Mathematical Modelling of Cervical Precancerous Lesion Grade Risk Scores: Linear Regression Analysis of Cellular Protein Biomarkers and Human Papillomavirus E6/E7 RNA Staining Patterns
Abstract
1. Introduction
2. Materials and Methods
2.1. Specimens
2.2. Tissue Microarray (TMA) Preparation
2.3. HR-HPV E6/E7 RNA Chromogenic ISH
2.4. IHC Staining
2.5. Mathematical Models (MM) and Risk Score Development
2.6. Statistical Analysis
3. Results
3.1. Baseline Characteristics
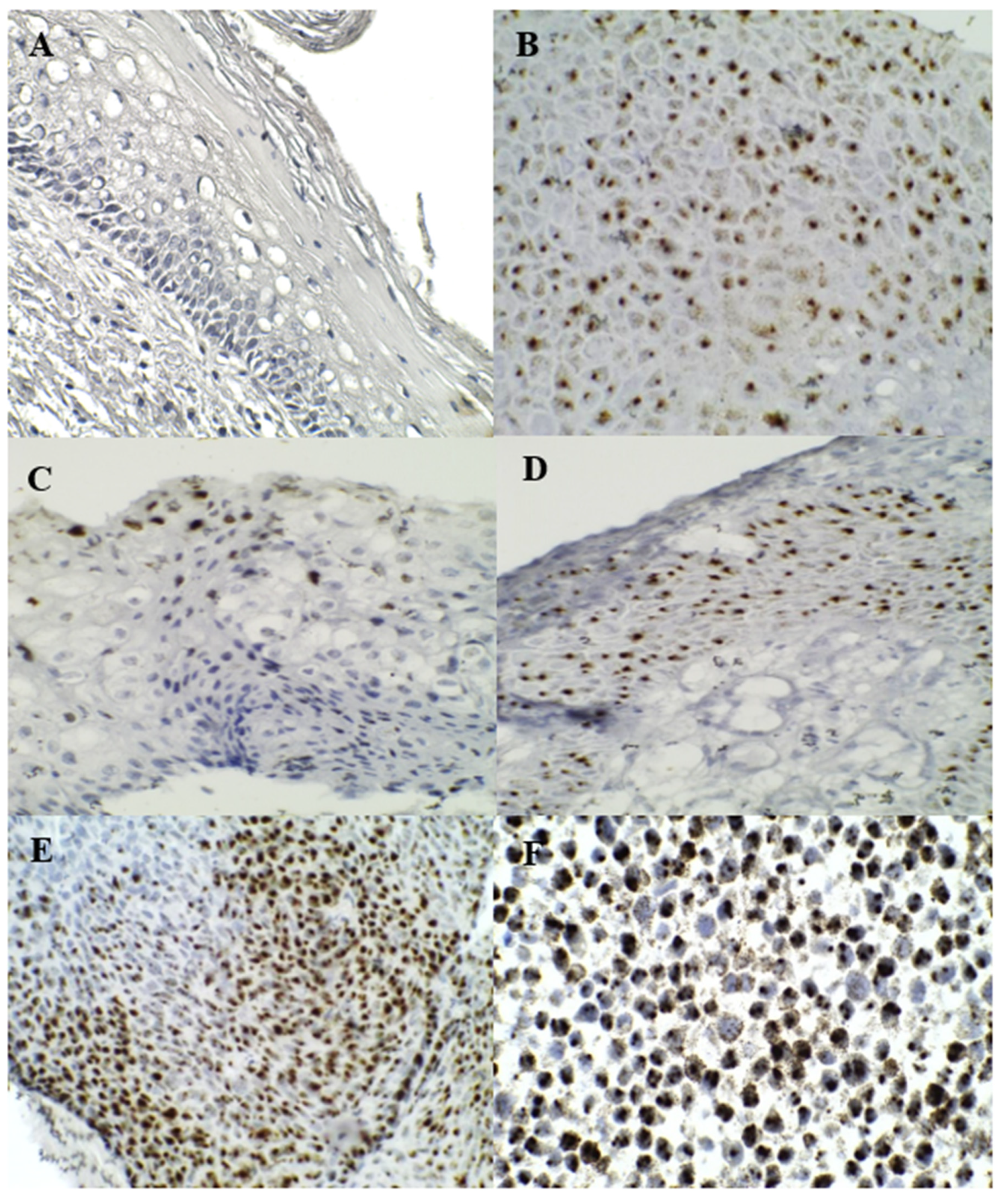
3.2. HR–HPV E6/E7 RNA Chromogenic ISH
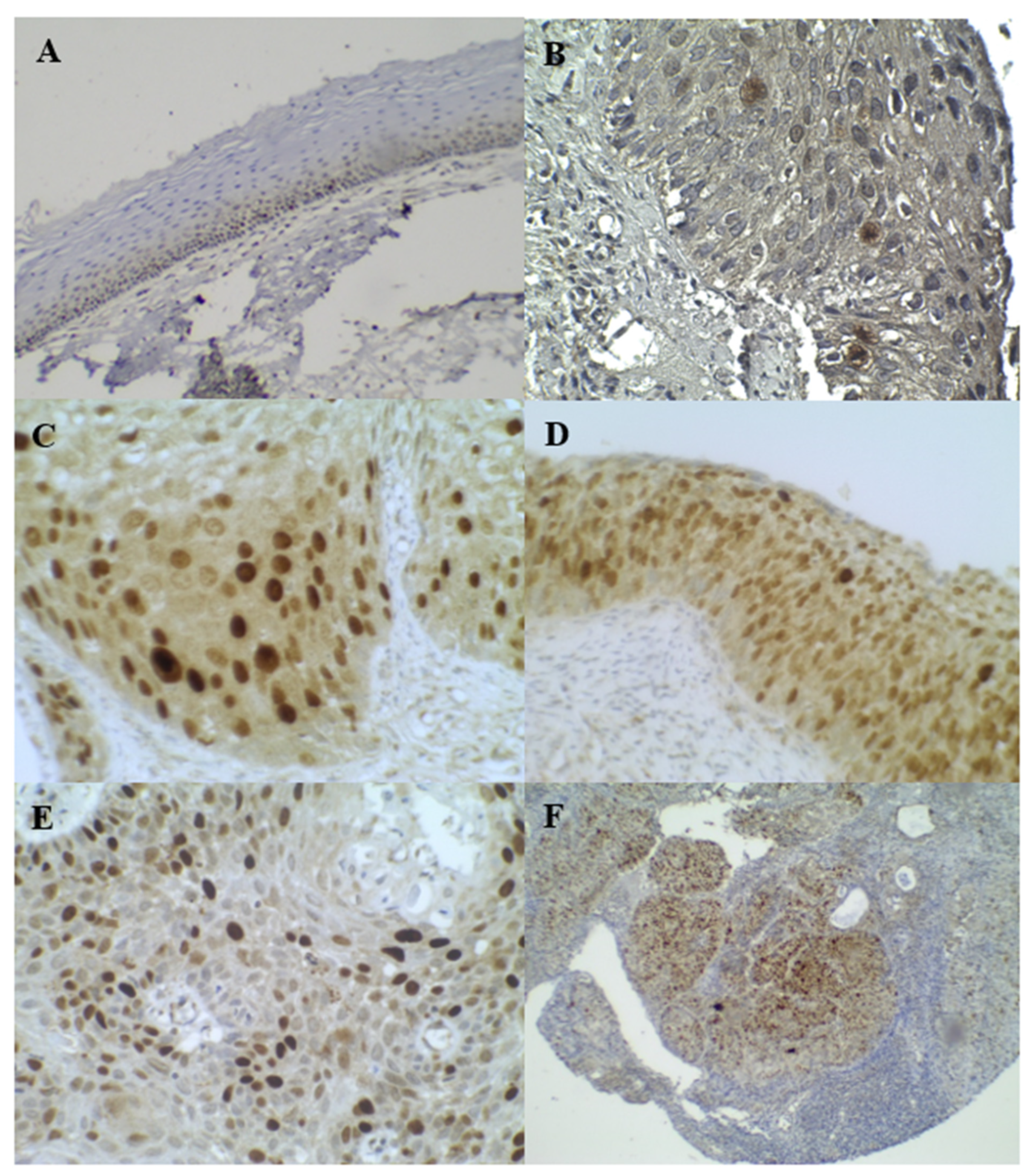
3.3. p16INK4A and Ki–67 Immunostaining
3.4. Cortactin Immunostaining
3.5. Mathematical Models (MM)
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Rotondo, J.C.; Bosi, S.; Bassi, C.; Ferracin, M.; Lanza, G.; Gafà, R.; Martini, F. Gene expression changes in pro-gression of cervical neoplasia revealed by microarray analysis of cervical neoplastic keratinocytes. J. Cell Physiol. 2015, 230, 806–812. [Google Scholar] [CrossRef]
- Mello, V.; Sundstrom, R.K. Cervical Intraepithelial Neoplasia. In StatPearls; StatPearls Publishing: Treasure Island, FL, USA, 2022. [Google Scholar]
- Øvestad, I.T.; Engesæter, B.; Halle, M.K.; Akbari, S.; Bicskei, B.; Lapin, M. High-Grade Cervical Intraepi-thelial Neoplasia (CIN) Associates with Increased Proliferation and Attenuated Immune Signaling. Int. J. Mol. Sci. 2021, 23, 373. [Google Scholar] [CrossRef]
- World Health Organization. WHO Guidelines for Treatment of Cervical Intraepithelial Neoplasia 2–3 and Adenocarcinoma In Situ: Cryotherapy, Large Loop Excision of the Transformation Zone, and Cold Knife Conization; World Health Organization: Geneva, Switzerland, 2014. [Google Scholar]
- Lyon, F. IARC Monographs on the Evaluation of Carcinogenic Risks to Humans: Human Papillomaviruses; IARC Working Group: Lyon, France, 2007. [Google Scholar]
- Schiffman, M. Integration of human papillomavirus vaccination, cytology, and human papillomavirus testing. Cancer 2007, 111, 145–153. [Google Scholar] [CrossRef]
- Schiffman, M.; Castle, P.E.; Jeronimo, J.; Rodriguez, A.C.; Wacholder, S. Human papillomavirus and cervical cancer. Lancet 2007, 370, 890–907. [Google Scholar] [CrossRef]
- Baseman, J.G.; Koutsky, L.A. The epidemiology of human papillomavirus infections. J. Clin. Virol. 2005, 32 (Suppl. S1), S16–S24. [Google Scholar] [CrossRef]
- McCredie, M.R.; Sharples, K.J.; Paul, C.; Baranyai, J.; Medley, G.; Jones, R.W.; Skegg, D.C. Natural history of cervical neoplasia and risk of invasive cancer in women with cervical intraepithelial neoplasia 3: A retrospective cohort study. Lancet Oncol. 2008, 9, 425–434. [Google Scholar] [CrossRef]
- Khan, M.J.; Castle, P.E.; Lorincz, A.T.; Wacholder, S.; Sherman, M.; Scott, D.R.; Schiffman, M. The elevated 10-year risk of cervical precancer and cancer in women with human papillomavirus (HPV) type 16 or 18 and the possible utility of type-specific HPV testing in clinical practice. J. Natl. Cancer Inst. 2005, 97, 1072–1079. [Google Scholar] [CrossRef] [PubMed]
- Duensing, S.; Munger, K. The human papillomavirus type 16 E6 and E7 oncoproteins independently induce numerical and structural chromosome instability. Cancer Res. 2002, 62, 7075–7082. [Google Scholar] [PubMed]
- Yu, L.L.; Chen, W.; Lei, X.Q.; Qin, Y.; Wu, Z.N.; Pan, Q.J.; Qiao, Y.L. Evaluation of p16/Ki-67 dual staining in detection of cervical precancer and cancers: A multicenter study in China. Oncotarget 2016, 7, 21181–21189. [Google Scholar] [CrossRef] [PubMed]
- Cuzick, J.; Arbyn, M.; Sankaranarayanan, R.; Tsu, V.; Ronco, G.; Mayrand, M.H.; Meijer, C.J. Overview of human papillomavirus-based and other novel options for cervical cancer screening in developed and developing countries. Vaccine 2008, 26 (Suppl. S10), K29–K41. [Google Scholar] [CrossRef]
- Gravitt, P.E.; Coutlée, F.; Iftner, T.; Sellors, J.W.; Quint, W.G.; Wheeler, C.M. New technologies in cervical cancer screening. Vaccine 2008, 26 (Suppl. S10), K42–K52. [Google Scholar] [CrossRef]
- Wentzensen, N.; von Knebel Doeberitz, M. Biomarkers in cervical cancer screening. Dis. Markers 2007, 23, 315–330. [Google Scholar] [CrossRef]
- Ghittoni, R.; Accardi, R.; Hasan, U.; Gheit, T.; Sylla, B.; Tommasino, M. The biological properties of E6 and E7 oncoproteins from human papillomaviruses. Virus Genes 2010, 40, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Sokolova, I.; Algeciras-Schimnich, A.; Song, M.; Sitailo, S.; Policht, F.; Kipp, B.R.; Morrison, L. Chromosomal biomarkers for detection of human papillomavirus associated genomic instability in epithelial cells of cervical cytology specimens. J. Mol. Diagn. 2007, 9, 604–611. [Google Scholar] [CrossRef] [PubMed]
- Johansson, H.; Bjelkenkrantz, K.; Darlin, L.; Dilllner, J.; Forslund, O. Presence of High-Risk HPV mRNA in Relation to Future High-Grade Lesions among High-Risk HPV DNA Positive Women with Minor Cytological Abnormalities. PLoS ONE 2015, 10, e0124460. [Google Scholar] [CrossRef]
- Rotondo, J.C.; Oton-Gonzalez, L.; Mazziotta, C.; Lanzillotti, C.; Iaquinta, M.R.; Tognon, M. Simultaneous Detection and Viral DNA Load Quantification of Different Human Papillomavirus Types in Clinical Specimens by the High Analytical Droplet Digital PCR Method. Front. Microbiol. 2020, 11, 591452. [Google Scholar] [CrossRef]
- Carow, K.; Read, C.; Häfner, N.; Runnebaum, I.B.; Corner, A.; Dürst, M. A comparative study of digital PCR and real-time qPCR for the detection and quantification of HPV mRNA in sentinel lymph nodes of cervical cancer patients. BMC Res. Notes 2017, 10, 532. [Google Scholar] [CrossRef] [PubMed]
- Duvlis, S.; Jankovic, K.P.; Arsova, Z.S.; Memeti, S.; Popeska, Z.; Plaseska-Karanfilska, D. HPV E6/E7 mRNA Versus HPV DNA Biomarker in Cervical Cancer Screening of a Group of Macedonian Women. J. Med. Virol. 2015, 87, 1578–1586. [Google Scholar] [CrossRef]
- Bishop, J.A.; Ma, X.J.; Wang, H.; Luo, Y.; Illei, P.B.; Begum, S.; Westra, W.H. Detection of transcriptionally active high-risk HPV in patients with head and neck squamous cell carcinoma as visualized by a novel E6/E7 mRNA in situ hybridization method. Am. J. Surg. Pathol. 2012, 36, 1874–1882. [Google Scholar] [CrossRef]
- Brown, R.E.; Naqvi, S.; McGuire, M.F.; Buryanek, J.; Karni, R.J. Morphoproteomics, E6/E7 in-situ hybridization, and biomedical analytics define the etiopathogenesis of HPV-associated oropharyngeal carcinoma and provide targeted therapeutic options. J. Otolaryngol. Head Neck Surg. 2017, 46, 52. [Google Scholar] [CrossRef]
- Koliopoulos, G.; Arbyn, M.; Martin-Hirsch, P.; Kyrgiou, M.; Prendiville, W.; Paraskevaidis, E. Diagnostic accuracy of human papillomavirus testing in primary cervical screening: A systematic review and meta-analysis of non-randomized studies. Gynecol. Oncol. 2007, 104, 232–246. [Google Scholar] [CrossRef]
- Sarwath, H.; Bansal, D.; Husain, N.E.; Mohamed, M.; Sultan, A.A.; Bedri, S. Introduction of p16INK4a as a surrogate biomarker for HPV in women with invasive cervical cancer in Sudan. Infect. Agent Cancer 2017, 12, 50. [Google Scholar] [CrossRef]
- Sarma, U.; Biswas, I.; Das, A.; Das, G.C.; Saikia, C.; Sarma, B. p16INK4a Expression in Cervical Lesions Correlates with Histologic Grading—A Tertiary Level Medical Facility Based Retrospective Study. Asian Pac. J. Cancer Prev. 2017, 18, 2643–2647. [Google Scholar]
- Singh, C.; Manivel, J.C.; Truskinovsky, A.M.; Savik, K.; Amirouche, S.; Holler, J.; Pambuccian, S.E. Variability of Pathologists’ Utilization of p16 and Ki-67 Immunostaining in the Diagnosis of Cervical Biopsies in Routine Pathology Practice and Its Impact on the Frequencies of Cervical Intraepithelial Neoplasia Diagnoses and Cytohistologic Correlations. Arch. Pathol. Lab. Med. 2014, 138, 76–87. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, S.A.; Obaseki, D.E.; Mayun, A.A.; Mohammed, A.; Rafindadi, A.H.; Abdul, M.A. The Role of Biomarkers (p16INK4a and Ki-67) in Cervical Cancer Screening: An Appraisal. Ann. Trop. Pathol. 2017, 8, 1–4. [Google Scholar] [CrossRef]
- Cuschieri, K.; Wentzensen, N. Human papillomavirus mRNA and p16 detection as biomarkers for the improved diagnosis of cervical neoplasia. Cancer Epidemiol. Biomarkers Prev. 2008, 17, 2536–2545. [Google Scholar] [CrossRef]
- Carreon, J.D.; Sherman, M.E.; Guillen, D.; Solomon, D.; Herrero, R.; Jerónimo, J.; Schiffman, M. CIN 2 is a much less reproducible and less valid diagnosis than CIN 3: Results from a histological review of population-based cervical samples. Int. J. Gynecol. Pathol. 2007, 26, 441–446. [Google Scholar] [CrossRef] [PubMed]
- Jeronimo, J.; Schiffman, M. Colposcopy at a crossroads. Am. J. Obstet. Gynecol. 2006, 195, 349–353. [Google Scholar] [CrossRef] [PubMed]
- Zappacosta, R.; Colasante, A.; Viola, P.; D’Antuono, T.; Lattanzio, G.; Capanna, S.; Rosini, S. Chromogenic in situ hybridization and p16/Ki67 dual staining on formalin-fixed paraffin-embedded cervical specimens: Correlation with HPV-DNA test, E6/E7 mRNA test, and potential clinical applications. Biomed. Res. Int. 2013, 2013, 453606. [Google Scholar] [CrossRef]
- MacGrath, S.M.; Koleske, A.J. Cortactin in cell migration and cancer at a glance. J. Cell Sci. 2012, 125, 1621–1626. [Google Scholar] [CrossRef]
- Cosen-Binker, L.I.; Kapus, A. Cortactin. The Gray Eminence of the Cytoskeleton. Physiology 2006, 21, 352–361. [Google Scholar] [CrossRef] [PubMed]
- Weaver, A.M. Cortactin in tumor invasiveness. Cancer Lett. 2008, 265, 157–166. [Google Scholar] [CrossRef]
- Yin, M.; Ma, W.; An, L. Cortactin in cancer cell migration and invasion. Oncotarget 2017, 8, 88232–88243. [Google Scholar] [CrossRef]
- Buday, L.; Downward, J. Roles of cortactin in tumor pathogenesis. Biochim. Biophys. Acta 2007, 1775, 263–273. [Google Scholar] [CrossRef]
- Gibcus, J.H.; Mastik, M.F.; Menkema, L.; De Bock, G.H.; Kluin, P.M.; Schuuring, E.; Van Der Wal, J.E. Cortactin expression predicts poor survival in laryngeal carcinoma. Br. J. Cancer 2008, 98, 950–955. [Google Scholar] [CrossRef] [PubMed]
- Belfatto, A.; Riboldi, M.; Ciardo, D.; Cattani, F.; Cecconi, A.; Lazzari, R.; Cerveri, P. Kinetic Models for Predicting Cervical Cancer Response to Radiation Therapy on Individual Basis Using Tumor Regression Measured In Vivo With Volumetric Imaging. Technol. Cancer Res. Treat. 2016, 15, 146–158. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Liu, K.Y.; Wong, S.T. Cancer classification and prediction using logistic regression with Bayesian gene selection. J. Biomed. Inform. 2004, 37, 249–259. [Google Scholar] [CrossRef]
- Vickers, A.J. Prediction models in cancer care. CA Cancer J. Clin. 2011, 61, 315–326. [Google Scholar] [CrossRef]
- Jiang, L.; Yin, K.; Wen, Q.; Chen, C.; Ge, M.H.; Tan, Z. Predictive Risk-scoring Model For Central Lymph Node Metastasis and Predictors of Recurrence in Papillary Thyroid Carcinoma. Sci. Rep. 2020, 10, 710. [Google Scholar] [CrossRef]
- Enderling, H.; Chaplain, M.A.; Anderson, A.R.; Vaidya, J.S. A mathematical model of breast cancer development, local treatment and recurrence. J. Theor. Biol. 2007, 246, 245–259. [Google Scholar] [CrossRef]
- Murphy, H.; Jaafari, H.; Dobrovolny, H.M. Differences in predictions of ODE models of tumor growth: A cautionary example. BMC Cancer 2016, 16, 163. [Google Scholar] [CrossRef]
- Lin, M.; Ye, M.; Zhou, J.; Wang, Z.P.; Zhu, X. Recent Advances on the Molecular Mechanism of Cervical Carcinogenesis Based on Systems Biology Technologies. Comput. Struct. Biotechnol. J. 2019, 17, 241–250. [Google Scholar] [CrossRef]
- Charan, J.; Biswas, T. How to calculate sample size for different study designs in medical research? Indian J. Psychol. Med. 2013, 35, 121–126. [Google Scholar] [CrossRef]
- Bumrungthai, S.; Munjal, K.; Nandekar, S.; Cooper, K.; Ekalaksananan, T.; Pientong, C.; Evans, M.F. Epidermal growth factor receptor pathway mutation and expression profiles in cervical squamous cell carcinoma: Therapeutic implications. J. Transl. Med. 2015, 13, 244. [Google Scholar] [CrossRef]
- Devasena, K.; Shana, J. Building Machine Learning Model for Predicting Breast Cancer Using different Regression Techniques. In IOP Conference Series: Materials Science and Engineering; IOP Publishing: Bristol, UK, 2021; Volume 1166, p. 12029. [Google Scholar] [CrossRef]
- Xu, J.; Xue, D. Cell Carcinosis Prediction using Linear Regression with Nuclear Statistics. In Proceedings of the 2022 International Conference on Big Data, Information and Computer Network (BDICN), Sanya, China, 20–22 January 2022; pp. 276–282. [Google Scholar] [CrossRef]
- Murugan, S.; Kumar, B.M.; Amudha, S. Classification and Prediction of Breast Cancer using Linear Regression, Decision Tree and Random Forest. In Proceedings of the 2017 International Conference on Current Trends in Computer, Electrical, Electronics and Communication (CTCEEC), Mysore, India, 8–9 September 2017; pp. 763–766. [Google Scholar] [CrossRef]
- Shi, Q.; Xu, L.; Yang, R.; Meng, Y.; Qiu, L. Ki-67 and P16 proteins in cervical cancer and precancerous lesions of young women and the diagnostic value for cervical cancer and precancerous lesions. Oncol. Lett. 2019, 18, 1351–1355. [Google Scholar] [CrossRef]
- Murphy, N.; Ring, M.; Killalea, A.G.; Uhlmann, V.; O’Donovan, M.; Mulcahy, F.; O’Leary, J.J. p16INK4A as a marker for cervical dyskaryosis: CIN and cGIN in cervical biopsies and ThinPrep smears. J. Clin. Pathol. 2003, 56, 56–63. [Google Scholar] [CrossRef]
- Leite, P.M.B.O.; Tafuri, L.; Costa, M.Z.B.O.; Lima, M.I.B.M.; Simões, R.T. Evaluation of the p16 and Ki-67 Biomarkers as Predictors of the Recurrence of Premalignant Cervical Cancer Lesions after LEEP Conization. Rev. Bras. Ginecol. Obstet. 2017, 39, 288–293. [Google Scholar] [CrossRef]
- Chuma, M.; Sakamoto, M.; Yasuda, J.; Fujii, G.; Nakanishi, K.; Tsuchiya, A.; Hirohashi, S. Overexpression of cortactin is involved in motility and metastasis of hepatocellular carcinoma. J. Hepatol. 2004, 41, 629–636. [Google Scholar] [CrossRef] [PubMed]
- Bissinger, O.; Kolk, A.; Drecoll, E.; Straub, M.; Lutz, C.; Wolff, K.D.; Götz, C. EGFR and Cortactin: Markers for potential double target therapy in oral squamous cell carcinoma. Exp. Ther. Med. 2017, 14, 4620–4626. [Google Scholar] [PubMed]
- Timpson, P.; Wilson, A.S.; Lehrbach, G.M.; Sutherland, R.L.; Musgrove, E.A.; Daly, R.J. Aberrant Expression of Cortactin in Head and Neck Squamous Cell Carcinoma Cells Is Associated with Enhanced Cell Proliferation and Resistance to the Epidermal Growth Factor Receptor Inhibitor Gefitinib. Cancer Res. 2007, 67, 9304–9314. [Google Scholar] [CrossRef] [PubMed]
- Miyamoto, S.; Hasegawa, J.; Morioka, M.; Hirota, Y.; Kushima, M.; Sekizawa, A. The association between p16 and Ki-67 immunohistostaining and the progression of cervical intraepithelial neoplasia grade 2. Int. J. Gynaecol. Obstet. 2016, 134, 45–48. [Google Scholar] [CrossRef] [PubMed]
- Lin, J.; Albers, A.E.; Qin, J.; Kaufmann, A.M. Prognostic Significance of Overexpressed p16INK4a in Patients with Cervical Cancer: A Meta-Analysis. PLoS ONE 2014, 9, e106384. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.Z.; Garcia, M.V.; Li, T.Y.; Khor, L.Y.; Gajapathy, R.S.; Spittle, C.; Wu, H. Cytoskeleton alterations in melanoma: Aberrant expression of cortactin, an actin-binding adapter protein, correlates with melanocytic tumor progression. Mod. Pathol. 2010, 23, 187–196. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Minaguchi, T.; Lachkar, B.; Zhang, S.; Xu, C.; Tenjimbayashi, Y.; Satoh, T. Separate analysis of human papillomavirus E6 and E7 messenger RNAs to predict cervical neoplasia progression. PLoS ONE 2018, 13, e0193061. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Ren, C.; Yang, L.; Zhang, X.; Liu, L.; Wang, Z. Performance of p16/Ki67 immunostaining, HPV E6/E7 mRNA testing, and HPV DNA assay to detect high-grade cervical dysplasia in women with ASCUS. BMC Cancer 2019, 19, 271. [Google Scholar] [CrossRef] [PubMed]
- Kourou, K.; Exarchos, T.P.; Exarchos, K.P.; Karamouzis, M.V.; Fotiadis, D.I. Machine learning applications in cancer prognosis and prediction. Comput. Struct. Biotechnol. J. 2014, 13, 8–17. [Google Scholar] [CrossRef] [PubMed]
- Schlecht, N.F.; Platt, R.W.; Duarte-Franco, E.; Costa, M.C.; Sobrinho, J.P.; Prado, J.C.; Franco, E.L. Human papillomavirus infection and time to progression and regression of cervical intraepithelial neoplasia. J. Natl. Cancer Inst. 2003, 95, 1336–1343. [Google Scholar] [CrossRef] [PubMed]
- Bruno, M.T.; Scalia, G.; Cassaro, N.; Costanzo, M.; Boemi, S. Conservative management of CIN 2 p16 positive lesions in women with multiple HPV infection. BMC Infect. Dis. 2020, 20, 801. [Google Scholar] [CrossRef] [PubMed]
- Austin, R.M.; Onisko, A.; Zhao, C. Are CIN 3 risk or CIN 3+ risk measures reliable surrogates for invasive cervical cancer risk? J. Am. Soc. Cytopathol. 2020, 9, 602–606. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.; Li, T.; Han, Y.; Jiang, M.; Yu, Y.; Xu, H.; Chen, W. Development of models for cervical cancer screening: Construction in a cross-sectional population and validation in two screening cohorts in China. BMC Med. 2021, 19, 197. [Google Scholar] [CrossRef] [PubMed]
- Deo, R.C. Machine Learning in Medicine. Circulation 2015, 132, 1920–1930. [Google Scholar] [CrossRef] [PubMed]
- Erickson, B.J.; Korfiatis, P.; Akkus, Z.; Kline, T.L. Machine Learning for Medical Imaging. Radiographics 2017, 37, 505–515. [Google Scholar] [CrossRef] [PubMed]


| Group 1 (233 Cases) | Group 2 (130 Cases) | Total (363 Cases) | p-Value | ||
|---|---|---|---|---|---|
| Age (years) | Mean | 46.14 | 45.89 | 46.05 | 0.835 * |
| SD | 10.81 | 11.21 | 10.93 | ||
| Age groups (years) | 19–30 | 11 (4.72%) | 10 (7.69%) | 21(5.78%) | |
| 31–40 | 56 (24.03%) | 28 (21.53%) | 84 (23.14%) | ||
| 41–50 | 94 (40.34%) | 43 (33.07%) | 137 (37.74%) | ||
| 51–60 | 54 (23.17%) | 38 (29.23%) | 92 (25.34%) | ||
| >60 | 19 (8.15%) | 11(8.46%) | 29 (7.98%) | ||
| Pathological grades | Normal | 156 (66.95%) | 55 (42.30%) | 211 (58.12%) | |
| LSIL | 34 (14.59%) | 31 (23.84%) | 65 (17.90%) | ||
| HSIL | 24 (10.30%) | 34 (26.15%) | 58 (15.97%) | ||
| SCC | 19 (8.15%) | 10 (7.69%) | 29 (7.98%) |
| Biomarkers | Pattern of Expression | Interpretation |
|---|---|---|
| p16INK4A | (1) Staining was assessed as strong positive (block positive) according to the amount of uniform strong positive staining in the cytoplasm and nucleus in ~1/3 to 3/3 thickness, signal strength (which would appear as a dark brown color), and diffusion (the signal involved >50% of the epithelium). | “Positive” |
| (2) Positive ambiguous results were further grouped into three patterns: (2.1) Strong/basal (strong, diffuse, continuous staining of the lower third of the epithelium without upward extension). (2.2) Weak/diffuse (weak, diffuse, discontinuous staining reaching at least two third of the epithelium). (2.3) Strong/focal (strong, focal, and discontinuous staining located at any level of the epithelium). | “Positive” | |
| (3) Negative results were defined as either the total absence of staining or weak, focal, and discontinuous staining. | “Negative” | |
| Ki–67 | Negative Ki–67 staining was defined as either the total absence of staining or weak basal staining. | “Negative” |
| Cortactin | (1) Negative, weak cytoplasmic and/or nuclear staining. (2) Weak focal staining in the cytoplasm or nucleus (heterogeneous). | “Negative” “Normal cortactin expression” |
| (3) Uniformly strong cytoplasmic staining, “positive cytoplasmic overexpression,” or strong focal staining in the cytoplasm or nucleus (heterogeneous). (4) Uniform strong cytoplasmic staining, focal nuclear staining, and “positive nuclear and cytoplasmic overexpression”. (5) Strong cytoplasmic membrane staining. | “Positive” |
| Biomarkers | N | Normal (211 Cases) | LSIL (65 Cases) | HSIL (58 Cases) | SCC (29 Cases) | Total (363 Cases) | p–Value | |
|---|---|---|---|---|---|---|---|---|
| p16INK4A | P/N | P | 9 (4.2%) | 7 (10.8%) | 40 (69.0%) | 28 (96.6%) | 103 (28.4%) | 0.000 * |
| N | 202 (95.2%) | 58 (89.2%) | 18 (31.0%) | 1 (3.4%) | 260 (71.6%) | |||
| AS | Mean | 0.35 | 1.46 | 4.62 | 6.24 | 1.70 | 0.000 ** | |
| SD | 1.15 | 2.25 | 2.98 | 1.38 | 2.70 | |||
| Ki-67 | P/N | P | 52 (24.6%) | 18 (27.7%) | 50 (86.2%) | 27 (93.1%) | 147 (40.5%) | 0.000 * |
| N | 159 (75.4%) | 47 (72.3%) | 8 (13.8%) | 2 (6.9%) | 216 (59.5%) | |||
| AS | Mean | 1.1226 | 1.2923 | 5.2241 | 5.9655 | 2.1923 | 0.000 ** | |
| SD | 1.84 | 2.10 | 2.19 | 1.90 | 2.68 | |||
| Cortactin | P/N | P | 84 (39.8%) | 50 (76.9%) | 46 (79.3%) | 24 (82.8%) | 204 (56.2%) | 0.000 * |
| N | 127 (60.2%) | 15 (23.1%) | 12 (20.7%) | 5 (17.2%) | 159 (43.8%) | |||
| AS | Mean | 2.74 | 4.40 | 5.09 | 5.65 | 3.64 | 0.000 ** | |
| SD | 2.36 | 2.45 | 2.27 | 2.68 | 2.62 | |||
| RNA E6/E7 | N | Normal (154 cases) | LSIL (62 cases) | HSIL (75 cases) | SCC (29 cases) | Total (320 cases) | p–Value | |
| P/N | P | 71 (46.1%) | 49 (79.0%) | 69 (92.0%) | 28 (96.6%) | 217 (67.8%) | 0.000 * | |
| N | 83 (53.9%) | 13 (21.0%) | 6 (8.0%) | 1 (3.4%) | 103 (32.2%) |
| Sensitivity | Specificity | PPV | NPV | |
|---|---|---|---|---|
| LSIL+ * vs. Normal | ||||
| p16INK4A | 49 | 96 | 89 | 72 |
| Ki-67 | 63 | 75 | 65 | 74 |
| Cotactin | 79 | 60 | 59 | 80 |
| RNA E6/E7 | 88 | 54 | 67 | 81 |
| HSIL+ ** vs. Normal | ||||
| p16INK4A | 78 | 94 | 81 | 93 |
| Ki-67 | 89 | 75 | 52 | 95 |
| Cotactin | 80 | 51 | 34 | 89 |
| RNA E6/E7 | 93 | 44 | 45 | 93 |
| Model | Unstandardized Coefficients | Sig. | Collinearity Statistics | |||
|---|---|---|---|---|---|---|
| B | Std. Error | Tolerance | VIF | |||
| Model 1 | (Constant) | 1.150 | 0.101 | 0.000 | ||
| p16INK4A AS | 0.197 | 0.038 | 0.000 | 0.574 | 1.742 | |
| Ki–67AS | 0.269 | 0.039 | 0.000 | 0.574 | 1.742 | |
| Model 2 | (Constant) | 0.358 | 0.235 | 0.130 | ||
| Ki–67AS | 0.267 | 0.037 | 0.000 | 0.574 | 1.742 | |
| p16INK4A P/N | 0.172 | 0.037 | 0.000 | 0.555 | 1.801 | |
| CortactinP/N | 0.570 | 0.154 | 0.000 | 0.938 | 1.066 | |
| Model 3 | (Constant) | −0.346 | 0.314 | 0.272 | ||
| Ki–67 AS | 0.245 | 0.037 | 0.000 | 0.556 | 1.799 | |
| p16INK4A AS | 0.152 | 0.036 | 0.000 | 0.539 | 1.854 | |
| CortactinP/N | 0.557 | 0.150 | 0.000 | 0.937 | 1.067 | |
| RNA E6/E7 P/N | 0.518 | 0.159 | 0.001 | 0.841 | 1.189 | |
| Model 4 | (Constant) | 0.535 | 0.506 | 0.292 | ||
| Ki–67AS | 0.387 | 0.074 | 0.000 | 0.134 | 7.446 | |
| p16INK4A AS | 0.142 | 0.036 | 0.000 | 0.531 | 1.883 | |
| CortactinP/N | 0.530 | 0.148 | 0.000 | 0.931 | 1.074 | |
| RNA E6/E7 P/N | 0.506 | 0.157 | 0.002 | 0.840 | 1.190 | |
| Ki-67 P/N | −0.786 | 0.356 | 0.029 | 0.166 | 6.040 | |
| Model 5 | (Constant) | 0.920 | 0.534 | 0.087 | ||
| Ki–67AS | 0.387 | 0.073 | 0.000 | 0.134 | 7.446 | |
| p16INK4A AS | 0.139 | 0.036 | 0.000 | 0.530 | 1.886 | |
| CortactinP/N | 0.539 | 0.147 | 0.000 | 0.930 | 1.075 | |
| RNA E6/E7 P/N | 0.517 | 0.155 | 0.001 | 0.839 | 1.191 | |
| Ki–67P/N | −0.747 | 0.353 | 0.036 | 0.165 | 6.057 | |
| Age groups | −0.153 | 0.073 | 0.039 | 0.987 | 1.014 | |
| Model | Equations |
|---|---|
| Model 1 | Y = 1.150 + 0.197 (p16INK4A AS) + 0.269 (Ki–67AS) |
| Model 2 | Y = −0.358+ 0.267 (Ki–67AS) + 0.172 (p16INK4A P/N) + 0.570 (CortactinP/N) |
| Model 3 | Y = −0.346 + 0.245 (Ki–67AS) + 0.152 (p16INK4AAS) + 0.557 (CortactinP/N) + 0.518 (RNA E6/E7 P/N) |
| Model 4 | Y = 0.535 + 0.387 (Ki–67AS) + 0.142 (p16INK4A AS) + 0.530 (CortactinP/N) + 0.506 (RNA E6/E7 P/N) − 0.786 (Ki–67 P/N) |
| Model 5 | Y = 0.920 + 0.387 (Ki–67 AS) + 0.139 (p16INK4A AS) + 0.539 (Cortactin P/N) + 0.517 (RNA E6/E7 P/N) − 0.747 (Ki–67 P/N) − 0.153 (Age groups) |
| Test Sample Set | Confirmed Sample Set | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| N | Mean | SD | p–Value | N | Mean | SD | p–Value | ||
| Model 1 | Normal | 156 | 1.4913 | 0.53391 | 0.000 | 55 | 1.6146 | 0.5864 | 0.000 |
| LSIL | 34 | 1.6402 | 0.87461 | 31 | 1.9450 | 0.8526 | |||
| HSIL | 24 | 3.4301 | 1.01324 | 34 | 3.4906 | 1.0748 | |||
| SCC | 19 | 3.9961 | 0.39674 | 10 | 3.9619 | 0.7745 | |||
| Total | 233 | 1.9170 | 1.06639 | 130 | 2.3646 | 1.2106 | |||
| Model 2 | Normal | 156 | 0.8589 | 0.49971 | 0.000 | 55 | 1.0939 | 0.7041 | 0.000 |
| LSIL | 34 | 1.0008 | 0.63920 | 31 | 1.3892 | 0.7130 | |||
| HSIL | 24 | 2.3430 | 0.73615 | 34 | 2.3541 | 0.7170 | |||
| SCC | 19 | 2.6392 | 0.33303 | 10 | 2.5678 | 0.7772 | |||
| Total | 233 | 1.1770 | 0.82221 | 130 | 1.6073 | 0.9170 | |||
| Model 3 | Normal | 156 | 1.2044 | 0.65480 | 0.000 | 55 | 1.2069 | 0.8686 | 0.000 |
| LSIL | 34 | 1.4843 | 1.10780 | 31 | 2.1960 | 0.9162 | |||
| HSIL | 24 | 3.5374 | 1.01427 | 34 | 3.3083 | 1.1946 | |||
| SCC | 19 | 3.9516 | 0.41838 | 10 | 3.9686 | 0.7087 | |||
| Total | 233 | 1.7096 | 1.22683 | 130 | 2.2048 | 1.3778 | |||
| Model 4 | Normal | 156 | 1.4316 | 0.73041 | 0.000 | 55 | 1.7865 | 0.9271 | 0.000 |
| LSIL | 34 | 1.6542 | 0.98381 | 31 | 2.0258 | 1.1043 | |||
| HSIL | 24 | 3.6226 | 1.23028 | 34 | 3.8366 | 1.1726 | |||
| SCC | 19 | 4.2301 | 0.52538 | 10 | 4.2166 | 1.2110 | |||
| Total | 233 | 1.9180 | 1.25429 | 130 | 2.5667 | 1.4364 | |||
| Model 5 | Normal | 156 | 1.2014 | 0.62799 | 0.000 | 55 | 1.1862 | 0.8520 | 0.000 |
| LSIL | 34 | 1.4728 | 1.08656 | 31 | 2.2305 | 0.9044 | |||
| HSIL | 24 | 3.5554 | 1.10289 | 34 | 3.3733 | 1.2589 | |||
| SCC | 19 | 4.0282 | 0.45701 | 10 | 3.9481 | 0.7920 | |||
| Total | 233 | 1.7140 | 1.24210 | 130 | 2.2197 | 1.4076 | |||
| Risk Biomolecules Prediction | Precancerous Lesion Prediction | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| LSIL Group (Normal + LSIL) vs. HSIL | LSIL Group (Normal + LSIL) vs. HSIL | |||||||||
| Sensitivity | Specificity | PPV | NPV | OR | Sensitivity | Specificity | PPV | NPV | OR | |
| Model 3 | 68 | 84 | 62 | 87 | 10.8 | 59 | 97 | 87 | 86 | 39.5 |
| Model 4 | 85 | 73 | 56 | 93 | 15.9 | 71 | 94 | 83 | 89 | 38.8 |
| Model 5 | 68 | 83 | 61 | 87 | 9.9 | 56 | 97 | 86 | 84 | 35.0 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bumrungthai, S.; Ekalaksananan, T.; Kleebkaow, P.; Pongsawatkul, K.; Phatnithikul, P.; Jaikan, J.; Raumsuk, P.; Duangjit, S.; Chuenchai, D.; Pientong, C. Mathematical Modelling of Cervical Precancerous Lesion Grade Risk Scores: Linear Regression Analysis of Cellular Protein Biomarkers and Human Papillomavirus E6/E7 RNA Staining Patterns. Diagnostics 2023, 13, 1084. https://doi.org/10.3390/diagnostics13061084
Bumrungthai S, Ekalaksananan T, Kleebkaow P, Pongsawatkul K, Phatnithikul P, Jaikan J, Raumsuk P, Duangjit S, Chuenchai D, Pientong C. Mathematical Modelling of Cervical Precancerous Lesion Grade Risk Scores: Linear Regression Analysis of Cellular Protein Biomarkers and Human Papillomavirus E6/E7 RNA Staining Patterns. Diagnostics. 2023; 13(6):1084. https://doi.org/10.3390/diagnostics13061084
Chicago/Turabian StyleBumrungthai, Sureewan, Tipaya Ekalaksananan, Pilaiwan Kleebkaow, Khajohnsilp Pongsawatkul, Pisit Phatnithikul, Jirad Jaikan, Puntanee Raumsuk, Sureewan Duangjit, Datchani Chuenchai, and Chamsai Pientong. 2023. "Mathematical Modelling of Cervical Precancerous Lesion Grade Risk Scores: Linear Regression Analysis of Cellular Protein Biomarkers and Human Papillomavirus E6/E7 RNA Staining Patterns" Diagnostics 13, no. 6: 1084. https://doi.org/10.3390/diagnostics13061084
APA StyleBumrungthai, S., Ekalaksananan, T., Kleebkaow, P., Pongsawatkul, K., Phatnithikul, P., Jaikan, J., Raumsuk, P., Duangjit, S., Chuenchai, D., & Pientong, C. (2023). Mathematical Modelling of Cervical Precancerous Lesion Grade Risk Scores: Linear Regression Analysis of Cellular Protein Biomarkers and Human Papillomavirus E6/E7 RNA Staining Patterns. Diagnostics, 13(6), 1084. https://doi.org/10.3390/diagnostics13061084






