Machine Learning Algorithms Based on Time Series Pre-Clustering for Nocturnal Glucose Prediction in People with Type 1 Diabetes
Abstract
1. Introduction
2. Materials and Methods
2.1. Dataset
2.2. Data Preprocessing
2.3. Algorithms
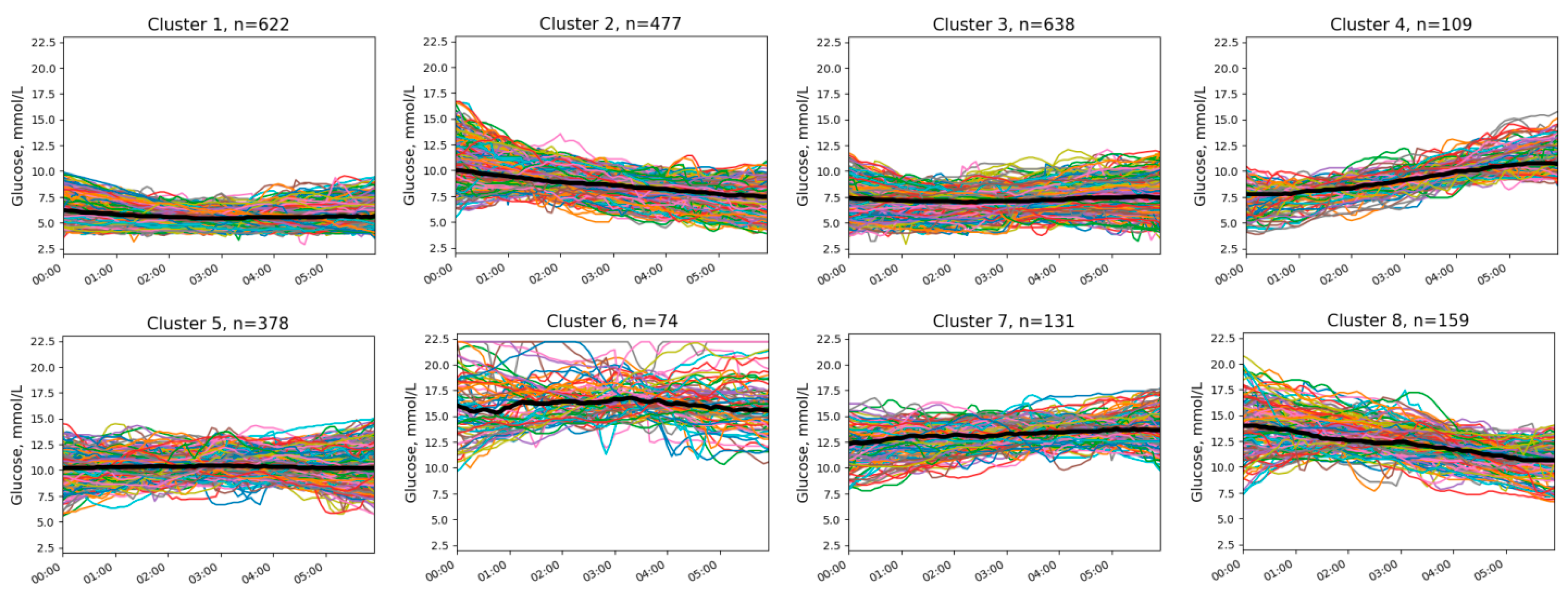
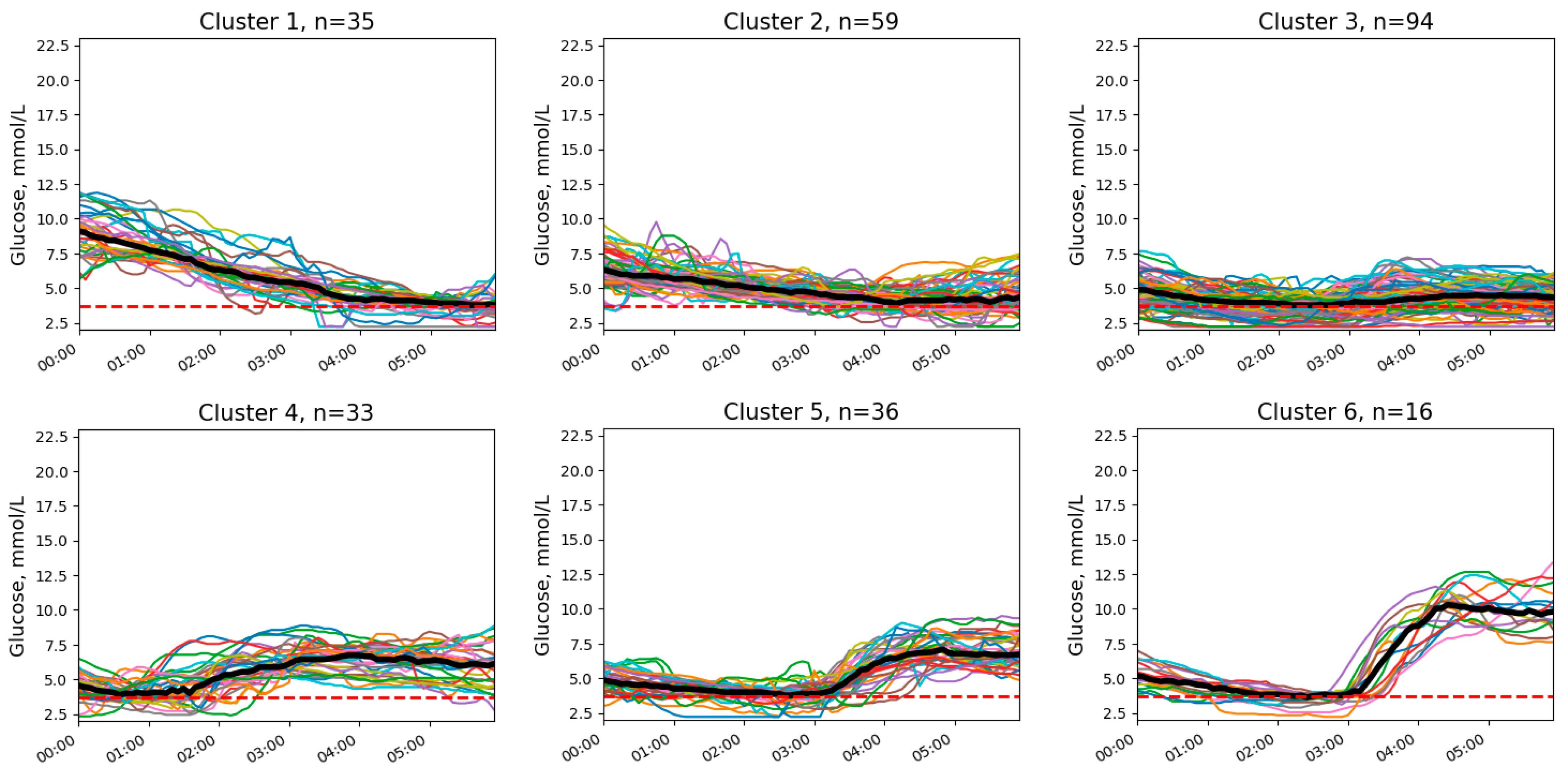
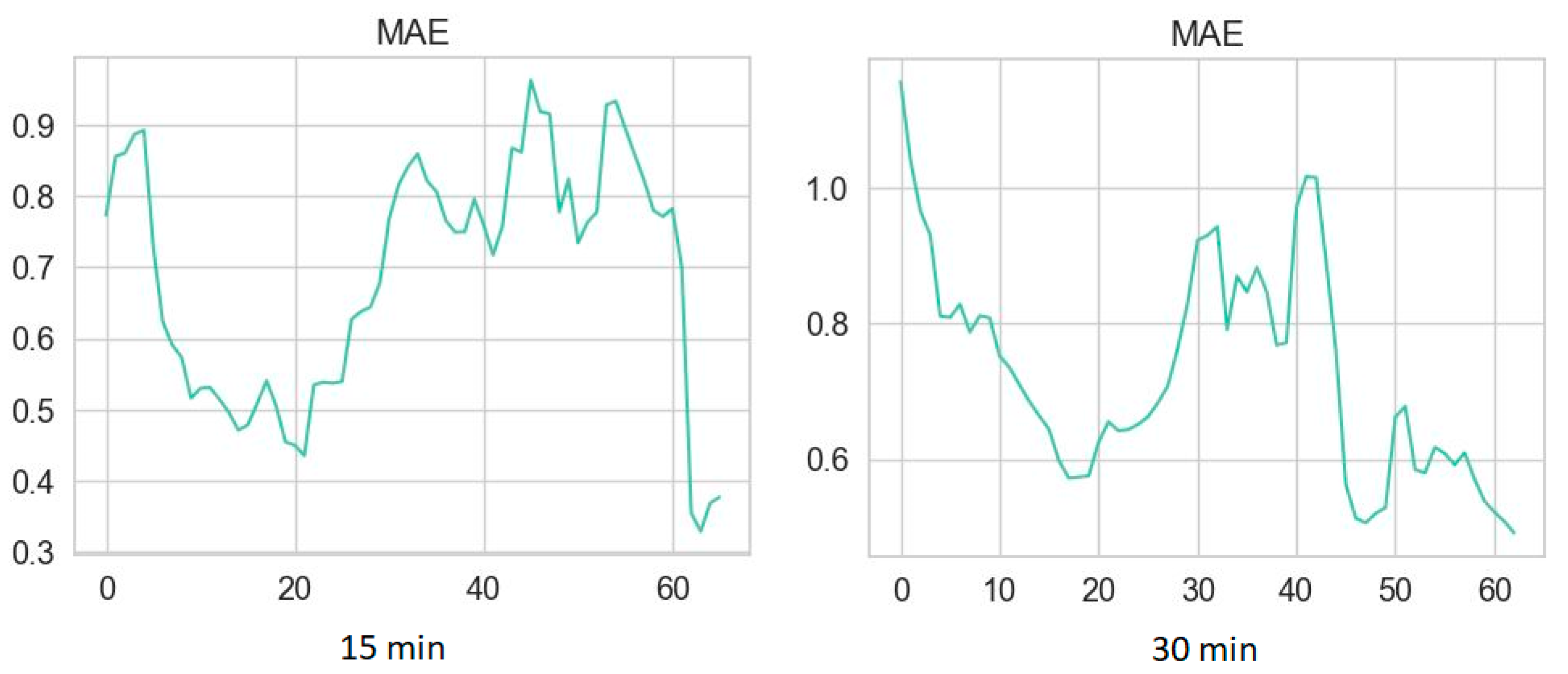
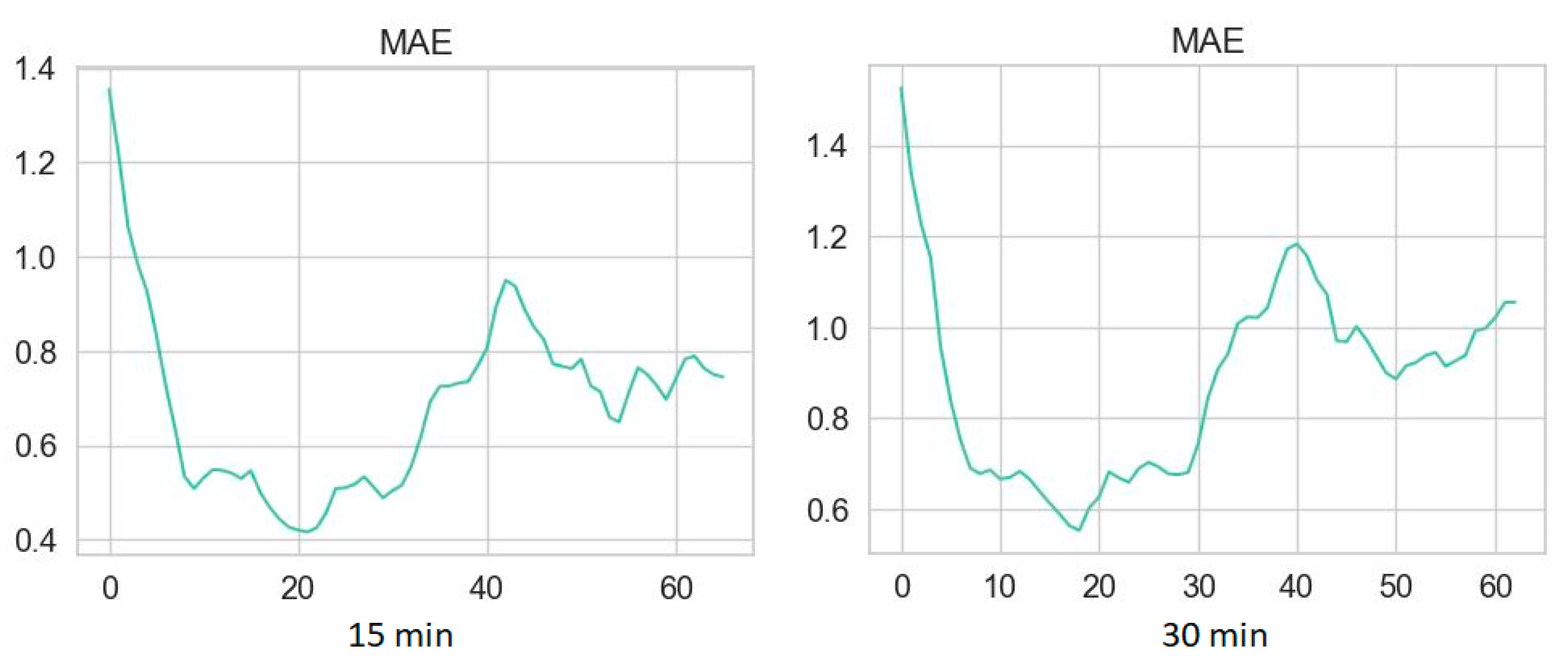
2.3.1. Clustering Procedure
2.3.2. Forecasting Using Medoids of Time Series Clusters (MTSCs)
- Apply clustering algorithm for and find the optimal number of clusters ;
- Evaluate the obtained clustering structure;
- For each cluster, specify a typical object (centroid, medoid, etc.) ;
- For , find the nearest (in the sense of a given distance) typical object , where is a metric function between observations;
- As a prediction for , take the next components of the selected typical object .
2.3.3. Forecasting Using Weighted Medoids of Time Series Clusters (WMTSCs)
2.3.4. Combined Algorithms Based on the Cluster Analysis and Supervised ML
- 1.
- Apply clustering algorithm for and find the optimal number of clusters ;
- 2.
- Evaluate the obtained clustering structure;
- 3.
- For each cluster , train ML model for prediction at the determined prediction horizon. Cluster is a training dataset for ;
- 4.
- For each cluster , specify a typical observation (centroid, medoid, etc.) ;
- 5.
- For , find the nearest cluster, i.e., the nearest typical object , where is a metric function between objects;
- 6.
- Predict with a model trained on the nearest cluster found in step 5.
2.3.5. Baseline Algorithms
- An algorithm that uses averaging to improve the predictive accuracy. In this study, we used an RF algorithm. In the RF, each tree in the ensemble is built from a sample drawn with replacement (i.e., a bootstrap sample) from a training set. This algorithm takes the average prediction of each tree in the ensemble as a result;
- An algorithm that builds an additive model in a forward stage-wise fashion. In each stage, a regression tree is fit onto the negative gradient of the given loss function to minimize it. We used a gradient boosting trees (GBTs) model.
2.4. Assessment of the Performance of Algorithms
- Root Mean Squared Error (RMSE): ;
- Mean Absolute Error (MAE): ;
- Mean Absolute Percentage Error (MAPE): .
2.5. Coding
3. Results
3.1. Clusters of Nocturnal Glucose Dynamics
3.2. Performance Metrics of Glucose Prediction Algorithms
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Ogle, G.D.; Wang, F.; Gregory, G.A.; Maniam, J.; T1D Index Consortium. Type 1 Diabetes Estimates in Children and Adults. 2022. Available online: https://diabetesatlas.org/atlas/t1d-index-2022/ (accessed on 17 September 2024).
- Chung, W.K.; Erion, K.; Florez, J.C.; Hattersley, A.T.; Hivert, M.F.; Lee, C.G.; McCarthy, M.I.; Nolan, J.J.; Norris, J.M.; Pearson, E.R.; et al. Precision medicine in diabetes: A Consensus Report from the American Diabetes Association (ADA) and the European Association for the Study of Diabetes (EASD). Diabetologia 2020, 63, 1671–1693. [Google Scholar] [CrossRef] [PubMed]
- Carr, A.L.J.; Evans-Molina, C.; Oram, R.A. Precision medicine in type 1 diabetes. Diabetologia 2022, 65, 1854–1866. [Google Scholar] [CrossRef] [PubMed]
- Peacock, S.; Frizelle, I.; Hussain, S. A Systematic Review of Commercial Hybrid Closed-Loop Automated Insulin Delivery Systems. Diabetes Ther. 2023, 14, 839–855. [Google Scholar] [CrossRef] [PubMed]
- Kulzer, B.; Heinemann, L. Predicting Glucose Values: A New Era for Continuous Glucose Monitoring. J. Diabetes Sci. Technol. 2024, 18, 1000–1003. [Google Scholar] [CrossRef]
- Siamashvili, M.; Davis, H.A.; Davis, S.N. Nocturnal hypoglycemia in type 1 and type 2 diabetes: An update on prevalence, prevention, pathophysiology and patient awareness. Expert Rev. Endocrinol. Metab. 2021, 16, 281–293. [Google Scholar] [CrossRef]
- Graveling, A.J.; Frier, B.M. The risks of nocturnal hypoglycaemia in insulin-treated diabetes. Diabetes Res. Clin. Pract. 2017, 133, 30–39. [Google Scholar] [CrossRef]
- Schultes, B.; Jauch-Chara, K.; Gais, S.; Hallschmid, M.; Reiprich, E.; Kern, W.; Oltmanns, K.M.; Peters, A.; Fehm, H.L.; Born, J. Defective awakening response to nocturnal hypoglycemia in patients with type 1 diabetes mellitus. PLoS Med. 2007, 4, e69. [Google Scholar] [CrossRef]
- Choudhary, P.; Amiel, S.A. Hypoglycaemia in type 1 diabetes: Technological treatments, their limitations and the place of psychology. Diabetologia 2018, 61, 761–769. [Google Scholar] [CrossRef]
- Woldaregay, A.Z.; Årsand, E.; Walderhaug, S.; Albers, D.; Mamykina, L.; Botsis, T.; Hartvigsen, G. Data-driven modeling and prediction of blood glucose dynamics: Machine learning applications in type 1 diabetes. Artif. Intell. Med. 2019, 98, 109–134. [Google Scholar] [CrossRef]
- Felizardo, V.; Garcia, N.M.; Pombo, N.; Megdiche, I. Data-based algorithms and models using diabetics real data for blood glucose and hypoglycaemia prediction—A systematic literature review. Artif. Intell. Med. 2021, 118, 102120. [Google Scholar] [CrossRef]
- Tsichlaki, S.; Koumakis, L.; Tsiknakis, M. Type 1 Diabetes Hypoglycemia Prediction Algorithms: Systematic Review. JMIR Diabetes 2022, 7, e34699. [Google Scholar] [CrossRef] [PubMed]
- Liu, K.; Li, L.; Ma, Y.; Jiang, J.; Liu, Z.; Ye, Z.; Liu, S.; Pu, C.; Chen, C.; Wan, Y. Machine Learning Models for Blood Glucose Level Prediction in Patients with Diabetes Mellitus: Systematic Review and Network Meta-Analysis. JMIR Med. Inform. 2023, 11, e47833. [Google Scholar] [CrossRef] [PubMed]
- Dave, D.; DeSalvo, D.J.; Haridas, B.; McKay, S.; Shenoy, A.; Koh, C.J.; Lawley, M.; Erraguntla, M. Feature-Based Machine Learning Model for Real-Time Hypoglycemia Prediction. J. Diabetes Sci. Technol. 2021, 15, 842–855. [Google Scholar] [CrossRef]
- Berikov, V.B.; Kutnenko, O.A.; Semenova, J.F.; Klimontov, V.V. Machine Learning Models for Nocturnal Hypoglycemia Prediction in Hospitalized Patients with Type 1 Diabetes. J. Pers. Med. 2022, 12, 1262. [Google Scholar] [CrossRef]
- Bertachi, A.; Viñals, C.; Biagi, L.; Contreras, I.; Vehí, J.; Conget, I.; Giménez, M. Prediction of Nocturnal Hypoglycemia in Adults with Type 1 Diabetes under Multiple Daily Injections Using Continuous Glucose Monitoring and Physical Activity Monitor. Sensors 2020, 20, 1705. [Google Scholar] [CrossRef]
- Kladov, D.E.; Berikov, V.B.; Semenova, J.F.; Klimontov, V.V. Nocturnal Glucose Patterns with and without Hypoglycemia in People with Type 1 Diabetes Managed with Multiple Daily Insulin Injections. J. Pers. Med. 2023, 13, 1454. [Google Scholar] [CrossRef] [PubMed]
- Cassisi, C.; Montaldo, P.; Aliotta, M.; Cannata, A.; Pulvirenti, A. Similarity Measures and Dimensionality Reduction Techniques for Time Series Data Mining. In Advances in Data Mining Knowledge Discovery and Applications; Karahoca, A., Ed.; InTechOpen: Rijeka, Croatia, 2012; pp. 71–96. [Google Scholar] [CrossRef]
- Rousseeuw, P. Silhouettes: A Graphical Aid to the Interpretation and Validation of Cluster Analysis. J. Comput. Appl. Math. 1987, 20, 53–65. [Google Scholar] [CrossRef]
- Ward, J.H., Jr. Hierarchical grouping to optimize an objective function. J. Am. Stat. Assoc. 1963, 58, 236–244. [Google Scholar] [CrossRef]
- Kirilyuk, I.L.; Senko, O.V. Assessing the validity of clustering of panel data by Monte Carlo methods (using as example the data of the Russian regional economy). Comput. Res. Model. 2020, 12, e1501–e1513. [Google Scholar] [CrossRef]
- Mujahid, O.; Contreras, I.; Vehi, J. Machine learning techniques for hypoglycemia prediction: Trends and challenges. Sensors 2021, 21, 546. [Google Scholar] [CrossRef]
- Li, J.; Ma, X.; Tobore, I.; Liu, Y.; Kandwal, A.; Wang, L.; Lu, J.; Lu, W.; Bao, Y.; Zhou, J.; et al. A Novel CGM Metric-Gradient and Combining Mean Sensor Glucose Enable to Improve the Prediction of Nocturnal Hypoglycemic Events in Patients with Diabetes. J. Diabetes Res. 2020, 2020, 8830774. [Google Scholar] [CrossRef] [PubMed]
- Zhu, T.; Li, K.; Herrero, P.; Georgiou, P. Personalized Blood Glucose Prediction for Type 1 Diabetes Using Evidential Deep Learning and Meta-Learning. IEEE Trans. Biomed. Eng. 2023, 70, 193–204. [Google Scholar] [CrossRef]
- Kozinetz, R.M.; Berikov, V.B.; Semenova, J.F.; Klimontov, V.V. Machine Learning and Deep Learning Models for Nocturnal High- and Low-Glucose Prediction in Adults with Type 1 Diabetes. Diagnostics 2024, 14, 740. [Google Scholar] [CrossRef]
- Kalita, D.; Sharma, H.; Panda, J.K.; Mirza, K.B. Platform for precise, personalised glucose forecasting through continuous glucose and physical activity monitoring and deep learning. Med. Eng. Phys. 2024, 132, 104241. [Google Scholar] [CrossRef]
- Zhu, T.; Kuang, L.; Piao, C.; Zeng, J.; Li, K.; Georgiou, P. Population-Specific Glucose Prediction in Diabetes Care with Transformer-Based Deep Learning on the Edge. IEEE Trans. Biomed. Circuits Syst. 2024, 18, 236–246. [Google Scholar] [CrossRef]
- Han, Y.; Kim, D.Y.; Woo, J.; Kim, J. Glu-Ensemble: An ensemble deep learning framework for blood glucose forecasting in type 2 diabetes patients. Heliyon 2024, 10, e29030. [Google Scholar] [CrossRef] [PubMed]
- Deng, Y.; Lu, L.; Aponte, L.; Angelidi, A.M.; Novak, V.; Karniadakis, G.E.; Mantzoros, C.S. Deep transfer learning and data augmentation improve glucose levels prediction in type 2 diabetes patients. NPJ Digit Med. 2021, 4, 109. [Google Scholar] [CrossRef] [PubMed]
- Wadghiri, M.Z.; Idri, A.; El Idrissi, T.; Hakkoum, H. Ensemble blood glucose prediction in diabetes mellitus: A review. Comput. Biol. Med. 2022, 147, 105674. [Google Scholar] [CrossRef]
- Yang, H.; Chen, Z.; Huang, J.; Li, S. AWD-stacking: An enhanced ensemble learning model for predicting glucose levels. PLoS ONE 2024, 19, e0291594. [Google Scholar] [CrossRef]
- Ibrahim, M.; Beneyto, A.; Contreras, I.; Vehi, J. An ensemble machine learning approach for the detection of unannounced meals to enhance postprandial glucose control. Comput. Biol. Med. 2024, 171, 108154. [Google Scholar] [CrossRef]
- Parcerisas, A.; Contreras, I.; Delecourt, A.; Bertachi, A.; Beneyto, A.; Conget, I.; Viñals, C.; Giménez, M.; Vehi, J. A Machine Learning Approach to Minimize Nocturnal Hypoglycemic Events in Type 1 Diabetic Patients under Multiple Doses of Insulin. Sensors 2022, 22, 1665. [Google Scholar] [CrossRef] [PubMed]
- Battaglia, M.; Ahmed, S.; Anderson, M.S.; Atkinson, M.A.; Becker, D.; Bingley, P.J.; Bosi, E.; Brusko, T.M.; DiMeglio, L.A.; Evans-Molina, C.; et al. Introducing the Endotype Concept to Address the Challenge of Disease Heterogeneity in Type 1 Diabetes. Diabetes Care 2020, 43, 5–12. [Google Scholar] [CrossRef] [PubMed]
- Redondo, M.J.; Morgan, N.G. Heterogeneity and endotypes in type 1 diabetes mellitus. Nat. Rev. Endocrinol. 2023, 19, 542–554. [Google Scholar] [CrossRef] [PubMed]
- Fagherazzi, G.; Aguayo, G.A.; Zhang, L.; Hanaire, H.; Picard, S.; Sablone, L.; Vergès, B.; Hamamouche, N.; Detournay, B.; Joubert, M.; et al. SFDT1 study team. Heterogeneity of glycaemic phenotypes in type 1 diabetes. Diabetologia 2024, 67, 1567–1581. [Google Scholar] [CrossRef]
- Cichosz, S.L.; Hejlesen, O.; Jensen, M.H. Identification of individuals with diabetes who are eligible for continuous glucose monitoring forecasting. Diabetes Metab. Syndr. Clin. Res. Rev. 2024, 18, 102972. [Google Scholar] [CrossRef]
- Langarica, S.; de la Vega, D.; Cariman, N.; Miranda, M.; Andrade, D.C.; Nunez, F.; Rodriguez-Fernandez, M. Deep Learning-Based Glucose Prediction Models: A Guide for Practitioners and a Curated Dataset for Improved Diabetes Management. IEEE Open J. Eng. Med. Biol. 2024, 5, 467–475. [Google Scholar] [CrossRef]
| Parameter | Time Series Without NH | Time Series with NH |
|---|---|---|
| 3376 | 449 | |
| Delta (%) | 0.02 | 19.2 |
| 3318 | 363 |
| PH | Algorithm | RMSE | MAE | MAPE |
|---|---|---|---|---|
| 15 min | MTSCs | 1.643 | 1.221 | 0.148 |
| WMTSCs | 3.555 | 3.029 | 0.434 | |
| Holt model | 0.640 | 0.327 | 0.039 | |
| GBTs without pre-clustering | 0.600 | 0.324 | 0.037 | |
| RF with pre-clustering | 0.532 | 0.322 | 0.038 | |
| GBTs with pre-clustering | 0.542 | 0.305 | 0.034 | |
| 30 min | MTSCs | 1.742 | 1.276 | 0.154 |
| WMTSCs | 3.553 | 3.028 | 0.433 | |
| Holt model | 1.219 | 0.634 | 0.074 | |
| GBTs without pre-clustering | 0.770 | 0.450 | 0.052 | |
| RF with pre-clustering | 0.811 | 0.526 | 0.062 | |
| GBTs with pre-clustering | 0.771 | 0.422 | 0.051 |
| PH | Algorithm | RMSE | MAE | MAPE |
|---|---|---|---|---|
| 15 min | MTSCs | 1.916 | 1.406 | 0.274 |
| WMTSCs | 2.706 | 2.193 | 0.457 | |
| Holt model | 0.797 | 0.440 | 0.086 | |
| GBTs without clustering | 1.169 | 0.724 | 0.120 | |
| RF with pre-clustering | 0.755 | 0.488 | 0.091 | |
| GBTs with pre-clustering | 0.682 | 0.451 | 0.080 | |
| 30 min | MTSCs | 1.936 | 1.433 | 0.278 |
| WMTSCs | 2.706 | 2.194 | 0.457 | |
| Holt model | 1.461 | 0.890 | 0.154 | |
| GBTs without pre-clustering | 1.534 | 1.027 | 0.171 | |
| RF with pre-clustering | 1.201 | 0.787 | 0.150 | |
| GBTs with pre-clustering | 1.305 | 0.918 | 0.166 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kladov, D.E.; Berikov, V.B.; Semenova, J.F.; Klimontov, V.V. Machine Learning Algorithms Based on Time Series Pre-Clustering for Nocturnal Glucose Prediction in People with Type 1 Diabetes. Diagnostics 2024, 14, 2427. https://doi.org/10.3390/diagnostics14212427
Kladov DE, Berikov VB, Semenova JF, Klimontov VV. Machine Learning Algorithms Based on Time Series Pre-Clustering for Nocturnal Glucose Prediction in People with Type 1 Diabetes. Diagnostics. 2024; 14(21):2427. https://doi.org/10.3390/diagnostics14212427
Chicago/Turabian StyleKladov, Danil E., Vladimir B. Berikov, Julia F. Semenova, and Vadim V. Klimontov. 2024. "Machine Learning Algorithms Based on Time Series Pre-Clustering for Nocturnal Glucose Prediction in People with Type 1 Diabetes" Diagnostics 14, no. 21: 2427. https://doi.org/10.3390/diagnostics14212427
APA StyleKladov, D. E., Berikov, V. B., Semenova, J. F., & Klimontov, V. V. (2024). Machine Learning Algorithms Based on Time Series Pre-Clustering for Nocturnal Glucose Prediction in People with Type 1 Diabetes. Diagnostics, 14(21), 2427. https://doi.org/10.3390/diagnostics14212427







