An Efficient and Robust Method for Chest X-ray Rib Suppression That Improves Pulmonary Abnormality Diagnosis
Abstract
1. Introduction
2. Method
2.1. ST Smoothing
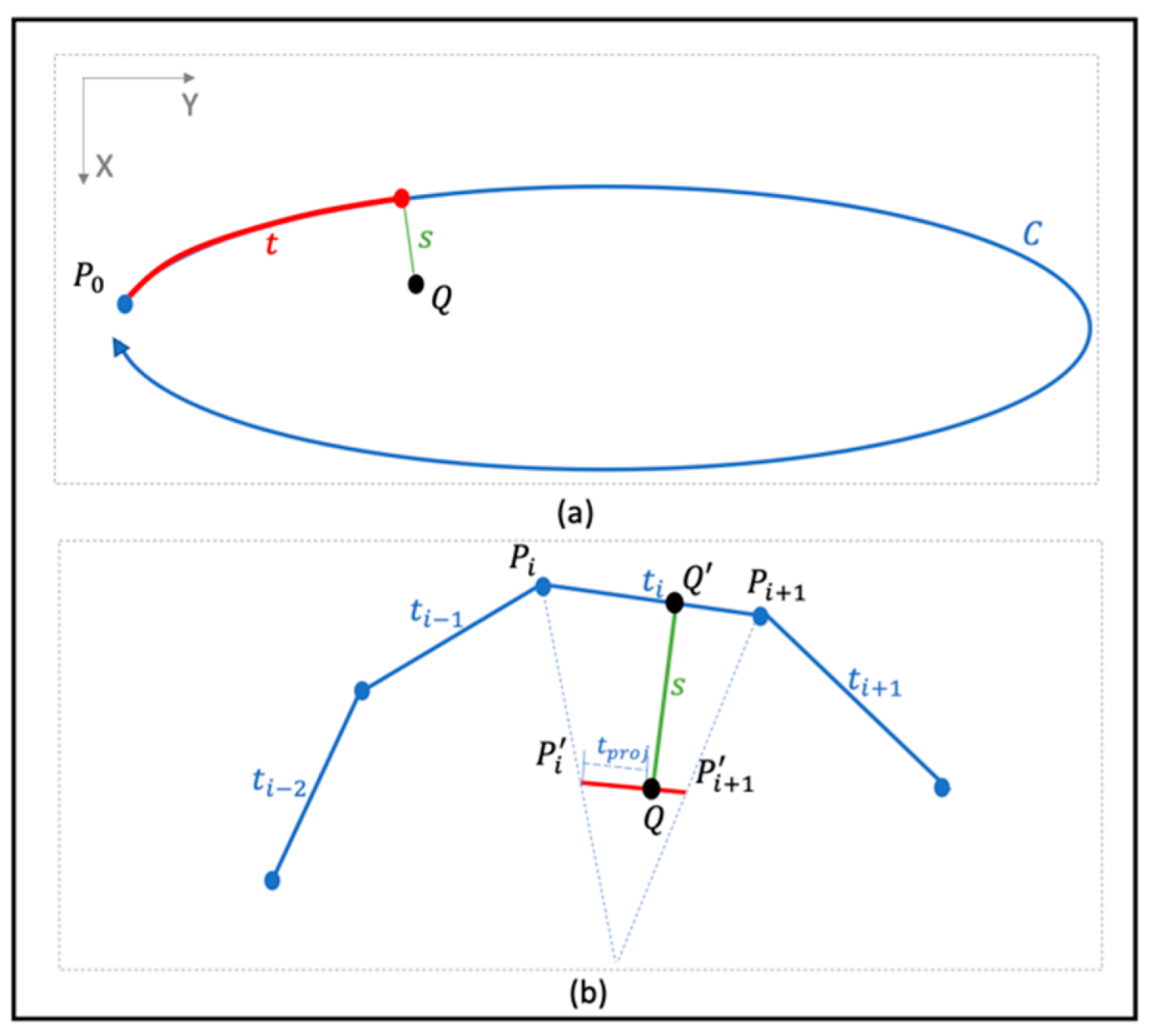
2.1.1. From Image Space to ST-Coordinate System
2.1.2. Rib Extraction via Partial Derivatives Smoothing in ST Space
Discrete Partial Derivative in ST Space
Smoothing, Reintegration, and Transformation Back to the XY Domain
2.1.3. Rib Removal and Border Blending
2.2. Data Cohort
2.2.1. VinDr-RibCXR Dataset
2.2.2. FX-RRCXR Dataset Preparation
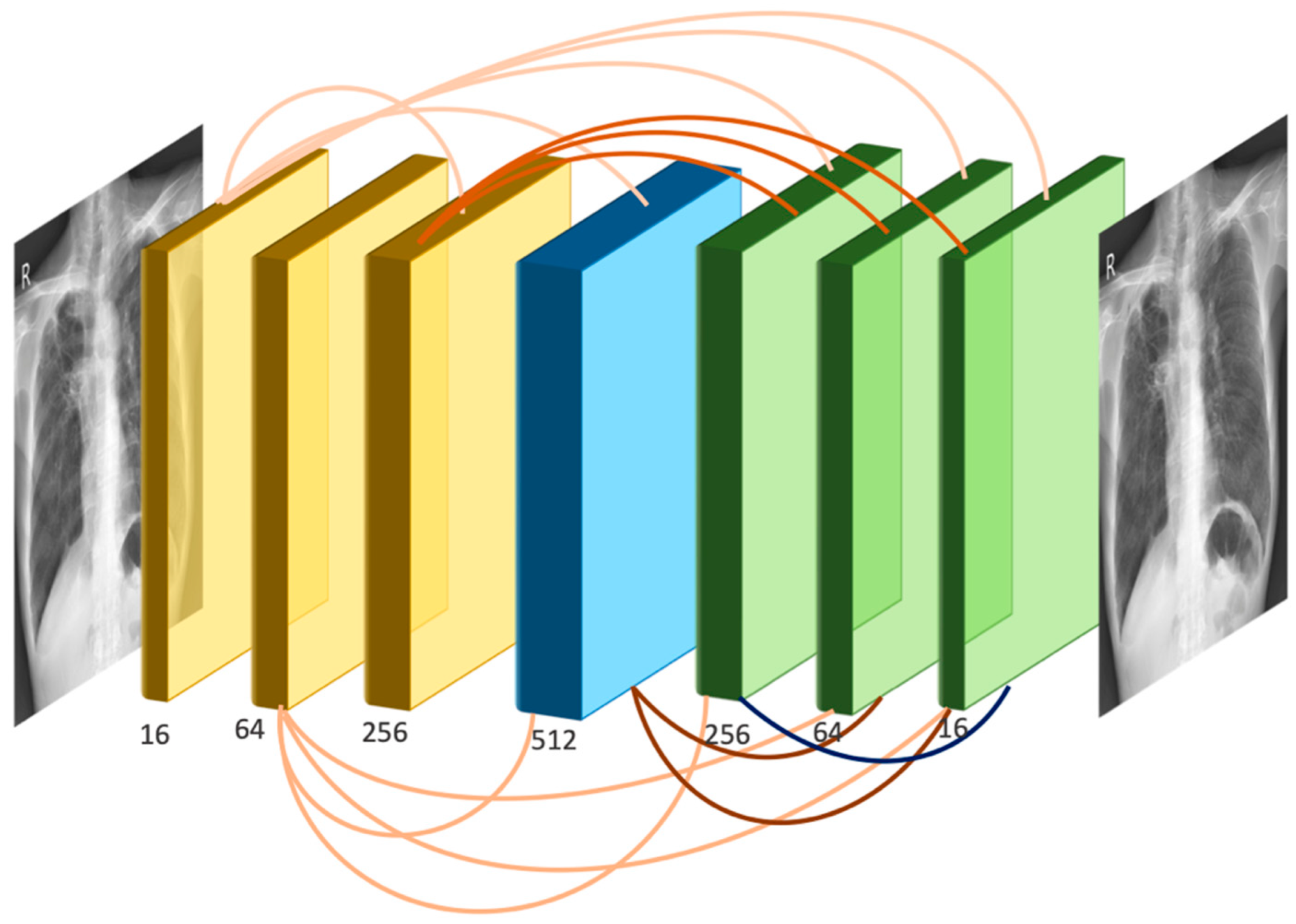
2.3. SADXNet
2.4. Downstream Clinical Task Validation
3. Results
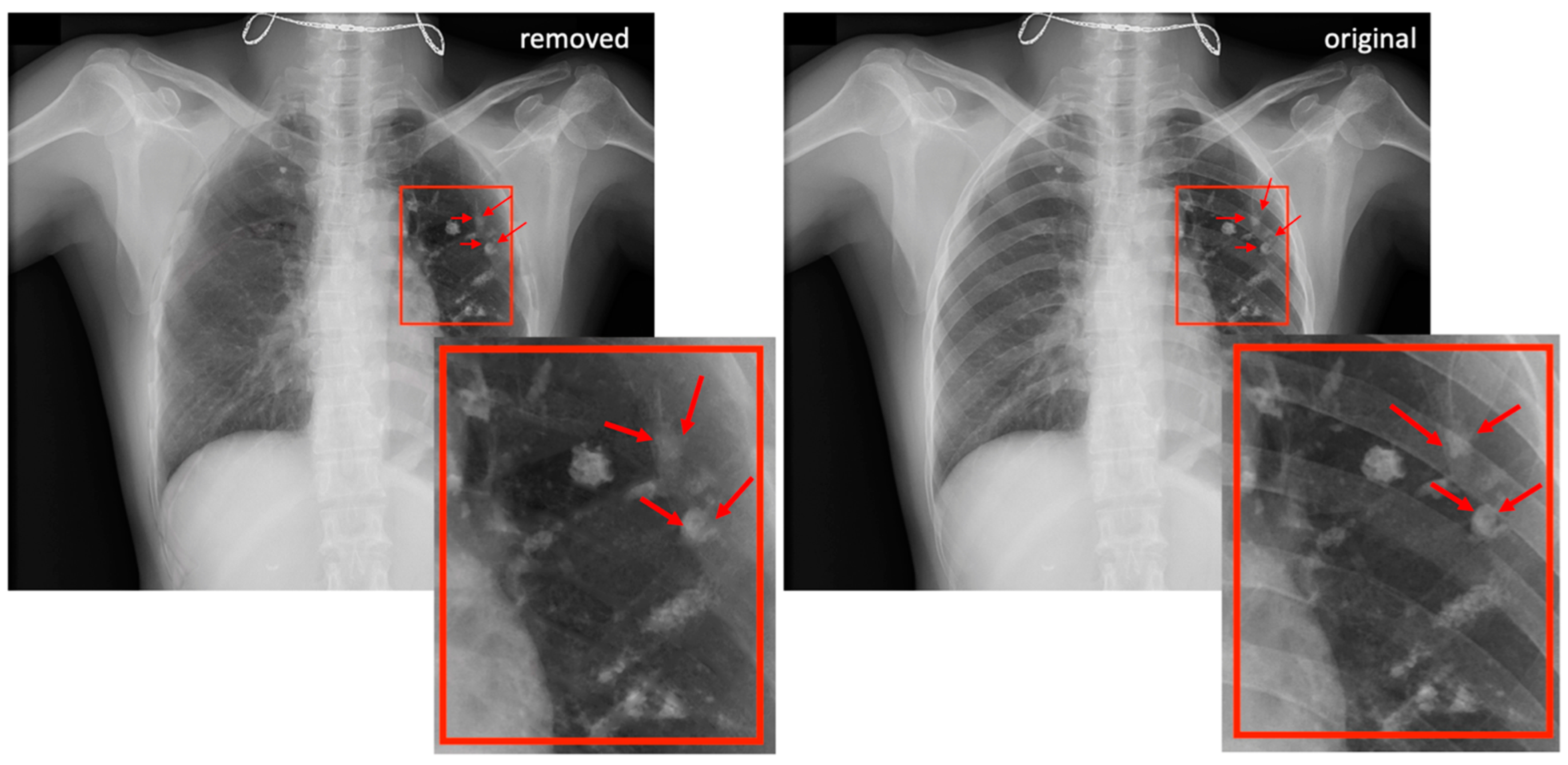
3.1. Rib Suppression
3.1.1. ST Smoothing
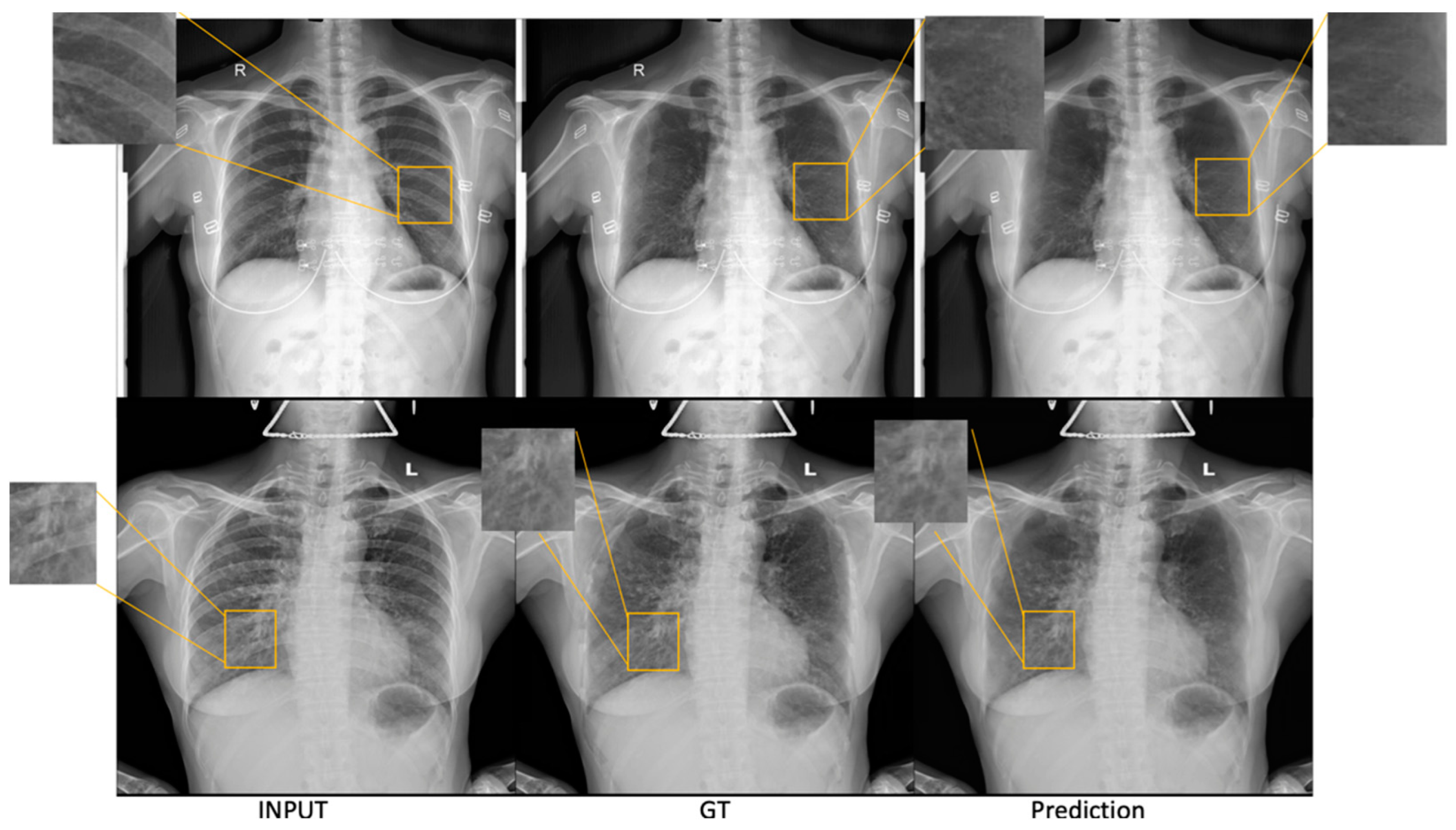
3.1.2. SADXNet
3.2. Downstream Clinical Task Validation
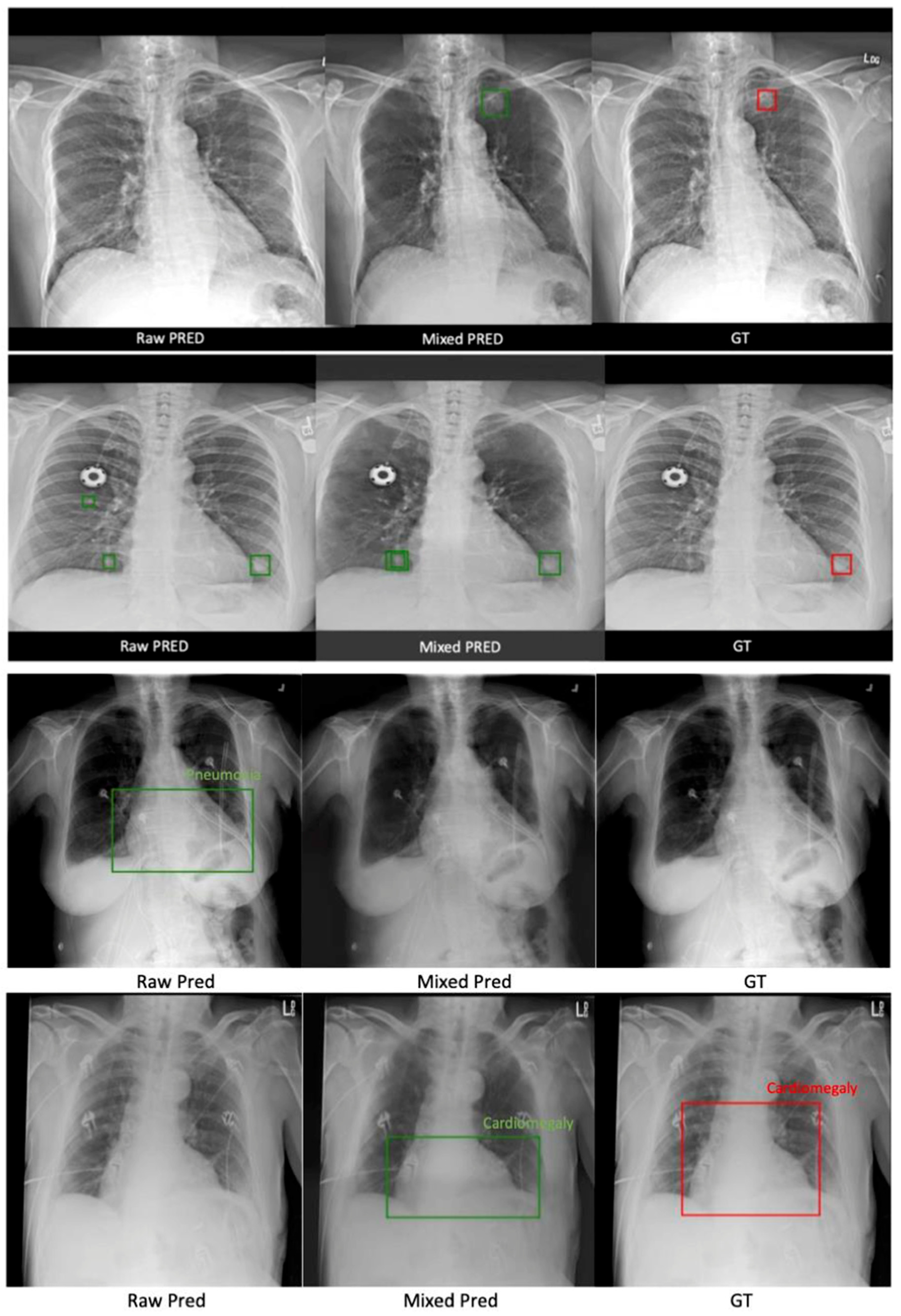
3.2.1. NOD21 Detection
3.2.2. ChestX-ray14 Classification and Localization
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- The Lancet. GBD 2017: A fragile world. Lancet 2018, 392, 1683. [Google Scholar] [CrossRef] [PubMed]
- Soriano, J.B.; Kendrick, P.J.; Paulson, K.R.; Gupta, V.; Abrams, E.M.; Adedoyin, R.A.; Adhikari, T.B.; Advani, S.M.; Agrawal, A.; Ahmadian, E.; et al. Prevalence and attributable health burden of chronic respiratory diseases, 1990–2017: A systematic analysis for the Global Burden of Disease Study 2017. Lancet Respir. Med. 2020, 8, 585–596. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Suzuki, K. Separation of bones from chest radiographs by means of anatomically specific multiple massive-training ANNs combined with total variation minimization smoothing. IEEE Trans. Med. Imaging 2014, 33, 246–257. [Google Scholar] [CrossRef]
- Hogeweg, L.; Sanchez, C.I.; van Ginneken, B. Suppression of Translucent Elongated Structures: Applications in Chest Radiography. IEEE Trans. Med. Imaging 2013, 32, 2099–2113. [Google Scholar] [CrossRef]
- Laskey, M.A. Dual-energy X-ray absorptiometry and body composition. Nutrition 1996, 12, 45–51. [Google Scholar] [CrossRef]
- Kelcz, F.; Zink, F.E.; Peppler, W.W.; Kruger, D.G.; Ergun, D.L.; Mistretta, C.A. Conventional chest radiography vs dual-energy computed radiography in the detection and characterization of pulmonary nodules. Am. J. Roentgenol. 1994, 162, 271–278. [Google Scholar] [CrossRef]
- Von Berg, J.; Young, S.; Carolus, H.; Wolz, R.; Saalbach, A.; Hidalgo, A.; Gimenez, A.; Franquet, T. A novel bone suppression method that improves lung nodule detection: Suppressing dedicated bone shadows in radiographs while preserving the remaining signal. Int. J. CARS 2016, 11, 641–655. [Google Scholar] [CrossRef] [PubMed]
- Rajaraman, S.; Zamzmi, G.; Folio, L.; Alderson, P.; Antani, S. Chest X-ray Bone Suppression for Improving Classification of Tuberculosis-Consistent Findings. Diagnostics 2021, 11, 840. [Google Scholar] [CrossRef]
- Han, L.; Lyu, Y.; Peng, C.; Zhou, S.K. GAN-based disentanglement learning for chest X-ray rib suppression. Med. Image Anal. 2022, 77, 102369. [Google Scholar] [CrossRef]
- Wu, D.; Liu, D.; Puskás, Z.; Lu, C.; Wimmer, A.; Tietjen, C.; Soza, G.; Zhou, S.K. A learning based deformable template matching method for automatic rib centerline extraction and labeling in CT images. In Proceedings of the 2012 IEEE Conference on Computer Vision and Pattern Recognition, Providence, RI, USA, 16–21 June 2012; pp. 980–987. [Google Scholar] [CrossRef]
- Hogeweg, L.; Sánchez, C.I.; de Jong, P.A.; Maduskar, P.; van Ginneken, B. Clavicle segmentation in chest radiographs. Med. Image Anal. 2012, 16, 1490–1502. [Google Scholar] [CrossRef]
- Simkó, G.; Orbán, G.; Máday, P.; Horváth, G. Elimination of clavicle shadows to help automatic lung nodule detection on chest radiographs. In Proceedings of the 4th European Conference of the International Federation for Medical and Biological Engineering, Antwerpen, Belgium, 23–27 November 2008; Sloten, J.V., Verdonck, P., Nyssen, M., Haueisen, J., Eds.; Springer: Berlin/Heidelberg, Germany, 2009; Volume 22, pp. 488–491. [Google Scholar] [CrossRef]
- Dargan, S.; Kumar, M.; Ayyagari, M.R.; Kumar, G. A Survey of Deep Learning and Its Applications: A New Paradigm to Machine Learning. Arch. Computat. Methods Eng. 2020, 27, 1071–1092. [Google Scholar] [CrossRef]
- Kido, S.; Kuriyama, K.; Kuroda, C.; Nakamura, H.; Ito, W.; Shimura, K.; Kato, H. Detection of simulated pulmonary nodules by single-exposure dual-energy computed radiography of the chest: Effect of a computer-aided diagnosis system (Part 2). Eur. J. Radiol. 2002, 44, 205–209. [Google Scholar] [CrossRef]
- Kido, S.; Nakamura, H.; Ito, W.; Shimura, K.; Kato, H. Computerized detection of pulmonary nodules by single-exposure dual-energy computed radiography of the chest (part 1). Eur. J. Radiol. 2002, 44, 198–204. [Google Scholar] [CrossRef]
- Suzuki, K.; Abe, H.; MacMahon, H.; Doi, K. Image-processing technique for suppressing ribs in chest radiographs by means of massive training artificial neural network (MTANN). IEEE Trans. Med. Imaging 2006, 25, 406–416. [Google Scholar] [CrossRef] [PubMed]
- Manji, F.; Wang, J.; Norman, G.; Wang, Z.; Koff, D. Comparison of dual energy subtraction chest radiography and traditional chest X-rays in the detection of pulmonary nodules. Quant. Imaging Med. Surg. 2016, 6, 1–5. [Google Scholar] [CrossRef]
- Li, Z.; Li, H.; Han, H.; Shi, G.; Wang, J.; Zhou, S.K. Encoding CT Anatomy Knowledge for Unpaired Chest X-ray Image Decomposition. In Lecture Notes in Computer Science, Proceedings of the Medical Image Computing and Computer Assisted Intervention—MICCAI 2019, Shenzhen, China, 13–17 October 2019; Shen, D., Liu, T., Peters, T.M., Staib, L.H., Essert, C., Zhou, S., Yap, P.-T., Khan, A., Eds.; Springer International Publishing: Berlin/Heidelberg, Germany, 2019; Volume 11769, pp. 275–283. [Google Scholar] [CrossRef]
- Nguyen, H.C.; Le, T.T.; Pham, H.H.; Nguyen, H.Q. VinDr-RibCXR: A Benchmark Dataset for Automatic Segmentation and Labeling of Individual Ribs on Chest X-rays. arXiv 2021. [Google Scholar] [CrossRef]
- Sogancioglu, E.; Murphy, K.; Van Ginneken, B. NODE21. Zenodo. 2021. Available online: https://zenodo.org/record/5548363#.ZFV7lOzML0o (accessed on 9 April 2023).
- Wang, X.; Peng, Y.; Lu, L.; Lu, Z.; Bagheri, M.; Summers, R.M. ChestX-Ray8: Hospital-Scale Chest X-Ray Database and Benchmarks on Weakly-Supervised Classification and Localization of Common Thorax Diseases. In Proceedings of the 2017 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Honolulu, HI, USA, 21–26 July 2017; pp. 3462–3471. [Google Scholar] [CrossRef]
- He, K.; Gkioxari, G.; Dollár, P.; Girshick, R. Mask R-CNN. arXiv 2018. [Google Scholar] [CrossRef]
- Huang, G.; Liu, Z.; van der Maaten, L.; Weinberger, K.Q. Densely Connected Convolutional Networks. arXiv 2016. [Google Scholar] [CrossRef]
- Long, J.; Shelhamer, E.; Darrell, T. Fully Convolutional Networks for Semantic Segmentation. arXiv 2014. [Google Scholar] [CrossRef]
- Zhao, H.; Gallo, O.; Frosio, I.; Kautz, J. Loss Functions for Image Restoration with Neural Networks. IEEE Trans. Comput. Imaging 2017, 3, 47–57. [Google Scholar] [CrossRef]
- Betancur, J.; Commandeur, F.; Motlagh, M.; Sharir, T.; Einstein, A.J.; Bokhari, S.; Fish, M.B.; Ruddy, T.D.; Kaufmann, P.; Sinusas, A.J.; et al. Deep Learning for Prediction of Obstructive Disease from Fast Myocardial Perfusion SPECT. JACC Cardiovasc. Imaging 2018, 11, 1654–1663. [Google Scholar] [CrossRef] [PubMed]

| Modality | Training Input | Validation Input | AUC | FN | FP | TP |
|---|---|---|---|---|---|---|
| Mask R-CNN | Raw | Raw | 94.76% | 48 | 1273 | 372 |
| Rib suppressed | Rib suppressed | 95.32% | 45 | 1193 | 375 | |
| Raw + Rib suppressed | Raw | 97.99% | 32 | 1070 | 388 | |
| Rib suppressed | 97.31% | 33 | 1082 | 387 |
| Modality | Training Input | Validation Input | AUC | FN | FP | TP |
|---|---|---|---|---|---|---|
| Mask R-CNN | Raw | Raw | 80.54% | 137 | 3029 | 245 |
| Rib suppressed | Rib suppressed | 81.55% | 138 | 2909 | 244 | |
| Rib suppressed | Raw | 87.16% | 116 | 2644 | 275 | |
| Rib suppressed | 86.89% | 124 | 2701 | 267 |
| Input: Tr/Val | Raw/Raw | ST/ST | Raw + ST/Raw | Raw + ST/ST | |
|---|---|---|---|---|---|
| Disease | |||||
| Atelectasis | 79.21% | 77.35% | 85.27% | 84.89% | |
| Cardiomegaly | 83.47% | 82.03% | 93.84% | 92.67% | |
| Effusion | 84.98% | 84.72% | 91.02% | 91.02% | |
| Infiltration | 69.84% | 70.00% | 75.21% | 74.32% | |
| Mass | 77.23% | 77.38% | 87.83% | 85.79% | |
| Nodule | 76.32% | 75.37% | 82.33% | 81.29% | |
| Pneumonia | 73.89% | 73.24% | 79.43% | 78.33% | |
| Pneumothorax | 82.43% | 82.32% | 91.37% | 90.62% | |
| Consolidation | 74.57% | 75.23% | 82.77% | 81.98% | |
| Edema | 88.14% | 87.28% | 95.32% | 95.33% | |
| Emphysema | 90.68% | 89.47% | 96.21% | 95.79% | |
| Fibrosis | 82.99% | 83.21% | 87.45% | 86.76% | |
| Pleural Thickening | 73.37% | 72.45% | 80.34% | 79.85% | |
| Hernia | 88.03% | 89.21% | 97.24% | 96.79% | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xu, D.; Xu, Q.; Nhieu, K.; Ruan, D.; Sheng, K. An Efficient and Robust Method for Chest X-ray Rib Suppression That Improves Pulmonary Abnormality Diagnosis. Diagnostics 2023, 13, 1652. https://doi.org/10.3390/diagnostics13091652
Xu D, Xu Q, Nhieu K, Ruan D, Sheng K. An Efficient and Robust Method for Chest X-ray Rib Suppression That Improves Pulmonary Abnormality Diagnosis. Diagnostics. 2023; 13(9):1652. https://doi.org/10.3390/diagnostics13091652
Chicago/Turabian StyleXu, Di, Qifan Xu, Kevin Nhieu, Dan Ruan, and Ke Sheng. 2023. "An Efficient and Robust Method for Chest X-ray Rib Suppression That Improves Pulmonary Abnormality Diagnosis" Diagnostics 13, no. 9: 1652. https://doi.org/10.3390/diagnostics13091652
APA StyleXu, D., Xu, Q., Nhieu, K., Ruan, D., & Sheng, K. (2023). An Efficient and Robust Method for Chest X-ray Rib Suppression That Improves Pulmonary Abnormality Diagnosis. Diagnostics, 13(9), 1652. https://doi.org/10.3390/diagnostics13091652






