CBCT Images to an STL Model: Exploring the “Critical Factors” to Binarization Thresholds in STL Data Creation
Abstract
1. Introduction
2. Materials and Methods
2.1. Definition
2.2. CBCT Scanning and Imaging to STL Model Creation
2.3. Converting the DICOM Image Dataset to 256 Grayscale and Making a Histogram
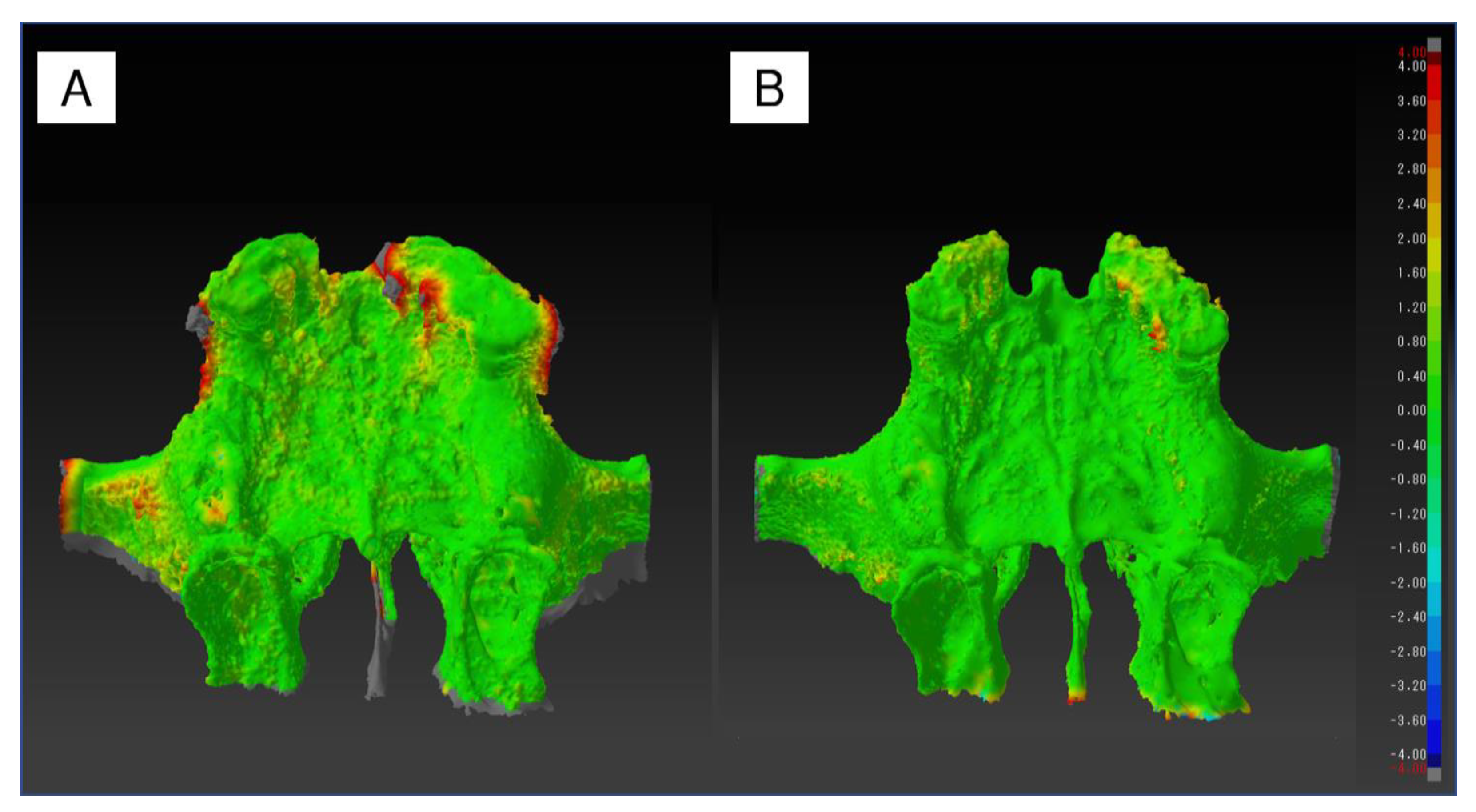
2.4. STL Model Superimposition, Comparison, and 3D Model Fabrication
3. Results
3.1. Visibility of the Exported Images
3.2. Differences in the Shape of Each STL Model
3.3. Histogram of the Voxel Intensity Distribution
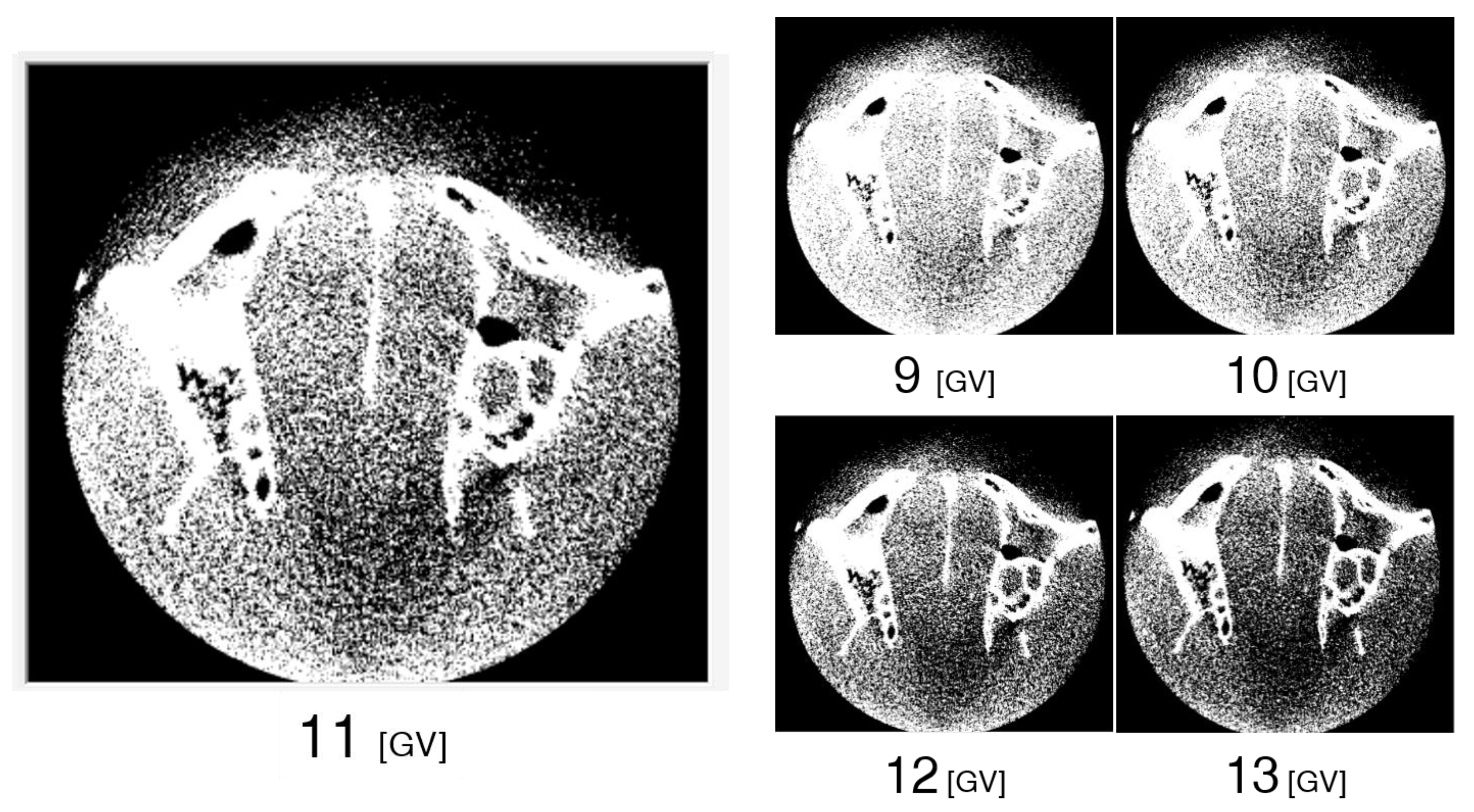
3.4. Differences in Images for Each GV
3.5. Shape Error of the STL Models
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Tack, P.; Victor, J.; Gemmel, P.; Annemans, L. 3D-printing techniques in a medical setting: A systematic literature review. Biomed. Eng. Online 2016, 15, 115. [Google Scholar] [CrossRef] [PubMed]
- Zheng, B.; Wang, X.; Zheng, Y.; Feng, J. 3D-printed model improves clinical assessment of surgeons on anatomy. J. Robot Surg. 2019, 3, 61–67. [Google Scholar] [CrossRef] [PubMed]
- Narita, M.; Takaki, T.; Shibahara, T.; Iwamoto, M.; Yakushiji, T.; Kamio, T. Utilization of desktop 3D printer-fabricated “Cost-Effective” 3D models in orthognathic surgery. Maxillofac. Plast. Reconstr. Surg. 2020, 42, 24. [Google Scholar] [CrossRef] [PubMed]
- Tomohisa, O.; Kamio, T.; Maeda, Y.; Tsubosaki, K.; Kato, T.; Iwata, H. Application of Medical Imaging and 3D Printing Technology in Teaching the Handling of Novel Medicine in Periodontal Surgery. Cureus 2022, 14, e29271. [Google Scholar] [CrossRef]
- Kamio, T.; Hayashi, K.; Onda, T.; Takaki, T.; Shibahara, T.; Yakushiji, T.; Shibui, T.; Kato, H. Utilizing a low-cost desktop 3D printer to develop a “one-stop 3D printing lab” for oral and maxillofacial surgery and dentistry fields. 3D Print Med. 2018, 4, 6. [Google Scholar] [CrossRef] [PubMed]
- Arai, Y.; Tammisalo, E.; Iwai, K.; Hashimoto, K.; Shinoda, K. Development of a compact computed tomographic apparatus for dental use. Dentomaxillofac. Radiol. 1999, 28, 245–248. [Google Scholar] [CrossRef] [PubMed]
- Kamio, T.; Suzuki, M.; Asaumi, R.; Kawai, T. DICOM segmentation and STL creation for 3D printing: A process and software package comparison for osseous anatomy. 3D Print Med. 2020, 6, 17. [Google Scholar] [CrossRef]
- Katsumata, A.; Hirukawa, A.; Okumura, S.; Naitoh, M.; Fujishita, M.; Ariji, E.; Langlais, R.P. Effects of image artifacts on gray-value density in limited-volume cone-beam computerized tomography. Oral Surg. Oral Med. Oral Pathol. Oral Radiol. Endod. 2007, 104, 829–836. [Google Scholar] [CrossRef]
- Doi, A.; Suzuki, S.; Yamasa, F.; Itoh, F.; Matsui, K.; Sachio, M.; Itoh, S. Volume Extractor Ver.3.0- 3D Image Processing and 3D Model Reconstruction. IIEE J. 2008, 37, 1037–1043. [Google Scholar]
- Tanimoto, S. Collaborative research on polygon engineering with RIKEN. Unisys Technol. Rev. 2012, 32, 283–292. (In Japanese). Available online: https://pr.biprogy.com/tec_info/tr114/11413.pdf (accessed on 19 January 2023).
- Kamio, T.; Onda, T. Fused Deposition Modeling 3D Printing in Oral and Maxillofacial Surgery: Problems and Solutions. Cureus 2022, 14, e28906. [Google Scholar] [CrossRef] [PubMed]
- Adolphs, N.; Liu, W.; Keeve, E.; Hoffmeister, B. Craniomaxillofacial surgery planning based on 3D models derived from Cone-Beam CT data. Comput. Aided Surg. 2013, 18, 101–108. [Google Scholar] [CrossRef] [PubMed]
- Rong, Y.; Smilowitz, J.; Tewatia, D.; Tome, W.A.; Paliwal, B. Dose calculation on kV cone beam CT images: An investigation of the Hu-density conversion stability and dose accuracy using the site-specific calibration. Med. Dosim. 2010, 35, 195–207. [Google Scholar] [CrossRef] [PubMed]
- Molteni, R. Prospects and challenges of rendering tissue density in Hounsfield units for cone beam computed tomography. Oral Surg. Oral Med. Oral Pathol. Oral Radiol. 2013, 116, 105–119. [Google Scholar] [CrossRef] [PubMed]
- Pauwels, R.; Jacobs, R.; Singer, S.R.; Mupparapu, M. CBCT-based bone quality assessment: Are Hounsfield units applicable? Dentomaxillofac. Radiol. 2015, 44, 20140238. [Google Scholar] [CrossRef]
- Pauwels, R.; Araki, K.; Siewerdsen, J.H.; Thongvigitmanee, S.S. Technical aspects of dental CBCT: State of the art. Dentomaxillofac. Radiol. 2015, 44, 20140224. [Google Scholar] [CrossRef] [PubMed]
- Ronkainen, A.P.; Al-Gburi, A.; Liimatainen, T.; Matikka, H. A dose-neutral image quality comparison of different CBCT and CT systems using paranasal sinus imaging protocols and phantoms. Eur. Arch. Otorhinolaryngol. 2022, 279, 4407–4414. [Google Scholar] [CrossRef] [PubMed]
- Jin, J.Y.; Ren, L.; Liu, Q.; Kim, J.; Wen, N.; Guan, H.; Movsas, B.; Chetty, I.J. Combining scatter reduction and correction to improve image quality in cone-beam computed tomography (CBCT). Med. Phys. 2010, 37, 5634–5644. [Google Scholar] [CrossRef]
- Zheng, J.; Zhang, D.; Huang, K.; Sun, Y. A CBCT series slice image segmentation method. J. Xray. Sci. Technol. 2018, 26, 815–832. [Google Scholar] [CrossRef]
- Wang, L.; Li, J.P.; Ge, Z.P.; Li, G. CBCT image based segmentation method for tooth pulp cavity region extraction. Dentomaxillofac. Radiol. 2019, 48, 20180236. [Google Scholar] [CrossRef]
- Li, C.; Lin, L.; Zheng, Z.; Chung, C.H. A User-Friendly Protocol for Mandibular Segmentation of CBCT Images for Superimposition and Internal Structure Analysis. J. Clin. Med. 2021, 10, 127. [Google Scholar] [CrossRef] [PubMed]
- Borohovitz, C.L.; Abraham, Z.; Redmond, W.R. The diagnostic advantage of a CBCT-derived segmented STL rendition of the teeth and jaws using an AI algorithm. J. Clin. Orthod. 2021, 55, 361–369. [Google Scholar] [PubMed]
- Lo Giudice, A.; Ronsivalle, V.; Grippaudo, C.; Lucchese, A.; Muraglie, S.; Lagravère, M.O.; Isola, G. One Step before 3D Printing—Evaluation of Imaging Software Accuracy for 3-Dimensional Analysis of the Mandible: A Comparative Study Using a Surface-to-Surface Matching Technique. Materials 2020, 13, 2798. [Google Scholar] [CrossRef] [PubMed]
- van Eijnatten, M.; Koivisto, J.; Karhu, K.; Forouzanfar, T.; Wolff, J. The impact of manual threshold selection in medical additive manufacturing. Int. J. Comput. Assist. Radiol. Surg. 2017, 12, 607–615. [Google Scholar] [CrossRef]
- van Eijnatten, M.; Berger, F.H.; de Graaf, P.; Koivisto, J.; Forouzanfar, T.; Wolff, J. Influence of CT parameters on STL model accuracy. Rapid Prototyp. J. 2017, 23, 678–685. [Google Scholar] [CrossRef]
- Wu, A.M.; Shao, Z.X.; Wang, J.S.; Yang, X.D.; Weng, W.Q.; Wang, X.Y.; Xu, H.Z.; Chi, Y.L.; Lin, Z.K. The accuracy of a method for printing three-dimensional spinal models. PLoS ONE 2015, 10, e0124291. [Google Scholar] [CrossRef]
- Rangel, F.A.; Maal, T.J.; Bronkhorst, E.M.; Breuning, K.H.; Schols, J.G.; Bergé, S.J.; Kuijpers-Jagtman, A.M. Accuracy and reliability of a novel method for fusion of digital dental casts and Cone Beam Computed Tomography scans. PLoS ONE 2013, 8, e59130. [Google Scholar] [CrossRef]
- Kane, B.; Shah, K.C. A dual scan approach to creating an accurate dental surface for virtual implant planning: A dental technique. J. Prosthet. Dent. 2021, 126, 464–470. [Google Scholar] [CrossRef]
- Michelinakis, G.; Apostolakis, D.; Pavlakis, E.; Kourakis, G.; Papavasiliou, G. Accuracy of IOS in Full-Arch Dentate Patients Compared to CBCT Cast-Scanning. An In-Vivo Study. Eur. J. Prosthodont. Restor. Dent. 2019, 27, 122–130. [Google Scholar]


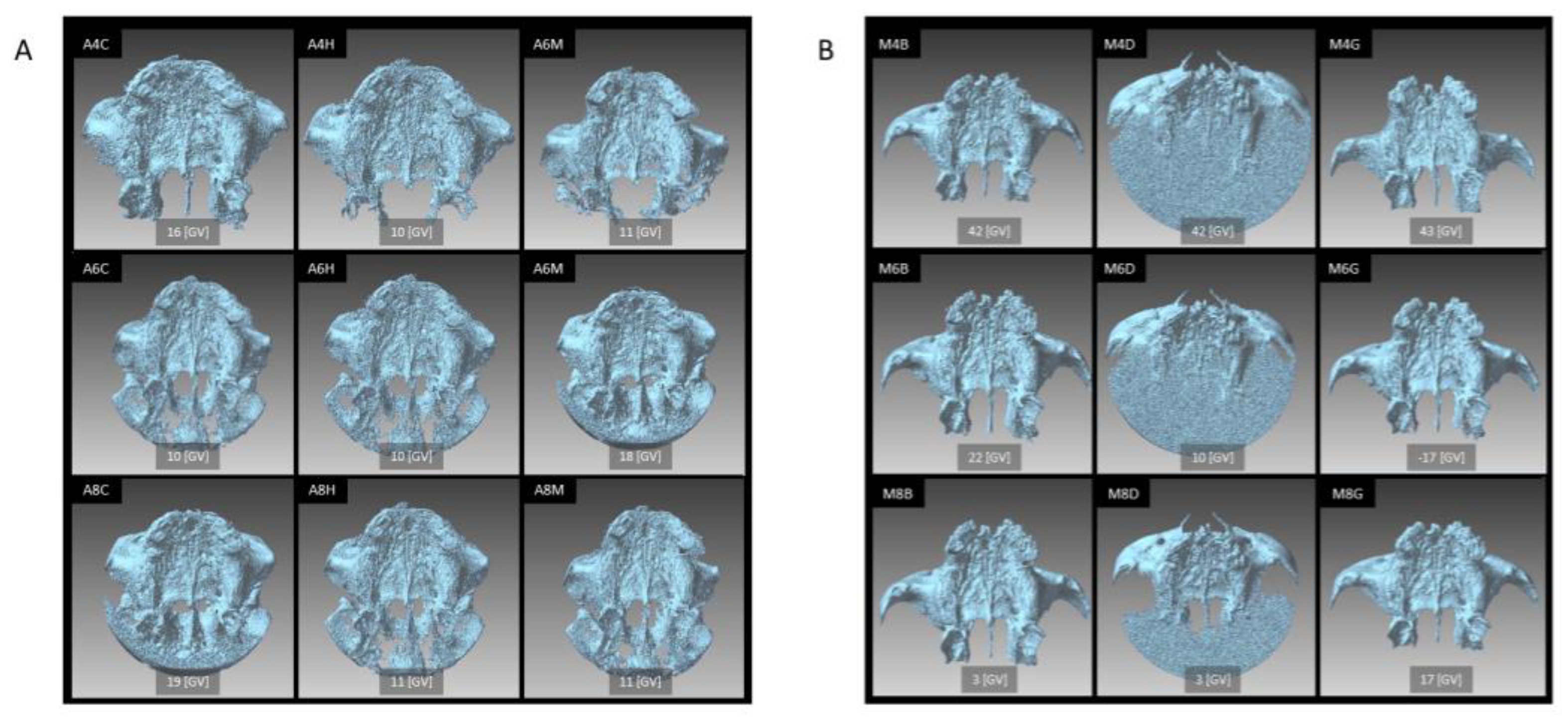
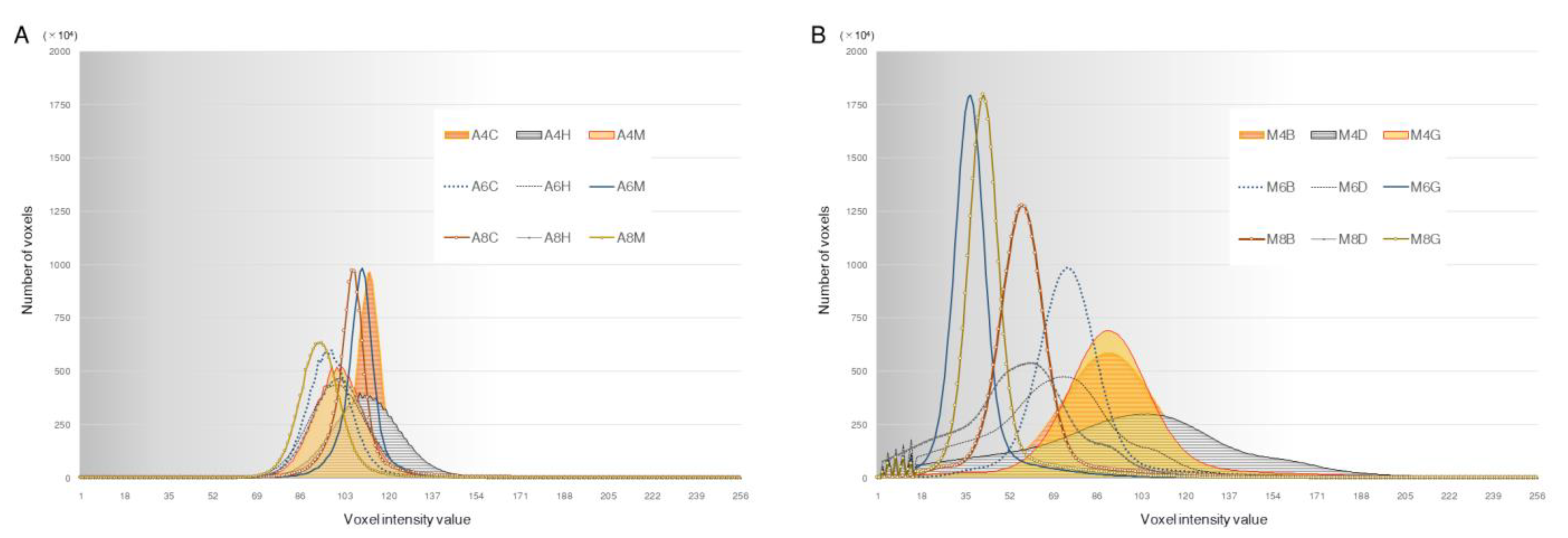
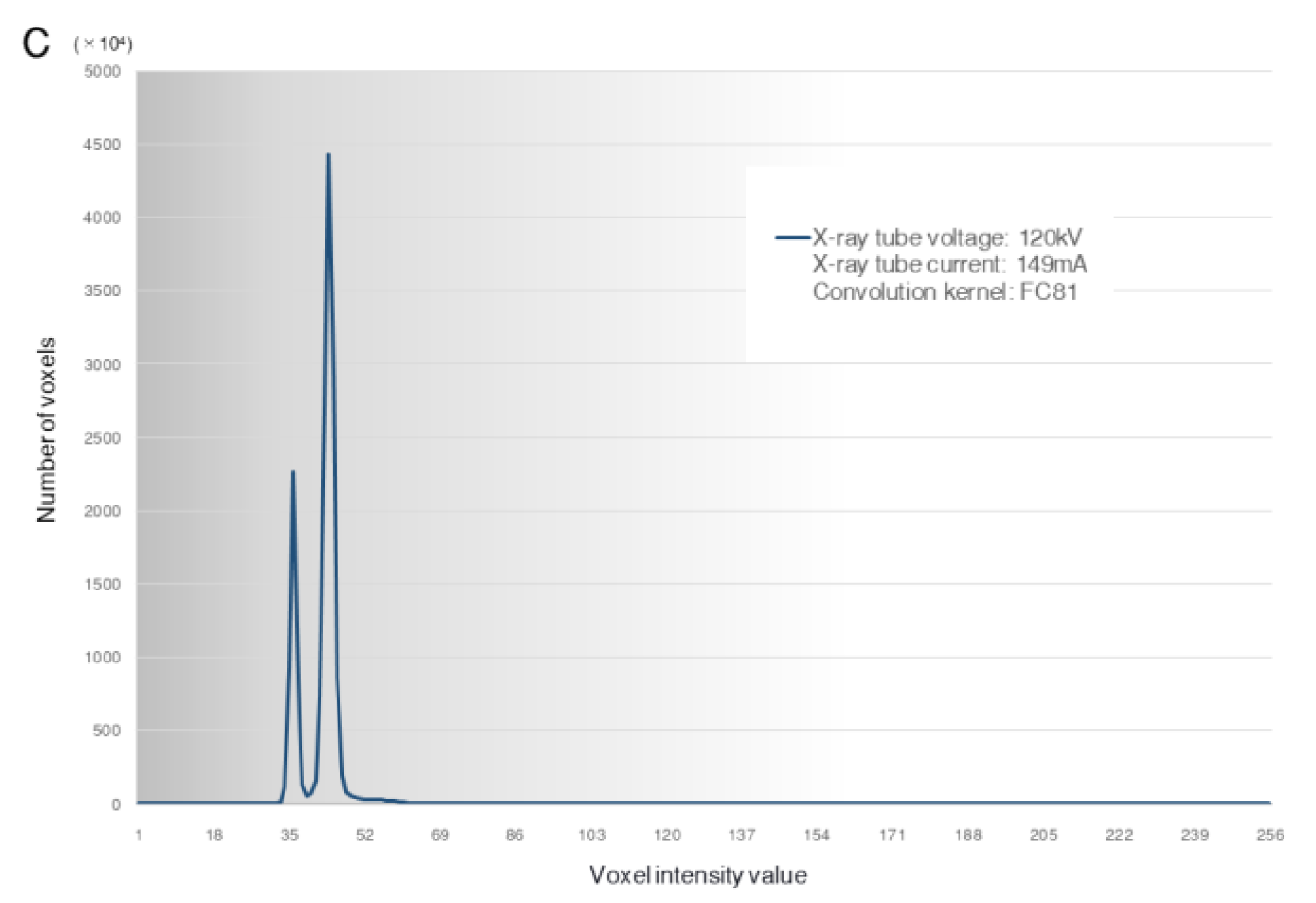

| CBCT Unit | X-ray Tube Voltage | Scanning Mode Field of View Voxel Size | Image Processing Software (Version) | X-ray Tube Current | Image Reconstruction Filter | Abbreviation |
|---|---|---|---|---|---|---|
| SOLIO XZ II | 85 kV | I-MODE φ90 × 91 mm 0.177 mm | NEOPREMIUM2 (NeoExpCalc 1.0.17.0) | 4.0 mA | Scattered ray correction | A4C |
| Sharp | A4H | |||||
| Smooth | A4M | |||||
| 6.0 mA | Scattered ray correction | A6C | ||||
| Sharp | A6H | |||||
| Smooth | A6M | |||||
| 8.0 mA | Scattered ray correction | A8C | ||||
| Sharp | A8H | |||||
| Smooth | A8M | |||||
| 3D Accuitomo F17 | 85 kV | D140 × H100 Hi-Fi φ140 × 100 mm 0.200 mm | i-Dixel (3DXAPP 2.3.7.5) | 4.0 mA | Bone | M4B |
| Dental | M4D | |||||
| General | M4G | |||||
| 6.0 mA | Bone | M6B | ||||
| Dental | M6D | |||||
| General | M6G | |||||
| 8.0 mA | Bone | M8B | ||||
| Dental | M8D | |||||
| General | M8G |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kamio, T.; Kawai, T. CBCT Images to an STL Model: Exploring the “Critical Factors” to Binarization Thresholds in STL Data Creation. Diagnostics 2023, 13, 921. https://doi.org/10.3390/diagnostics13050921
Kamio T, Kawai T. CBCT Images to an STL Model: Exploring the “Critical Factors” to Binarization Thresholds in STL Data Creation. Diagnostics. 2023; 13(5):921. https://doi.org/10.3390/diagnostics13050921
Chicago/Turabian StyleKamio, Takashi, and Taisuke Kawai. 2023. "CBCT Images to an STL Model: Exploring the “Critical Factors” to Binarization Thresholds in STL Data Creation" Diagnostics 13, no. 5: 921. https://doi.org/10.3390/diagnostics13050921
APA StyleKamio, T., & Kawai, T. (2023). CBCT Images to an STL Model: Exploring the “Critical Factors” to Binarization Thresholds in STL Data Creation. Diagnostics, 13(5), 921. https://doi.org/10.3390/diagnostics13050921






