossRef]
- Khan, A.; Malik, H. Gastrointestinal Bleeding WCE images Dataset 2023. Available online: https://data.mendeley.com/datasets/8pbbjf274w/1 (accessed on 29 March 2023).
- Jain, S.; Seal, A.; Ojha, A.; Yazidi, A.; Bures, J.; Tacheci, I.; Krejcar, O. A deep CNN model for anomaly detection and localization in wireless capsule endoscopy images. Comput. Biol. Med. 2021, 137, 104789. [Google Scholar] [CrossRef]
- Iqbal, I.; Walayat, K.; Kakar, M.U.; Ma, J. Automated identification of human gastrointestinal tract abnormalities based on deep convolutional neural network with endoscopic images. Intell. Syst. Appl. 2022, 16, 200149. [Google Scholar] [CrossRef]
- Handa, P.; Goel, N.; Indu, S. Automatic intestinal content classification using transfer learning architectures. In Proceedings of the 2022 IEEE International Conference on Electronics, Computing and Communication Technologies (CONECCT), Bangalore, India, 8–10 July 2022; IEEE: New York, NY, USA, 2022; pp. 1–5. [Google Scholar]
- Sushma, B.; Aparna, P. Recent developments in wireless capsule endoscopy imaging: Compression and summarization techniques. Comput. Biol. Med. 2022, 149, 106087. [Google Scholar]
- Panetta, K.; Bao, L.; Agaian, S. Novel multi-color transfer algorithms and quality measure. IEEE Trans. Consum. Electron. 2016, 62, 292–300. [Google Scholar] [CrossRef]
- Nascimento, H.A.R.; Ramos, A.C.A.; Neves, F.S.; Deazevedovaz, S.L.; Freitas, D.Q. The ‘Sharpen’ filter improves the radiographic detection of vertical root fractures. Int. Endod. J. 2015, 48, 428–434. [Google Scholar] [CrossRef] [PubMed]
- Hentschel, C.; Lahei, D. Effective peaking filter and its implementation on a programmable architecture. IEEE Trans. Consum. Electron. 2001, 47, 33–39. [Google Scholar] [CrossRef]
- Walt, S.V.D.; Schönberger, J.L.; NunezIglesias, J.; Boulogne, F.; Warner, J.D.; Yager, N.; Gouillart, E.; Yu, T. scikit-image: Image processing in Python. PeerJ 2014, 2, e453. [Google Scholar] [CrossRef] [PubMed]
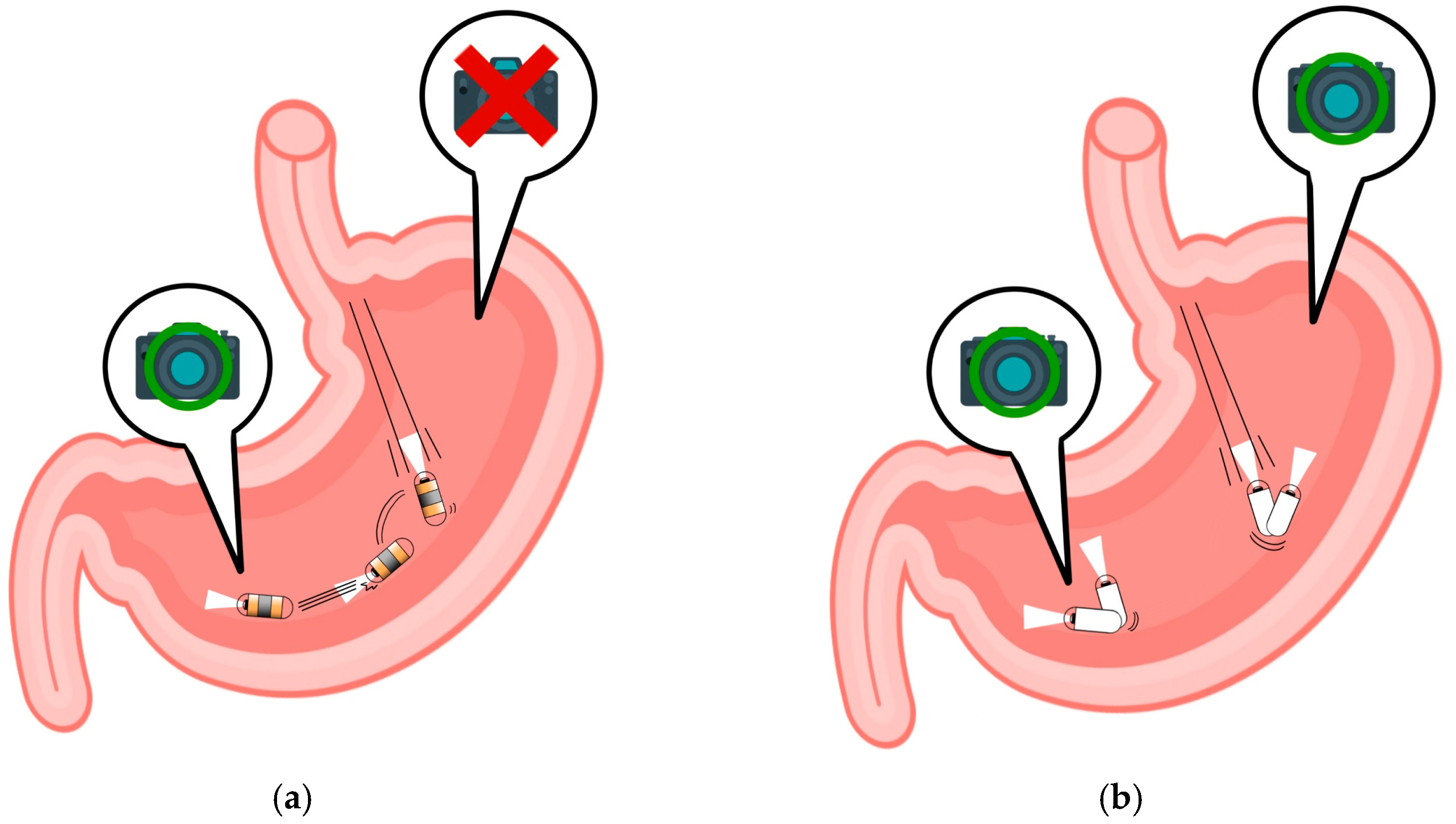







| Autor | Year | Landmark Regions | CNN | Applications | Accuracy |
|---|---|---|---|---|---|
| Rahman et al. [29] | 2016 | Upper stomach | - | MACE | 88–100% |
| Chen et al. [30] | 2017 | Esophagus, stomach, Small intestine, colon | N-CNN, O-CNN, etc. | WCE, HMM | 78–97% |
| Jang et al. [31] | 2020 | Stomach, small intestine, Large intestine | Purposed CNN | WCE, GTA | 95%, 71% |
| Adewole et al. [14] | 2021 | Esophagus, stomach, Small intestine, colon | VGG19, GoogleNet, ResNet50, AlexNet | VCE, Grad-CAM | 85.40% 99.10% |
| Wang et al. [32] | 2022 | Small intestine | VGG16, VGG19, DenseNet121, DenseNet201, AGDN, InceptionV3, etc. | WCE, CNN feature extraction module | 94.83% 85.99% |
| Alam et al. [33] | 2022 | Small intestine | Rat-CapsNet | WCE, VAM | 98.51%, 95.65% |
| Pascual et al. [34] | 2022 | Small intestine, large intestine | ResNet50 | WCE, SSL | 95.00% 92.77% |
| Athanasiou et al. [35] | 2023 | Esophagus, stomach, Small intestine, large intestine | Purposed 3 CNN | WCE, CAD | 95.56% |
| Laiz et al. [36] | 2023 | Small intestine, large intestine | ResNet50 | WCE, CMT Time Block | 91.36% 94.58% 99.09% |
| Vaghela et al. [37] | 2023 | Small intestine | DCAN-DenseNet | WCE, SR | 94.86% 93.78% |
| Model | Accuracy |
|---|---|
| ResNet50 | 92.03% |
| ResNet50V2 | 90.58% |
| InceptionV3 | 89.83% |
| DenseNet121 | 92.26% |
| DenseNet169 | 93.28% |
| DenseNet201 | 91.17% |
| Class | Images |
|---|---|
| Angulus | 493 |
| Antrum | 901 |
| Body A | 533 |
| Body B | 392 |
| Cardia and Fundus | 207 |
| Color Transfer Kvasir Class | Kvasir-Capsule Image (1) | Kvasir-Capsule Image (2) |
|---|---|---|
| Normal cecum | 0.90–1.18 | 0.89–1.19 |
| Normal pylorus | 0.78–1.21 | 0.82–1.24 |
| Normal z line | 0.77–1.24 | 0.81–1.21 |
| Dataset | Angulus | Antrum | Body A | Body B | Cardia and Fundus |
|---|---|---|---|---|---|
| L dataset | 0.92–1.20 | 0.90–1.17 | 0.93–1.20 | 0.93–1.20 | 0.93–1.20 |
| R dataset | 0.93–1.21 | 0.90–1.20 | 0.87–1.19 | 0.90–1.18 | 0.93–1.20 |
| G dataset | 0.78–1.16 | 0.76–1.10 | 0.76–1.13 | 0.84–1.15 | 0.82–1.19 |
| Dataset | 128 × 128 | 256 × 256 | 320 × 320 | 384 × 384 | 512 × 512 |
|---|---|---|---|---|---|
| L dataset | 93.20% | 92.89% | 92.26% | 92.57% | 92.18% |
| R dataset | 92.65% | 91.40% | 92.34% | 92.73% | 91.87% |
| G dataset | 93.20% | 92.73% | 92.18% | 91.71% | 91.32% |
| Dataset | 128 × 128 | 256 × 256 | 320 × 320 | 384 × 384 | 512 × 512 | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Acc | Pre | Rec | F1 | Acc | Pre | Rec | F1 | Acc | Pre | Rec | F1 | Acc | Pre | Rec | F1 | Acc | Pre | Rec | F1 | |
| L Sharpen | 93.04 | 93.38 | 92.56 | 92.96 | 92.57 | 93.00 | 92.42 | 92.70 | 92.73 | 93.29 | 92.42 | 92.70 | 93.04 | 93.60 | 92.65 | 93.12 | 92.81 | 93.15 | 92.50 | 92.82 |
| L Detail | 92.18 | 92.66 | 91.79 | 92.22 | 93.12 | 93.43 | 92.73 | 93.07 | 92.73 | 93.37 | 92.42 | 92.89 | 92.50 | 93.04 | 92.26 | 92.64 | 92.65 | 93.50 | 92.18 | 92.83 |
| L Sharpen & Detail | 93.20 | 93.47 | 92.96 | 93.21 | 93.34 | 93.64 | 93.20 | 93.69 | 93.35 | 93.92 | 93.04 | 93.47 | 93.28 | 93.75 | 92.50 | 93.12 | 92.96 | 93.73 | 92.26 | 92.98 |
| R Sharpen | 93.28 | 93.63 | 93.04 | 93.33 | 92.57 | 92.99 | 92.34 | 92.66 | 93.12 | 93.70 | 92.96 | 93.32 | 92.57 | 93.13 | 92.18 | 92.65 | 92.50 | 93.16 | 91.56 | 92.35 |
| R Detail | 92.65 | 92.99 | 92.26 | 92.62 | 91.32 | 91.67 | 91.17 | 91.41 | 92.34 | 93.20 | 92.10 | 92.64 | 92.73 | 93.29 | 92.42 | 92.85 | 91.40 | 93.29 | 92.42 | 92.85 |
| R Sharpen & Detail | 93.65 | 93.86 | 93.28 | 93.56 | 93.04 | 93.30 | 92.57 | 92.93 | 93.20 | 93.69 | 92.81 | 93.24 | 93.20 | 93.69 | 92.89 | 93.28 | 94.06 | 94.55 | 93.67 | 94.10 |
| G Sharpen | 92.96 | 93.26 | 93.23 | 93.24 | 92.65 | 93.28 | 92.26 | 92.76 | 92.34 | 92.68 | 92.10 | 92.38 | 92.26 | 93.22 | 92.50 | 92.85 | 93.20 | 93.40 | 92.89 | 93.14 |
| G Detail | 92.73 | 93.08 | 92.50 | 92.78 | 92.26 | 92.65 | 91.64 | 92.14 | 92.50 | 92.84 | 92.18 | 92.50 | 92.89 | 93.42 | 92.10 | 92.75 | 91.40 | 92.84 | 92.18 | 92.50 |
| G Sharpen & Detail | 93.43 | 93.66 | 93.43 | 93.54 | 92.89 | 93.24 | 92.73 | 92.98 | 93.04 | 93.31 | 92.73 | 93.01 | 93.04 | 93.45 | 92.57 | 93.00 | 93.46 | 93.68 | 92.65 | 93.16 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, H.-S.; Cho, B.; Park, J.-O.; Kang, B. Color-Transfer-Enhanced Data Construction and Validation for Deep Learning-Based Upper Gastrointestinal Landmark Classification in Wireless Capsule Endoscopy. Diagnostics 2024, 14, 591. https://doi.org/10.3390/diagnostics14060591
Kim H-S, Cho B, Park J-O, Kang B. Color-Transfer-Enhanced Data Construction and Validation for Deep Learning-Based Upper Gastrointestinal Landmark Classification in Wireless Capsule Endoscopy. Diagnostics. 2024; 14(6):591. https://doi.org/10.3390/diagnostics14060591
Chicago/Turabian StyleKim, Hyeon-Seo, Byungwoo Cho, Jong-Oh Park, and Byungjeon Kang. 2024. "Color-Transfer-Enhanced Data Construction and Validation for Deep Learning-Based Upper Gastrointestinal Landmark Classification in Wireless Capsule Endoscopy" Diagnostics 14, no. 6: 591. https://doi.org/10.3390/diagnostics14060591
APA StyleKim, H.-S., Cho, B., Park, J.-O., & Kang, B. (2024). Color-Transfer-Enhanced Data Construction and Validation for Deep Learning-Based Upper Gastrointestinal Landmark Classification in Wireless Capsule Endoscopy. Diagnostics, 14(6), 591. https://doi.org/10.3390/diagnostics14060591






