Classification Prediction of Alzheimer’s Disease and Vascular Dementia Using Physiological Data and ECD SPECT Images
Abstract
1. Introduction
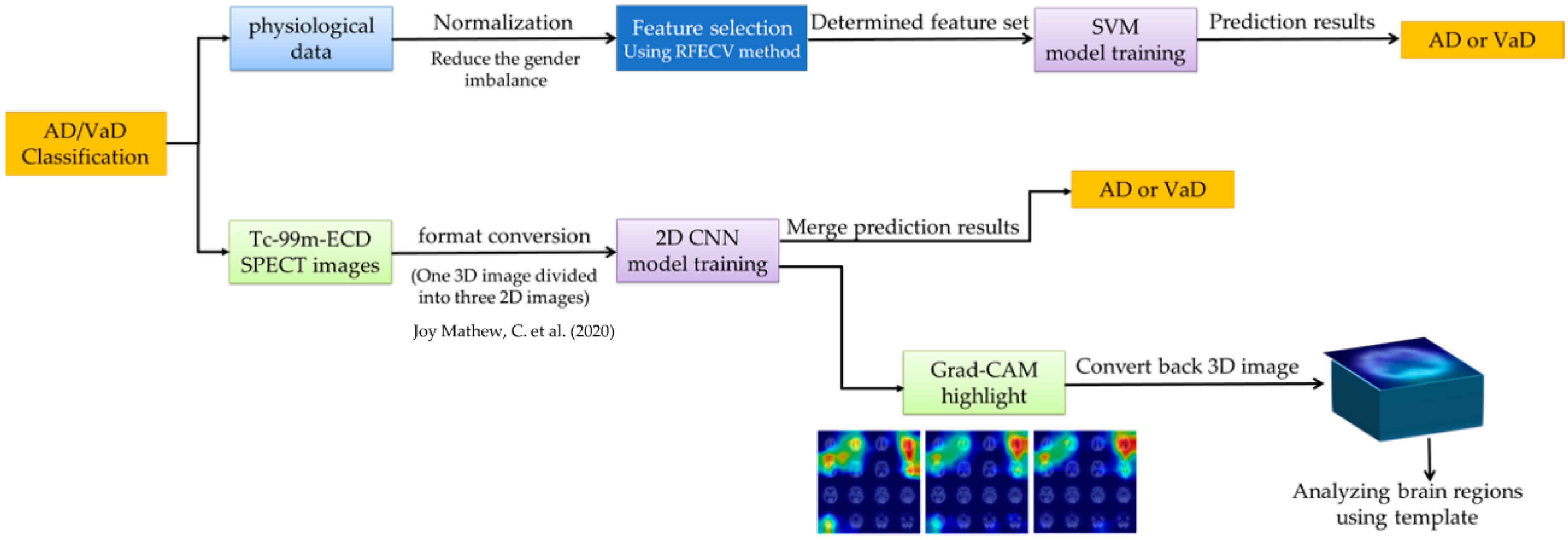
2. Materials and Methods
2.1. Subjects
2.2. Statistical Analysis and Normalization of Physiological Data
2.3. Machine Learning for Physiological Data
2.4. Image Acquisition and Processing
2.5. Deep Learning for ECD Image
2.6. Model Training and Evaluation
2.7. Interpreting Models with Grad-CAM
3. Results
3.1. Characteristics of Demographic Data and Physiological Data
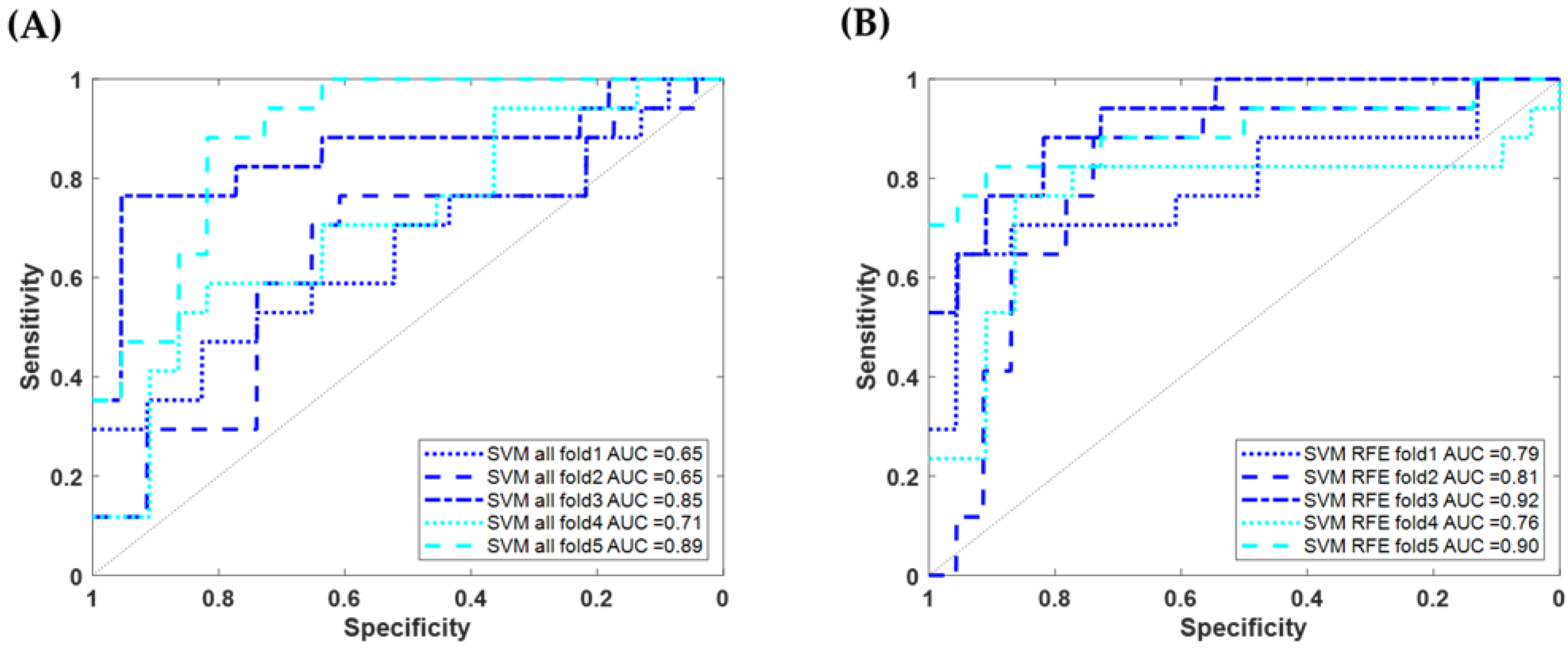
3.2. Using Machine Learning to Classify AD/VaD Using Physiological Data
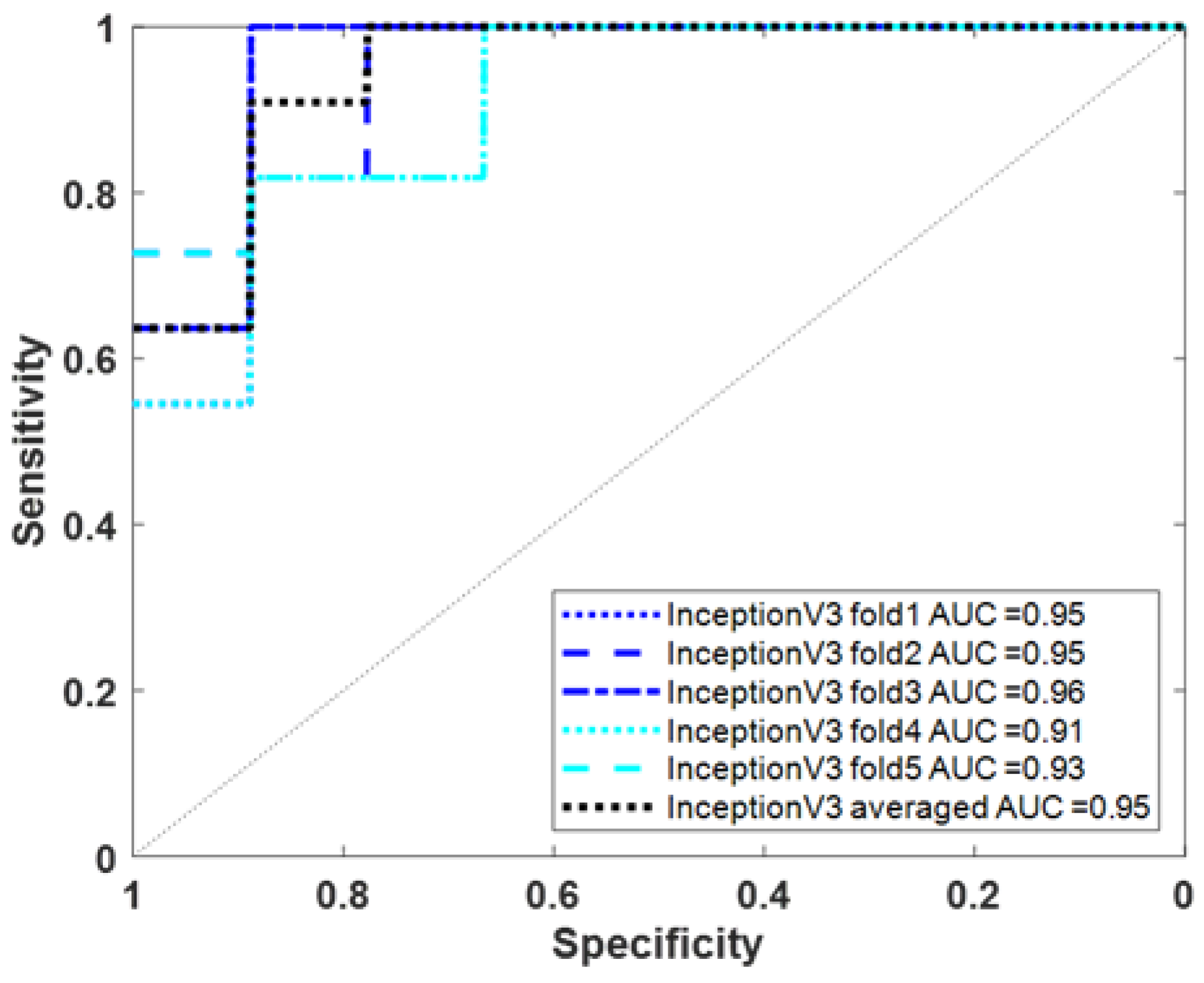
3.3. Using Deep Learning to Classify AD/VaD Using Tc-99m-ECD SPECT Images
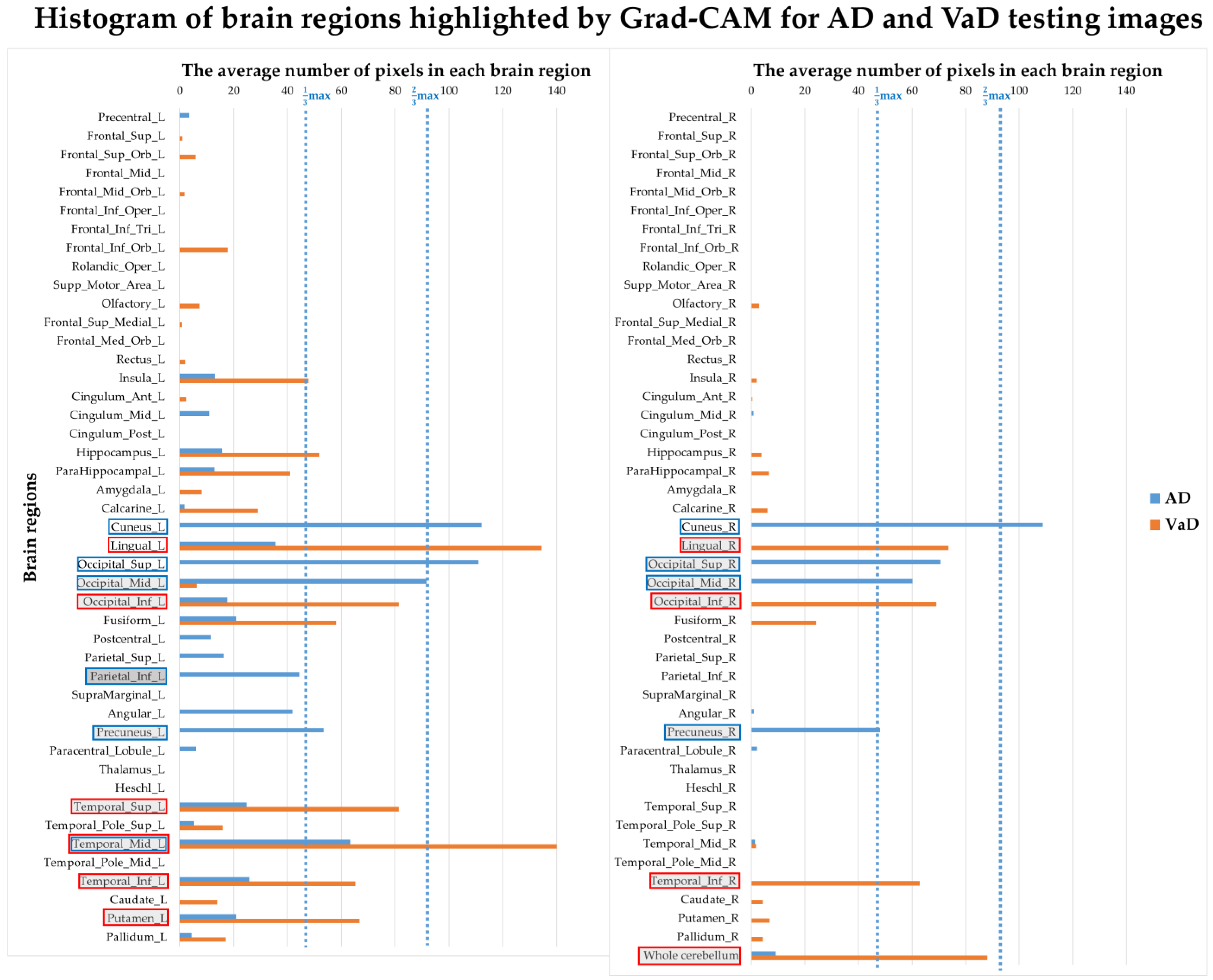
3.4. Correlation between Model Interpretation and Brain Regions
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Taiwan Alzheimer Disease Association. Handbook of Dementia Diagnosis and Treatment; Ministry of Health and Welfare: Taipei City, Taiwan, 2017; p. 16. [Google Scholar]
- Ramirez-Gomez, L.; Zheng, L.; Reed, B.; Kramer, J.; Mungas, D.; Zarow, C.; Vinters, H.; Ringman, J.M.; Chui, H. Neuropsychological Profiles Differentiate Alzheimer Disease from Subcortical Ischemic Vascular Dementia in an Autopsy-Defined Cohort. Dement. Geriatr. Cogn. Disord. 2017, 44, 1–11. [Google Scholar] [CrossRef]
- WHO. The World Health Report 2002: Reducing Risks to Health, Promoting Healthy Life; WHO: Geneva, Switzerland, 2002. [Google Scholar]
- Rizzi, L.; Rosset, I.; Roriz-Cruz, M. Global Epidemiology of Dementia: Alzheimer’s and Vascular Types. BioMed Res. Int. 2014, 2014, 908915. [Google Scholar] [CrossRef]
- Bennett, D. Public Health Importance of Vascular Dementia and Alzheimer’s Disease with Cerebrovascular Disease. Int. J. Clin. Pract. Suppl. 2001, 41–48. [Google Scholar]
- Román, G. Diagnosis of Vascular Dementia and Alzheimer’s Disease. Int. J. Clin. Pract. Suppl. 2001, 9–13. [Google Scholar]
- Erkinjuntti, T. Clinical Deficits of Alzheimer’s Disease with Cerebrovascular Disease and Probable VaD. Int. J. Clin. Pract. Suppl. 2001, 14–23. [Google Scholar]
- Riley, K.P.; Snowdon, D.A.; Markesbery, W.R. Alzheimer’s Neurofibrillary Pathology and the Spectrum of Cognitive Function: Findings from the Nun Study. Ann. Neurol. 2002, 51, 567–577. [Google Scholar] [CrossRef] [PubMed]
- Villain, N.; Fouquet, M.; Baron, J.-C.; Mézenge, F.; Landeau, B.; de La Sayette, V.; Viader, F.; Eustache, F.; Desgranges, B.; Chételat, G. Sequential Relationships between Grey Matter and White Matter Atrophy and Brain Metabolic Abnormalities in Early Alzheimer’s Disease. Brain 2010, 133, 3301–3314. [Google Scholar] [CrossRef] [PubMed]
- Jellinger, K.A. Pathology and Pathogenesis of Vascular Cognitive Impairment-a Critical Update. Front. Aging Neurosci. 2013, 5, 17. [Google Scholar] [CrossRef] [PubMed]
- O’Brien, J.T.; Thomas, A. Vascular Dementia. Lancet 2015, 386, 1698–1706. [Google Scholar] [CrossRef] [PubMed]
- Savoiardo, M.; Grisoli, M. Imaging Dementias. Eur. Radiol. 2001, 11, 484–492. [Google Scholar] [CrossRef]
- Vitali, P.; Migliaccio, R.; Agosta, F.; Rosen, H.J.; Geschwind, M.D. Neuroimaging in Dementia. Semin. Neurol. 2008, 28, 467–483. [Google Scholar] [CrossRef]
- Altamura, C.; Scrascia, F.; Quattrocchi, C.C.; Errante, Y.; Gangemi, E.; Curcio, G.; Ursini, F.; Silvestrini, M.; Maggio, P.; Zobel, B.B.; et al. Regional MRI Diffusion, White-Matter Hyperintensities, and Cognitive Function in Alzheimer’s Disease and Vascular Dementia. J. Clin. Neurol. 2016, 12, 201–208. [Google Scholar] [CrossRef] [PubMed]
- Goujon, A.; Mejdoubi, M.; Purcell, Y.; Banydeen, R.; Colombani, S.; Arrigo, A. Can MRI Water Apparent Diffusion Coefficient (ADC) Value Discriminate between Idiopathic Normal Pressure Hydrocephalus, Alzheimer’s Disease and Subcortical Vascular Dementia? J. Neuroradiol. 2018, 45, 15–22. [Google Scholar] [CrossRef] [PubMed]
- Schuff, N.; Matsumoto, S.; Kmiecik, J.; Studholme, C.; Du, A.; Ezekiel, F.; Miller, B.L.; Kramer, J.H.; Jagust, W.J.; Chui, H.C.; et al. Cerebral blood flow in ischemic vascular dementia and Alzheimer’s disease, measured by arterial spin-labeling magnetic resonance imaging. Alzheimers Dement. 2009, 5, 454–462. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.Z.; Zhang, J.J.; Liu, H.; Wu, G.Y.; Xiong, L.; Shu, M. Regional cerebral blood flow and cerebrovascular reactivity in Alzheimer’s disease and vascular dementia assessed by arterial spinlabeling magnetic resonance imaging. Curr. Neurovasc. Res. 2013, 10, 49–53. [Google Scholar] [CrossRef] [PubMed]
- Kiđemet-Piskač, S.; Babić Leko, M.; Blažeković, A.; Langer Horvat, L.; Klepac, N.; Sonicki, Z.; Kolenc, D.; Hof, P.R.; Boban, M.; Mimica, N.; et al. Evaluation of Cerebrospinal Fluid Phosphorylated Tau 231 as a Biomarker in the Differential Diagnosis of Alzheimer’s Disease and Vascular Dementia. CNS Neurosci. Ther. 2018, 24, 734–740. [Google Scholar] [CrossRef] [PubMed]
- Luo, J.; Chen, L.; Huang, X.; Xie, J.; Zou, C.; Pan, M.; Mo, J.; Zou, D. REPS1 as a Potential Biomarker in Alzheimer’s Disease and Vascular Dementia. Front. Aging Neurosci. 2022, 14, 894824. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Yu, C.; Tao, Y.; Yang, X.; Jiang, Q.; Yu, H.; Zhang, J. Transcriptome Analysis Reveals Potential Marker Genes for Diagnosis of Alzheimer’s Disease and Vascular Dementia. Front. Genet. 2022, 13, 1038585. [Google Scholar] [CrossRef] [PubMed]
- Ni, Y.C.; Tseng, F.P.; Pai, M.C.; Hsiao, I.T.; Lin, K.J.; Lin, Z.K.; Lin, C.Y.; Chiu, P.Y.; Hung, G.U.; Chang, C.C.; et al. The Feasibility of Differentiating Lewy Body Dementia and Alzheimer’s Disease by Deep Learning Using ECD SPECT Images. Diagnostics 2021, 11, 2091. [Google Scholar] [CrossRef]
- Ni, Y.C.; Tseng, F.P.; Pai, M.C.; Hsiao, I.T.; Lin, K.J.; Lin, Z.K.; Lin, W.B.; Chiu, P.Y.; Hung, G.U.; Chang, C.C.; et al. Detection of Alzheimer’s Disease Using ECD SPECT Images by Transfer Learning from FDG PET. Ann. Nucl. Med. 2021, 35, 889–899. [Google Scholar] [CrossRef]
- Kingma, D.P.; Ba, J. Adam: A Method for Stochastic Optimization. arXiv 2014, arXiv:1412.6980. [Google Scholar]
- Lin, R.T.; Lai, C.L.; Tai, C.T.; Liu, C.K.; Yen, Y.Y.; Howng, S.L. Prevalence and Subtypes of Dementia in Southern Taiwan: Impact of Age, Sex, Education, and Urbanization. J. Neurol. Sci. 1998, 160, 67–75. [Google Scholar] [CrossRef] [PubMed]
- Hosoki, S.; Tanaka, T.; Ihara, M. Diagnostic and Prognostic Blood Biomarkers in Vascular Dementia: From the Viewpoint of Ischemic Stroke. Neurochem. Int. 2021, 146, 105015. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.J.; Cho, K.R.; Jang, H.; Lee, N.K.; Jung, Y.H.; Kim, J.P.; Lee, J.I.; Chang, J.W.; Park, S.; Kim, S.T.; et al. Intracerebroventricular Injection of Human Umbilical Cord Blood Mesenchymal Stem Cells in Patients with Alzheimer’s Disease Dementia: A Phase I Clinical Trial. Alzheimer’s Res. Ther. 2021, 13, 154. [Google Scholar] [CrossRef]
- Wang, R.; Chen, Z.; Fu, Y.; Wei, X.; Liao, J.; Liu, X.; He, B.; Xu, Y.; Zou, J.; Yang, X.; et al. Plasma Cystatin C and High-Density Lipoprotein Are Important Biomarkers of Alzheimer’s Disease and Vascular Dementia: A Cross-Sectional Study. Front. Aging Neurosci. 2017, 9, 26. [Google Scholar] [CrossRef]
- Joy Mathew, C.; Jose, M.T.; Elshaikh, A.O.; Shah, L.; Lee, R.; Cancarevic, I. Is Hyperthyroidism a Possible Etiology of Early Onset Dementia? Cureus 2020, 12, e10603. [Google Scholar] [CrossRef]
- Durga, J.; van Boxtel, M.P.; Schouten, E.G.; Kok, F.J.; Jolles, J.; Katan, M.B.; Verhoef, P. Effect of 3-Year Folic Acid Supplementation on Cognitive Function in Older Adults in the FACIT Trial: A Randomised, Double Blind, Controlled Trial. Lancet 2007, 369, 208–216. [Google Scholar] [CrossRef]
- Wald, D.S.; Kasturiratne, A.; Simmonds, M. Effect of Folic Acid, with or without Other B Vitamins, on Cognitive Decline: Meta-Analysis of Randomized Trials. Am. J. Med. 2010, 123, 522–527. [Google Scholar] [CrossRef] [PubMed]
- Pávics, L.; Grünwald, F.; Reichmann, K.; Horn, R.; Kitschenberg, A.; Hartmann, A.; Menzel, C.; Schomburg, A.G.; Overbeck, B.; Csernay, L.; et al. Regional Cerebral Blood Flow Single-Photon Emission Tomography with 99mTc-HMPAO and the Acetazolamide Test in the Evaluation of Vascular and Alzheimer’s Dementia. Eur. J. Nucl. Med. 1999, 26, 239–245. [Google Scholar] [CrossRef]
- Tayler, H.; Miners, J.S.; Güzel, Ö.; MacLachlan, R.; Love, S. Mediators of Cerebral Hypoperfusion and Blood-Brain Barrier Leakiness in Alzheimer’s Disease, Vascular Dementia and Mixed Dementia. Brain Pathol. 2021, 31, e12935. [Google Scholar] [CrossRef]


| AD (n = 112) | VaD (n = 85) | p-Value (Ori) | p-Value (Norm) | |
|---|---|---|---|---|
| Basic information | ||||
| Age (y), mean (SD) | 74.5 (6.9) | 69.7 (7.0) | 0.000 *** | 0.000 *** |
| Gender (Male), n (%) | 53 (47.3) | 57 (67.1) | 0.008 | - |
| Education (y), mean (SD) | 8 (4.3) | 8.7 (5.2) | 0.012 | - |
| FM with dementia, n (%) | 27 (24.1) | 5 (5.9) | 0.000 *** | - |
| FM with PD, n (%) | 1 (0.9) | 2 (2.4) | 0.407 | - |
| FM with stroke, n (%) | 3 (2.7) | 6 (7.1) | 0.145 | - |
| Exercise habits, n (%) | 15 (13.4) | 15 (17.6) | 0.012 | - |
| Sleep disorder, n (%) | 27 (24.1) | 10 (11.8) | 0.028 | - |
| Height (cm), mean (SD) | 158.7 (7.5) | 163 (8) | 0.000 *** | 0.041 |
| Body weight (kg), mean (SD) | 59.2 (10.3) | 63.8 (9.5) | 0.001 | 0.022 |
| Blood pressure (mmHg) | ||||
| Systolic, mean (SD) | 130 (15.3) | 133 (16) | 0.184 | 0.152 |
| Diastolic, mean (SD) | 75.6 (9.9) | 74.2 (9.6) | 0.345 | 0.312 |
| Heartbeat (bpm), mean (SD) | 75.4 (11.7) | 79.6 (12.2) | 0.016 | 0.053 |
| Assessment scales | ||||
| CDR (>0.5), n (%) | 50 (44.6) | 33 (38.8) | 0.186 | - |
| MMSE, mean (SD) | 19.3 (5.2) | 20 (6.6) | 0.104 | 0.135 |
| CASI, mean (SD) | 64.7 (17.1) | 67.8 (21.2) | 0.047 | 0.075 |
| Laboratory examinations | ||||
| Blood count (k/µL) | ||||
| Hb, mean (SD) | 13.2 (1.5) | 13.4 (1.8) | 0.016 | 0.845 |
| WBC, mean (SD) | 6.1 (1.4) | 6.9 (1.8) | 0.000 *** | 0.003 |
| Plt, mean (SD) | 220.3 (71.1) | 223.2 (61.1) | 0.361 | 0.085 |
| Glucose AC (mg/dL), mean (SD) | 112.7 (31.2) | 119.6 (37.8) | 0.354 | 0.581 |
| HbA1c (%), mean (SD) | 6.2 (0.8) | 6.4 (1.2) | 0.967 | 0.872 |
| Triglyceride (mg/dL), mean (SD) | 121.3 (63.1) | 140.7 (74.5) | 0.074 | 0.057 |
| Cholesterol (mg/dL) | ||||
| Total cholesterol, mean (SD) | 184.4 (27.8) | 179.2 (39.6) | 0.151 | 0.471 |
| HDL, mean (SD) | 57.3 (15.1) | 50.2 (11.5) | 0.000 *** | 0.007 |
| LDL, mean (SD) | 114.3 (28.8) | 106.2 (33) | 0.148 | 0.287 |
| Liver fuction index (U/L) | ||||
| GOT, mean (SD) | 27.3 (10.4) | 26.6 (18.3) | 0.133 | 0.106 |
| GPT, mean (SD) | 24 (14.7) | 24.5 (13.9) | 0.693 | 0.929 |
| BUN (mg/dL), mean (SD) | 18.7 (9.7) | 19.2 (12.5) | 0.966 | 0.749 |
| Creatinine (mg/dL), mean (SD) | 1.1 (1.5) | 1.2 (1.2) | 0.007 | 0.027 |
| TSH (mU/L), mean (SD) | 3.1 (6.2) | 2.5 (4.1) | 0.276 | 0.709 |
| Free T4 (ng/dL), mean (SD) | 1.5 (1.3) | 1.6 (1.6) | 0.065 | 0.036 |
| VitB12 (pg/mL), mean (SD) | 959.8 (873.6) | 742.5 (481.9) | 0.500 | 0.861 |
| Folic acid (ng/mL), mean (SD) | 14.2 (8.5) | 11.1 (7.1) | 0.002 | 0.014 |
| Method | Sensitivity (%) | Specificity (%) | Precision (%) | Accuracy (%) | F1 Score (%) | AUC for AD/VaD (95% CI) | ||
|---|---|---|---|---|---|---|---|---|
| Physiological Data | SVM using 33 features | fold1 | 47.06 (8/17) | 82.61 (19/23) | 66.67 (8/12) | 67.50 (27/40) | 55.17 | 0.65 (0.47~0.81) |
| fold2 | 29.41 (5/17) | 82.61 (19/23) | 55.56 (5/9) | 60.00 (24/40) | 38.46 | 0.65 (0.44~0.80) | ||
| fold3 | 70.59 (12/17) | 95.45 (21/22) | 92.31 (12/13) | 84.62 (33/39) | 80.00 | 0.85 (0.62~0.95) | ||
| fold4 | 58.82 (10/17) | 68.18 (15/22) | 58.82 (10/17) | 64.10 (25/39) | 58.82 | 0.71 (0.49~0.85) | ||
| fold5 | 47.06 (8/17) | 90.91 (20/22) | 80.00 (8/10) | 71.79 (28/39) | 59.26 | 0.89 (0.76~0.97) | ||
| Averaged | 50.59 | 83.93 | 70.49 | 69.54 | 58.90 | 0.75 | ||
| SVM using 19 features | fold1 | 64.71 (11/17) | 86.96 (20/23) | 78.57 (11/14) | 77.50 (31/40) | 70.97 | 0.79 (0.59~0.92) | |
| fold2 | 64.71 (11/17) | 86.96 (20/23) | 78.57 (11/14) | 77.50 (31/40) | 70.97 | 0.81 (0.61~0.92) | ||
| fold3 | 76.47 (13/17) | 90.91 (20/22) | 86.67 (13/15) | 84.62 (33/39) | 81.25 | 0.92 (0.80~0.97) | ||
| fold4 | 76.47 (13/17) | 81.82 (18/22) | 76.47 (13/17) | 79.49 (31/39) | 76.47 | 0.76 (0.55~0.92) | ||
| fold5 | 76.47 (13/17) | 95.45 (21/22) | 92.86 (13/14) | 87.18 (34/39) | 83.87 | 0.90 (0.69~0.97) | ||
| Averaged | 71.76 | 88.39 | 82.43 | 81.22 | 76.73 | 0.836 | ||
| Method | Sensitivity (%) | Specificity (%) | Precision (%) | Accuracy (%) | F1 Score (%) | AUC for AD/VaD (95% CI) | ||
|---|---|---|---|---|---|---|---|---|
| ECD Image | InceptionV3 | fold1 | 63.64 (7/11) | 88.89 (8/9) | 87.50 (7/8) | 75.00 (15/20) | 73.68 | 0.95 (0.69~1.00) |
| fold2 | 90.91 (10/11) | 77.78 (7/9) | 83.33 (10/12) | 85.00 (17/20) | 86.96 | 0.95 (0.77~1.00) | ||
| fold3 | 100.00 (11/11) | 88.89 (8/9) | 91.67 (11/12) | 95.00 (19/20) | 95.65 | 0.96 (0.77~1.00) | ||
| fold4 | 90.91 (10/11) | 66.67 (6/9) | 76.92 (10/13) | 80.00 (16/20) | 83.33 | 0.91 (0.69~0.98) | ||
| fold5 | 72.73 (8/11) | 88.89 (8/9) | 88.89 (8/9) | 80.00 (16/20) | 80.00 | 0.93 (0.69~1.00) | ||
| Averaged | 81.82 | 88.89 | 90.00 | 85.00 | 85.71 | 0.95 (0.73~1.00) | ||
| Grad-CAM Map of Inception V3 Model | Image Analysis by SPM | |||
|---|---|---|---|---|
| Brain Region Importance Ranking | AD | VaD | AD | VaD |
| 1 | Cuneus_L | Temporal_Mid_L | Thalamus_L | Paracentral_Lobule_L |
| 2 | Occipital_Sup_L | Lingual_L | Thalamus_R | Postcentral_R |
| 3 | Cuneus_R | Whole cerebellum | Lingual_R | Precentral_L |
| 4 | Occipital_Mid_L | Temporal_Sup_L | Calcarine_R | Supp_Motor_Area_R |
| 5 | Occipital_Sup_R | Occipital_Inf_L | Hippocampus_R | Precentral_R |
| 6 | Temporal_Mid_L | Lingual_R | Precuneus_R | Frontal_Sup_R |
| 7 | Occipital_Mid_R | Occipital_Inf_R | ParaHippocampal_L | Supp_Motor_Area_L |
| 8 | Precuneus_L | Putamen_L | ParaHippocampal_R | Parietal_Sup_R |
| 9 | Precuneus_R | Temporal_Inf_L | Hippocampus_L | Paracentral_Lobule_R |
| 10 | Parietal_Inf_L | Temporal_Inf_R | Cuneus_R | Frontal_Sup_L |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ni, Y.-C.; Lin, Z.-K.; Cheng, C.-H.; Pai, M.-C.; Chiu, P.-Y.; Chang, C.-C.; Chang, Y.-T.; Hung, G.-U.; Lin, K.-J.; Hsiao, I.-T.; et al. Classification Prediction of Alzheimer’s Disease and Vascular Dementia Using Physiological Data and ECD SPECT Images. Diagnostics 2024, 14, 365. https://doi.org/10.3390/diagnostics14040365
Ni Y-C, Lin Z-K, Cheng C-H, Pai M-C, Chiu P-Y, Chang C-C, Chang Y-T, Hung G-U, Lin K-J, Hsiao I-T, et al. Classification Prediction of Alzheimer’s Disease and Vascular Dementia Using Physiological Data and ECD SPECT Images. Diagnostics. 2024; 14(4):365. https://doi.org/10.3390/diagnostics14040365
Chicago/Turabian StyleNi, Yu-Ching, Zhi-Kun Lin, Chen-Han Cheng, Ming-Chyi Pai, Pai-Yi Chiu, Chiung-Chih Chang, Ya-Ting Chang, Guang-Uei Hung, Kun-Ju Lin, Ing-Tsung Hsiao, and et al. 2024. "Classification Prediction of Alzheimer’s Disease and Vascular Dementia Using Physiological Data and ECD SPECT Images" Diagnostics 14, no. 4: 365. https://doi.org/10.3390/diagnostics14040365
APA StyleNi, Y.-C., Lin, Z.-K., Cheng, C.-H., Pai, M.-C., Chiu, P.-Y., Chang, C.-C., Chang, Y.-T., Hung, G.-U., Lin, K.-J., Hsiao, I.-T., Lin, C.-Y., & Yang, H.-C. (2024). Classification Prediction of Alzheimer’s Disease and Vascular Dementia Using Physiological Data and ECD SPECT Images. Diagnostics, 14(4), 365. https://doi.org/10.3390/diagnostics14040365





