To explore the predictive capacity of machine learning models in imbalanced metabolomics binary classification data sets, we adopted an evaluation methodology. Data sets reflect a high-dimensional data structure common to metabolomics studies. Given the challenges posed by the data set’s imbalance and complexity, we implemented an approach to model development and validation to ensure the reliability of our findings.
Initially, we developed eight machine learning models on the entire data set. These models were selected based on their ability to handle challenges in the data set and their various learning mechanisms, providing a broad range of evaluations. The development and evaluation of these models were carried out using a stratified 10-fold cross-validation technique. We selected 10 for k in the k-fold cross-validation as it offers a good balance between bias and variance in model evaluation, considering the sample size [
54]. Stratification in the 10-fold cross-validation involves dividing the data set into ten folds of equal size, ensuring that the distribution of labels is consistent across all folds, thereby maintaining the representativeness of the data set. In each iteration, nine folds were used to train the model, and the remaining fold served as a test set. This process was repeated ten times, each fold serving as a test set once, ensuring that every data point was used for both training and testing. The performance of each model was then evaluated based on the mean of the metrics obtained from the ten iterations, providing an aggregate measure of the model’s effectiveness across the entire data set.
In the field of machine learning, hyperparameters are variables that define the model’s structure or the characteristics of the learning algorithm, and are set before training begins. Unlike hyperparameters, other parameters like coefficients or weights change during training. The need and number of hyperparameters differ depending on the algorithm. To examine the impact of the model optimization, we enhanced our evaluation methodology by adopting a nested stratified cross-validation strategy for fine-tuning these hyperparameters.
Given the specific characteristics and challenges of our data sets, we opted for a limited search space tailored to each model to prevent overfitting, utilizing a grid search method for this purpose. In this approach, an additional stratified 5-fold cross-validation dedicated to hyperparameter tuning was conducted within each fold of the primary stratified 10-fold cross-validation. This nested setup enabled us to further partition the training folds from the outer loop into smaller sub-folds, helping to refine the model parameters and stabilize their performance across different folds. Here, four sub-folds were employed for training with different hyperparameter settings, while the fifth sub-fold acted as a validation set to evaluate these settings. The best-performing hyperparameter set on the validation sets from the five inner folds was then selected as the optimal configuration for each model. With these optimal hyperparameters, the model was trained anew on the complete training data set from the outer loop, prior to its assessment on the test fold.
3.2. Performance of Machine Learning Models
Table 3,
Table 4 and
Table 5 summarize the efficacy of different machine learning models across three metabolomics data sets, using five evaluation metrics. The values presented in these tables are the averages obtained from the 10-fold cross-validation process. First, we examine performance obtained by machine learning models in three metabolomics data sets based on each metric, so that we can identify which data set is most suitable for machine learning models to detect dry eye in cataract patients.
The best AUC result achieved by machine learning models on the merged data set was , surpassing its effectiveness on the ESI− and ESI+ data sets by and , respectively. The highest balanced accuracy recorded by machine learning models was in both the merged and ESI+ data sets, outperforming the ESI− data set by . Similarly, for the MCC metric, models performed better on the merged and ESI+ data sets, achieving a score of , which is an improvement of over the ESI− data set. In terms of specificity, models scored highest on the ESI− data set at , exceeding scores in the merged and ESI+ data sets by . For the F1-Score, the peak performance was observed in the ESI+ data set at , higher than in the merged and ESI− data sets by and .
These results suggest that models generally performed better on the merged data set in terms of AUC, balanced accuracy, and MCC, though they fell slightly short in specificity and F1-Score. Therefore, these outcomes indicate that the merged data set can enhance model performance across some metrics, and achieve more balanced performance across all metrics, suggesting a beneficial effect of merging the ESI+ and ESI− data sets on model efficacy.
Following, we explore the effectiveness of various machine learning models, focusing on the merged data set, as detailed in
Table 5, to find the most suitable models for identifying dry eye in cataract patients. The results in
Table 5 reveal that, compared to baseline approaches that involve random guesses or selecting the most frequent class (represented by Dummy Classifiers), almost all the models demonstrated better performance across multiple evaluation metrics.
According to
Table 5, the LR model has the highest AUC at
. The AUC metric, commonly used in medical machine learning applications for disease prediction problems, shows that the LR model effectively differentiates between the two classes. Compared to other tested machine learning methods, this model achieves a higher AUC. After LR, the RF and XGBoost models recorded the next highest scores of
and
, respectively. Due to the imbalance in the data set’s classes, other metrics besides the AUC were also considered to provide a clearer comparison of the models.
In terms of balanced accuracy, LR achieved the top score of , outperforming other models in the merged data set. This metric highlights the model’s effectiveness on an imbalanced data set by valuing the performance across both minority and majority classes equally. XGBoost and RF followed with scores of and , respectively.
For the MCC, LR again led with a score of , indicating its better performance in handling imbalanced data sets compared to other models. XGBoost and RF followed with scores of and , respectively.
For specificity, the LR and XGBoost achieved the highest score of , and MLP followed with a score of . The negative samples are the minority classes in the data sets, making this metric important for demonstrating the models’ ability to correctly detect negative samples.
Regarding the F1-score, the RF model outperformed the others with a score of , closely followed by LR and XGBoost with scores of and , respectively.
In summary, this comparative analysis indicates that the LR model achieved the highest performance in AUC, balanced accuracy, MCC, and specificity compared to other machine learning models in the merged data set. With a performance close to highest score in F1-score, it maintained balanced performance across all metrics compared to other machine learning models in this study on the merged metabolomics data set.
Previous research shows that the LR model was successful in biological and clinical contexts [
55,
56]. The results of this study demonstrated that the LR model outperforms other machine learning models and suggests that it is possible to achieve high performance even with a quite simple model. The more complex models, can quickly suffer from overfitting on the fairly small data set in this study. Additionally, LR is generally computationally less intensive and achieves convergence faster than more complex methods. This could be another reason for more stable model training and prediction, especially in this study’s data set, characterised by high-dimensional spaces and a limited number of samples. This is further discussed in
Section 3.3.
The next best machine learning models following the LR model on the merged data set are XGBoost and RF, which are ensemble models. They achieved satisfactory performance on some evaluation metrics, but underperformed on others, and overall did not achieve consistent performance across all evaluation metrics.
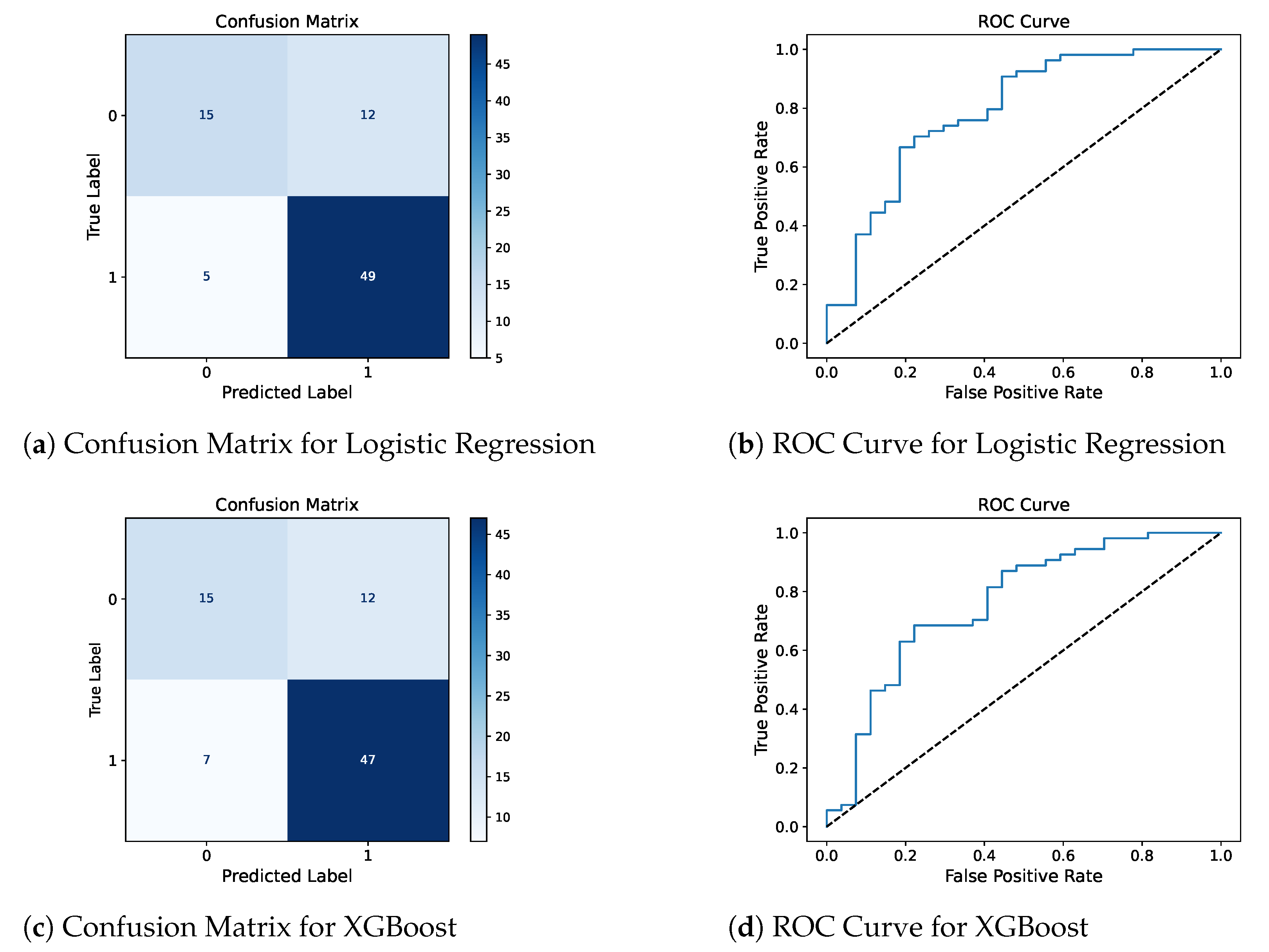
Confusion matrix and ROC curve for the two best models, LR and XGBoost, are presented in
Figure 2 to complement the reported metrics by providing detailed insights into the model’s classification results and their ability to distinguish between DED-positive and DED-negative patients.
In this study, the weakest results were observed with k-NN and SVM. Despite the theoretical benefits often associated with SVM, especially with an RBF kernel, in the field of bioinformatics and for high-dimensional data sets [
18,
57,
58], the results presented in
Table 5 demonstrate performance comparable to that of baseline dummy classifiers across several evaluation metrics on the merged data set. Both the k-NN and SVM models demonstrated suboptimal outcomes in this study. These results emphasize the need for a careful and nuanced approach when selecting models for metabolomics data sets.
Furthermore, integrating data sets from different ionization modes into a single data set has proven to improve model training by providing a wider range of features and patterns. This method enhances the data pool and helps models generalize better and increase prediction accuracy.
3.3. Model Performance Consistency
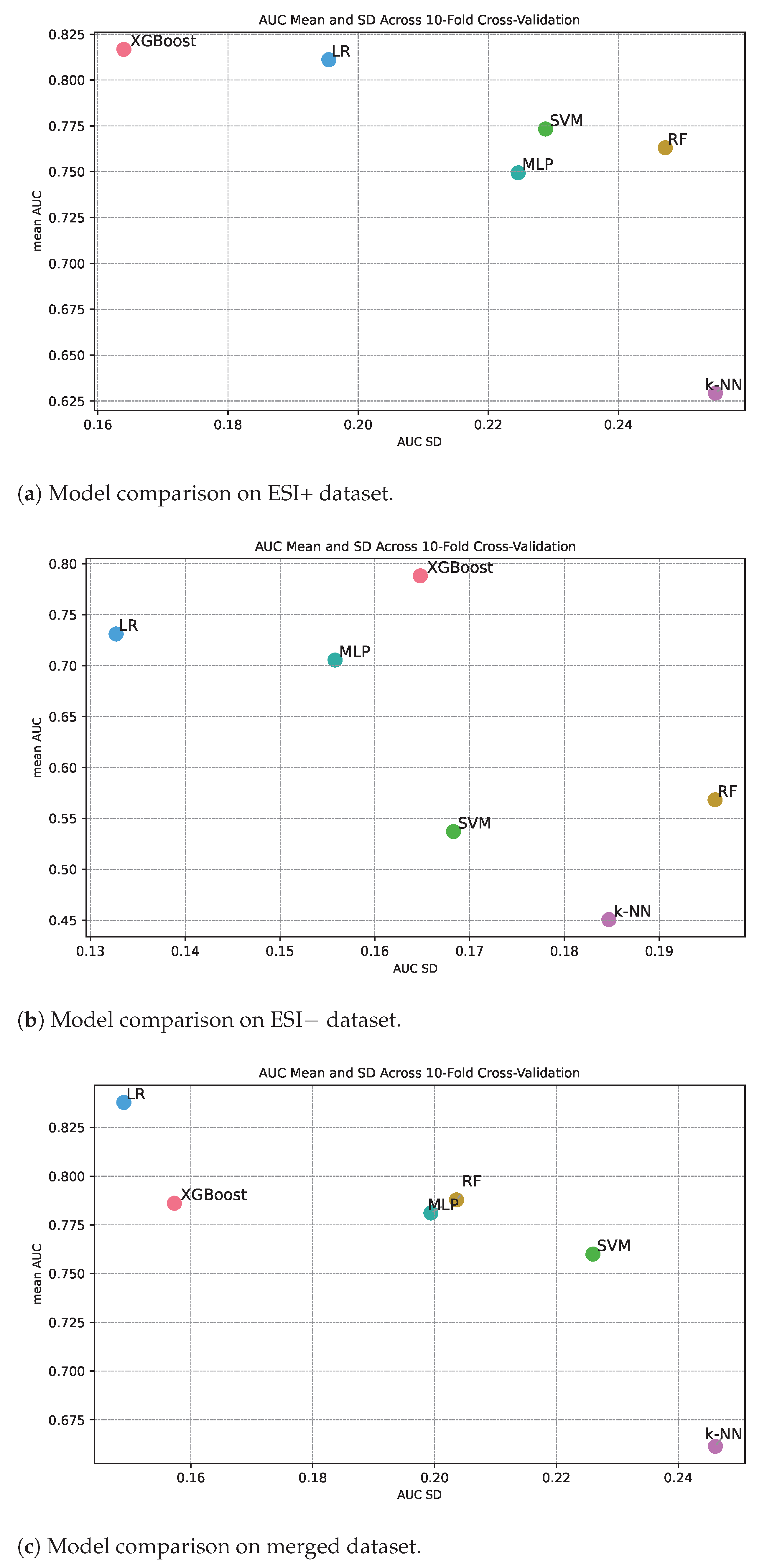
Figure 3 shows classification performance in terms of AUC along the y-axis and the standard deviation of the measured AUC from the different cross-validation folds along the x-axis. The standard deviation therefore gives an indication of the level of consistency in performance for different machine learning methods under repeated training of the algorithm. If a method has high classification performance over the folds, we also expect high classification constancy over the folds. Or said in another way, it is not possible to achieve high classification performance and at the same time have low consistency. Inspecting the three panels in
Figure 3, we see that this is the overall trend, but there are for sure also differences in consistency for methods with about the same classification performance. Intuitively, we can expect that algorithms that are simple to train with a convex loss landscape, such as logistic ridge regression (based on a linear classifier), might document higher constancy compared to models that have a more complex loss landscape. We see that logistic ridge regression documents high consistency relative to classification performance for all the three data sets (
Figure 3a–c).
Among the methods with a mean AUC over
for the ESI+ data set (
Figure 3a), we see that, in addition to logistic ridge regression, XGBoost also documents high constancy, while MLP, SVM, and RF document less consistency. Further, k-NN also documents poor constancy, which is as expected since the mean AUC is low.
Among the methods with a mean AUC over
for the ESI− data set (panel
Figure 3b), we see that only logistic ridge regression documents high consistency, while MLP and XGBoost document medium consistency. The other methods document poorer classification performance, and as expected, consistency is also low.
Among the methods with a mean AUC over
for the merged data (
Figure 3c), we see that in addition to logistic ridge regression, XGBoost also documents high consistency, while MLP, RF, and SVM document medium consistency.
To summarize, logistic ridge regression documents high constancy, XGBoost medium to high consistency in classification performance while the other methods document medium to low consistency.