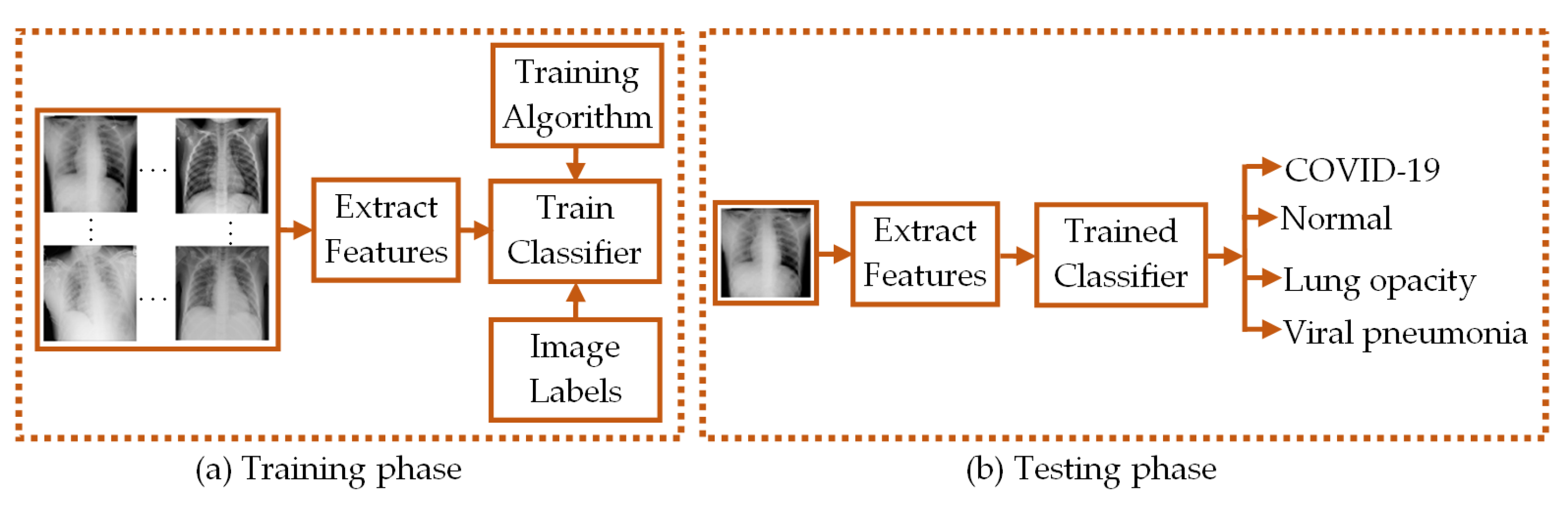
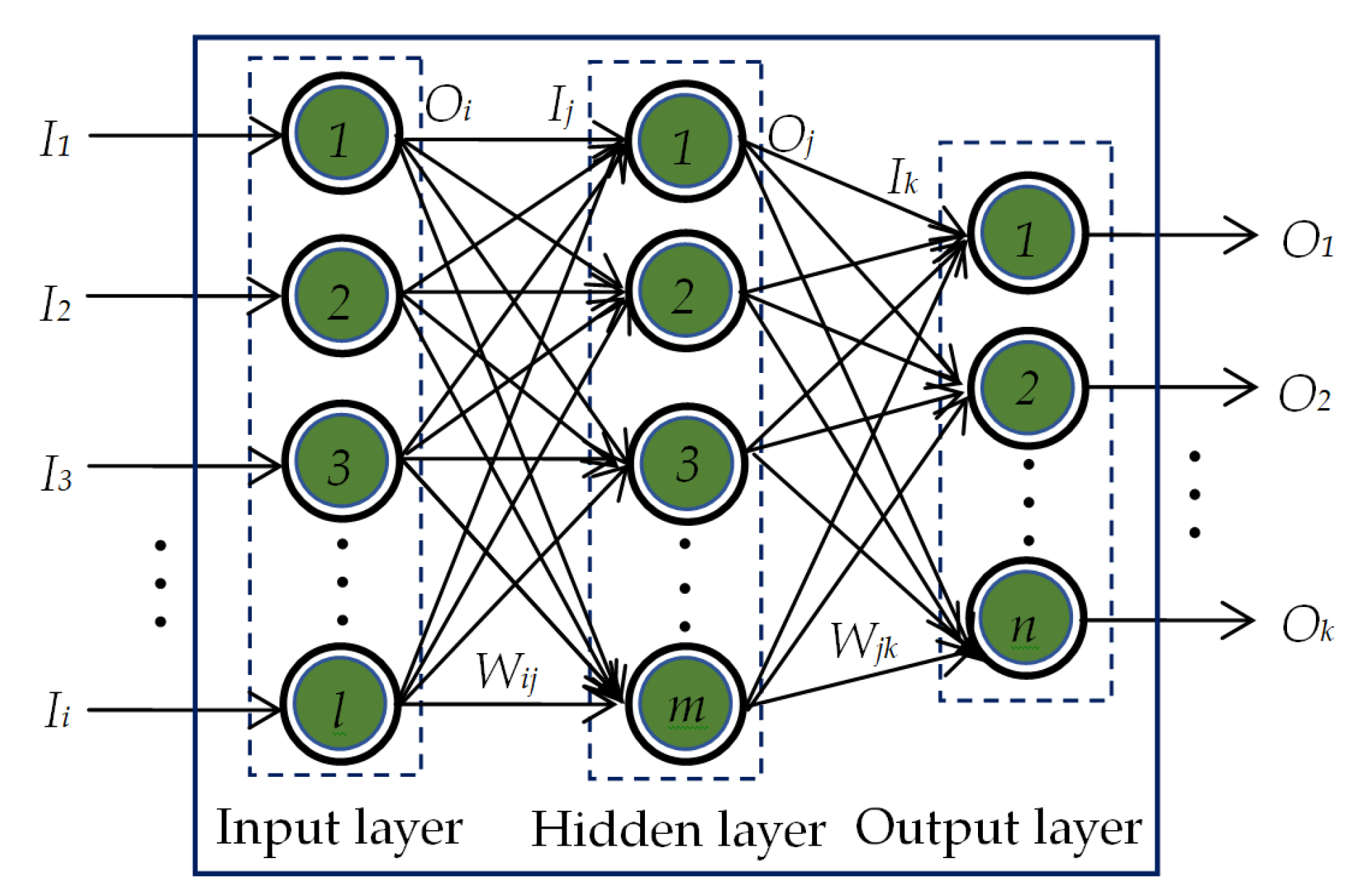
1. Introduction
The severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is responsible for the devastating coronavirus disease 2019 (COVID-19) outbreak which rapidly spread across the world and took over 6.55 million human lives to date [
1,
2,
3]. The SARS-CoV-2 is a single-stranded and approximately spherically shaped enveloped RNA virus with probable zoonotic origin belonging to the coronaviridae family [
4,
5,
6]. Although the SARS-CoV-2 infection can occur at all ages of human beings, its symptoms and acuteness vary with age from asymptomatic (in cases of children and young people) to severe respiratory malfunction and organ failure (above 60 y) [
2,
7]. The most common COVID-19 symptoms are fever, fatigue, and dry cough, whereas, headache, nausea, sore throat, sputum production, hemoptysis, diarrhea, anorexia, and chest pain are noted as less common features of it [
8,
9,
10,
11]. The gold standard of COVID-19 diagnosis is based on the detection of SARS-CoV-2 nucleic acid at a molecular level by real-time reverse transcription polymerase chain reaction (RT-PCR) [
12,
13]. However, the laboratory-grade accuracy and specificity of RT-PCR can be adversely affected by the variability of type, method of handling, and collection time of the sample with respect to the disease stage which may lead to a low detection rate below 50% [
14,
15,
16,
17]. As the early sign of acute COVID-19 reveals symptoms of pneumonia, radiological imaging techniques such as chest X-ray (CXR), computed tomography (CT), and magnetic resonance imaging (MRI) serve as indispensable tools to negate the false negative RT-PCR detection [
18,
19,
20,
21,
22,
23,
24,
25,
26,
27]. Although CXR is preferred over the sophisticated CT and MRI in terms of cost, equipment availability, measurement time, and radiation dose, the proper diagnosis of COVID-19 from it poses significant challenges in many parts of the world, arising from the scarcity of experienced radiologists for visual inspection of minute features such as ground-glass opacity and abnormalities of bilateral and interstitial nature in the radiographs relevant to COVID-19 pneumonia [
9,
11,
23,
28]. Moreover, the similarity of CXR images of other viral infections and respiratory diseases with COVID-19 makes it very challenging even for expert radiologists [
8,
21]. Artificial intelligence (AI)-based deep machine learning (ML) offers tremendous support in revealing subtle and hidden image features which may not be apparent in the original CXR and thereby facilitating the automation of image classification and COVID-19 disease diagnosis problem [
29]. Moreover, the abundance of publicly available CXR image datasets provided tremendous facilitation in training ML algorithms [
30].
Numerous attempts have been made to standardize the detection models for COVID-19 based on CXR images as a complement to RT-PCR using an AI-based DL approach [
31,
32,
33]. The success of CXR image-based COVID-19 diagnosis with DL potentially relies on the reliable detection of different abnormalities of interstitial and bilateral nature as well as ground-glass opacity irregularities in CXR radiographs [
34]. The local binary pattern (LBP) operator is a very popular feature extraction method for CXR radiographs due to its simplicity and low computational cost. The LBP operator divides the images into several parts and assigns binary patterns depending on the texture of the pixel neighbor to extract the feature. On the other hand, a sophisticated and computationally intense DL-based convolutional neural network (CNN), due to its inherent spatial feature extraction and classification capabilities, has emerged as a leading candidate for CXR-based COVID-19 detection [
35,
36,
37,
38]. With the aid of transfer learning, efficient training of deep CNN with a relatively small CXR dataset sparked its widespread usage in the ML research community to differentiate COVID-19 from non-COVID-19 pneumonia from CXR images [
39].
Many of the CXR image-based detection techniques use off-the-rack deep neural networks such as VGG-16, VGG-19, Inception-V3, MobileNet-V2, ResNet-18, ResNet-50, DenseNet-121, CapsNet, and EfficientNet. The CNN network prefers large datasets to avoid bias problem and hence the size of the dataset is crucial for the reliability of result presented in the different literature. For a small dataset containing 125 CXR images, 98.08% accuracy in COVID-19 classification was reported for binary classes in Ref. [
40]. Ismael et al., reported an accuracy of 92.63% in COVID-19 prediction working on a CXR image set containing 180 COVID-19 and 200 normal patients’ images, where they used a pre-trained ResNet50 model for feature extraction and support vector machine (SVM) with a linear kernel for classification [
41]. Afshar et al., used similar two-class CXR images with CapsNet to obtain an accuracy of 98.3% [
42]. Frid-Adar et al., achieved 98% accuracy using the ResNet50 over a two-class dataset of no lung opacity and lung opacity [
43]. For a large two-class CXR dataset with COVID-19 pneumonia (5805) and non–COVID-19 pneumonia (5300) images, Zhang et al., managed to report 92% accuracy using the DenseNet-121 [
44]. Vinod and Prabaharan used a pre-trained CNN in combination with a decision tree classifier to obtain a precision score of 88% on a CXR dataset containing ~300 two-class images [
45].
For a three-class classification problem, the achieved accuracy was 95% over a CXR dataset containing normal (104), non-COVID-19 pneumonia (80), and COVID-19 pneumonia (99) [
46]. Bukhari et al., reported overall accuracy of 98.24% using 278 images in ResNet-50 with three different image classes comprised of normal (93), COVID-19 (89), and non-COVID-19 pneumonia (96) [
47]. Tartaglione et al., achieved a maximum 85% accuracy over 5857 CXR images with three classes comprising normal (1583), bacterial pneumonia (2780), and viral pneumonia (1493) [
48]. For a different three-class CXR image dataset containing normal (1314), community-acquired pneumonia (2004), and COVID-19 pneumonia (204), the reported maximum accuracy was 98.71% [
49]. Kana et al., obtained 99% accuracy for a dataset with normal (2487), bacterial/viral pneumonia (2507), and COVID-19 (161) [
50]. On a similar three-class problem, 91.92% overall accuracy of prediction was achieved in Ref. [
44]. Bassi and Attux reported an accuracy of 99.94% for a small dataset with 439 images [
51]. The VGG-19 produced an accuracy of 89.3% for a dataset having normal (300), pneumonia (30), and COVID-19 (260) CXR images [
52]. Luz et al., performed classification with EfficientNet on a dataset containing 13,569 CXR images and obtained 93.9% accuracy [
53]. The use of a deep CNN DeTraC produced 93.1% accuracy in COVID-19 detection using a small dataset composed of normal (80), SARS (11), and COVID-19 (105) CXR images [
54]. Another variant of deep CNN, known as COVID-Net achieved a prediction accuracy of 83.5% on a larger dataset containing normal (358), non-COVID-19 pneumonia (8066), and COVID-19 pneumonia (5538) CXR images [
22]. The deep Bayes-SqueezeNet was capable of producing 98.3% COVID-19 prediction accuracy which utilized a dataset with normal (1583), non-COVID-19 pneumonia (4290), and COVID-19 (76) CXR images [
55]. Heidari et al., used three classes of CXR images with normal (2880), non-COVID-19 pneumonia (5179), and COVID-19 pneumonia (415) to obtain an accuracy of 94.5% [
56]. The classification accuracy can be increased to 97.8% with the VGG-16 CNN model with a similar-sized dataset [
57]. Vinod et al., used Covix-Net to yield 96.8% accuracy on a three-class CXR database containing a total of 9000 images divided into normal (3000), COVID-19 (3000), and pneumonia (3000) [
58].
In recent times, few investigations emerged with four-class classification capabilities in COVID-19 identification from CXR images. Hussain et al., used a small CXR dataset with normal (138), bacterial pneumonia (145), non-COVID-19 viral pneumonia (145), and COVID-19 (130) and managed to produce 79.52% accuracy [
59]. The deep CNN termed as CoroNet was able to produce 89.6% over a CXR dataset composed of normal (310), bacterial pneumonia (330), viral pneumonia (327), and COVID-19 (284) [
60]. The use of deep ResNet improved the accuracy of the four-class classification to 92.1% for a very small dataset consisting of 450 CXR images [
61]. Attaullah et al., combined patients’ symptoms with a total of 800 four-class CXR images and obtained an accuracy of 78.88% [
62]. The estimation of the uncertainty in the deep CNN with a Bayesian approach can improve the reliability of the accuracy measurements in a four-class classification problem [
63]. The pre-trained and fine-tuned ResNet-50 architecture have been shown to achieve 96.23% accuracy for four-class CXR dataset containing normal (1203), non-COVID-19 viral pneumonia (660), bacterial pneumonia (931), and COVID-19 pneumonia (68) [
64]. It is hard to find rigorous and extensive studies of COVID-19 identification within the framework of four-class classification problems using different ML algorithms on a relatively large dataset in the existing literature.
To that cause, here we used different ML algorithms on a comparatively large dataset comprising of 5360 CXR images containing four different classes, i.e., COVID-19, normal, lung opacity, and viral pneumonia, each of which contains 1340 images. The CXR image feature extractions were performed using local binary pattern (LBP) and pre-trained CNN. We used LBP-based PRN, LBP-based SVM, LBP-based DT, LBP-based RF, LBP-based KNN, and CNN-based SVM for image class identification. For the reliable performance analysis of LBP-PRN, a variety of six different training algorithms were used. The performance of SVM classifiers was assessed for nineteen different pre-trained CNNs in feature extraction. The classification performance provided by the ensemble configuration of the three best CNN-based SVM classifiers selected from these aforementioned nineteen different CNNs was also evaluated in this study. Overall, we believe that the results presented here have established an efficient and reliable CNN-based SVM framework for COVID-19 detection from CXR images.
3. Results and Discussion
In our demonstration of four-class (i.e., COVID-19, normal, lung opacity, and viral pneumonia) classification, we have extracted the feature vectors of CXR images by using the LBP operator as well as pre-trained CNNs. Then, the classification has been performed by PRN, SVM, DT, RF, and KNN classifiers based on those extracted feature vectors. We have also evaluated the classification performances of LBP-based PRN, LBP-based SVM, LBP-based DT, LBP-based RF, LBP-based KNN, CNN-based SVM, and ensemble-CNN-based SVM by adopting the process of five-fold cross-validation as depicted in
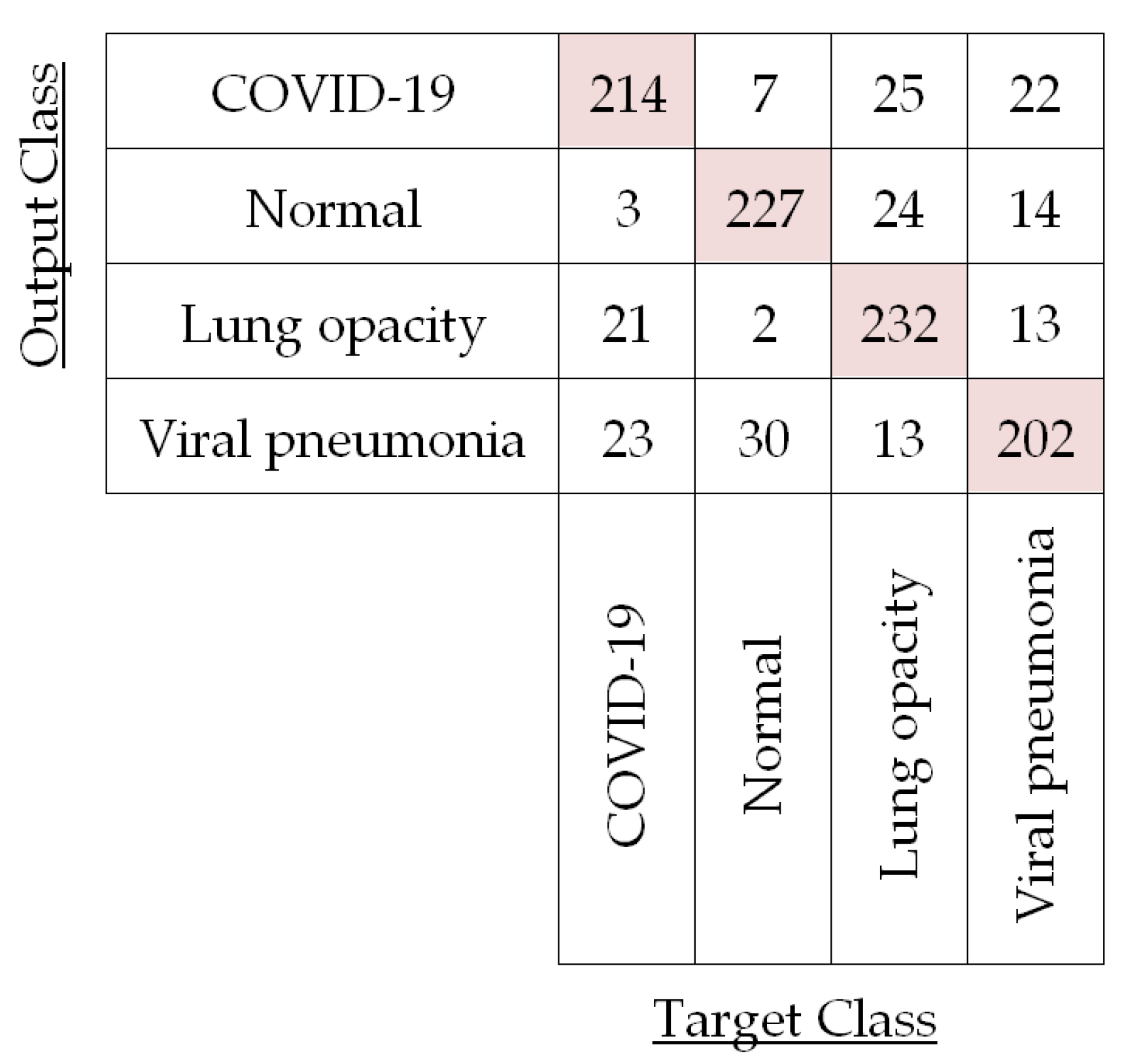
Figure 11. For instance, the confusion matrix for the testing images in a particular fold (i.e., Fold 1) is shown in
Figure 12 for LBP-based SVM.
The confusion matrix in
Figure 12 corroborates LBP-based classifiers’ success and is used to compute the performances of LBP-based SVM for each of the four different classes in Fold 1 by using Equations (10)–(13) as shown in
Table 4.
It is observed in
Table 4 that per-class classification accuracy for the test CXR images of each of the four different classes is around 90%. Now, we have averaged these per-class accuracies obtained for this fold to compute fold accuracy. For this Fold 1, LBP-based SVM yields 90.81% accuracy. We have also calculated the fold precision (81.62%), fold recall (81.68%), and fold specificity (93.90%) by averaging per-class precision, recall, and specificity, respectively, for Fold 1. In a similar fashion, we have calculated the per-fold accuracy, precision, recall, and specificity for the other four folds in the case of LBP-based SVM. The results for each of the five folds are listed in
Table 5.
The overall performance of this classification model is computed by averaging five per-fold performances as listed in
Table 5. Consequently, the overall accuracy, precision, recall, and specificity for using LBP-based SVM have been found to be 88.86%, 77.72%, 79.80%, and 92.58%, respectively. The overall F1 score for LBP-based SVM classification turned out to be 78.75% following Equation (14).
Next, we have applied a pattern recognition network (PRN) to classify four classes of CXR images utilizing the image feature vectors obtained from the LBP operator. In this classification process, we have also employed five-fold cross-validation to generalize the overall performance of LBP-based PRN. In this study, we have also explored the effects of applying six different training algorithms to train the PRN as listed in
Table 3. The overall classification performances of LBP-based PRN for adopting each of the six training algorithms are shown in
Figure 13.
The performances of LBP-based PRN vary with training algorithms used to train the PRN as seen in
Figure 13. It is evident that gradient descent (“traingd”) and gradient descent with momentum (“traingdm”) training algorithms failed to perform well when used with LBP-based PRN. It can also be observed in
Figure 13 that the performances of LBP-based PRN are comparable if such PRN is trained with variable learning rate gradient descent (“traingdx”), Levenberg–Marquardt (“trainlm”), resilient backpropagation (“trainrp”) and scaled conjugate gradient (“trainscg”) learning algorithms. However, the training of LBP-based PRN by adopting the Levenberg–Marquardt algorithm provides the best performances with accuracy, precision, recall, specificity, and F1 score of 88.61%, 77.28%, 79.60%, 92.44%, and 78.42%, respectively. It is worth mentioning that all six different algorithms mentioned in
Figure 13 have very similar runtime in the testing phase.
In a similar fashion, we have determined the classification performances of LBP-based DT, LBP-based RF, and LBP-based KNN. For instance, the overall accuracies provided by LBP-based DT, LBP-based RF, and LBP-based KNN are computed to be 83.77%, 87.43%, and 84.58%, respectively. The SVM yields the best accuracy in all cases of LBP-based machine learning classifiers used in this study. Consequently, we have only considered the SVM classifier for classifying CXR images in the next stage.
In this stage, we have extracted feature vectors from the CXR images by applying a deep learning algorithm where features are taken from the fully connected layer at the end of the CNN. To accomplish this feature extraction using CNN, we have employed a total of nineteen different pre-trained CNNs as listed in
Table 2. After extracting the feature vectors from the CXR images with CNNs, we have applied the SVM classifier in this stage for the four-class classification of CXR images as described previously. In this stage, we have also applied five-fold cross-validation to generalize the performance of CNN-based SVM classifiers. The overall classification performances of CNN-based SVM for adopting each of the nineteen different pre-trained CNNs along with the LBP-based different machine learning classifiers are shown in
Figure 14.
It is easily observed in
Figure 14 that each of the nineteen CNN-based SVM classifiers outperforms LBP-based different classifiers (i.e., DT, KNN, PRN, SVM, and RF). This is due to the fact that CNNs utilize a deep learning algorithm that is extremely powerful to extract feature vectors from the CXR images [
60,
61,
62,
63]. For instance, the lowest accuracy in
Figure 14 provided by the pre-trained CNN model of the NasNet-Mobile-based SVM is 92.74%, which is even better than that of LBP-based DT (83.77%), LBP-based KNN (84.58%), LBP-based PRN (88.61%), LBP-based SVM (88.86%), and LBP-based RF (87.43%) classifiers. However, the classification performances of SVM utilizing image feature vectors extracted with EfficientNet-b0 (model 13 in
Figure 14) are the best among the nineteen pre-trained CNN architectures as can be seen from
Figure 14. Such “EfficientNet-b0” pre-trained CNN-based SVM can achieve overall accuracy, precision, recall, specificity, and F1 score of 96.39%, 92.86%, 93.04%, 97.59%, and 92.95%, respectively.
To further improve the classification performance of CNN-based SVM, we have finally utilized ensemble-CNN-based SVM as described in
Section 2.3.6. To effectively utilize such ensemble configuration, we have selected the best three pre-trained CNNs (i.e., EfficientNet-b0, DenseNet-201, and DarkNet-53) among the nineteen different CNN architectures used in this study based on their classification metrics. The classification performances of ensemble-CNN-based SVM have also been plotted in
Figure 14 (model 25) for the purpose of comparison. The topmost classification performances provided by different feature extraction-based classifiers are listed in
Table 6.
It is seen in
Table 6 that the classification performances attained for using ensemble-CNN-based SVM are the highest among the 25 different classifiers adopted in this study. For instance, the ensemble-CNN-based SVM can improve the classification accuracy by ~1% as compared to the best CNN-based SVM (i.e., EfficientNet-b0-based SVM). These overall performances of EfficientNet-b0-based SVM and ensemble-CNN-based SVM are more promising compared to some recently published results as listed in
Table 7.
As observed in
Table 7, the performances attained by applying an Efficient-b0-based SVM classifier are much better than that achieved in Ref. [
60] for four-class classification using 1251 CXR images. The overall accuracy of the Efficient-b0-based SVM classifier is also comparable to that achieved in Ref. [
64]. It is to be noted that the dataset used in Ref. [
64] is imbalanced as there is a big difference in the number of images in each of the four classes (with only 68 CXR images in the COVID-19 class). However, the ensemble-CNN-based SVM classifier used in this study for four-class classification can provide much-improved classification performances as compared to other methods listed in
Table 7 with overall accuracy, precision, recall, specificity, and F1 score of 97.41%, 94.91%, 94.81%, 98.27%, and 94.86%, respectively. To the best of our knowledge, these classification performances rank the best among all other reported values for four-class classification of COVID-19, normal, lung opacity, and viral pneumonia CXR images in the existing literature.
Now we focus on relative runtime comparison analysis among different classifiers that use different feature extraction algorithms as shown in
Figure 14. It is evident that LBP-based DT requires the lowest runtime. Thus, the relative runtime of a particular technique is normalized with respect to the runtime taken by LBP-based DT. The relative runtimes of different LBP-based machine learning classifiers are nearly uniform with LBP-based RF being the slowest. However, the relative runtimes of CNN-based SVMs vary in accordance with the depth of the layered architecture of the pre-trained CNNs. Among them, the SqueezeNet-based SVM yields the lowest runtime while the NasNet-Large-based SVM requires the highest relative runtime as can be seen in
Figure 14. However, the relative runtime of the single CNN (i.e., EfficieNet-b0)-based SVM which provides the best classification performances is moderately low as compared to other CNN architectures used in this study. To be specific, the “EfficieNet-b0”-based SVM is 4.63 times slower as compared to LBP-based DT. However, such “EfficieNet-b0”-based SVM can provide significantly improved classification performances compared to that of LBP-based machine learning classifiers as shown in
Figure 14. It is also observed in
Figure 14 and
Table 6 that the ensemble-CNNs-based SVM provides the highest classification performance (e.g., 97.41% accuracy) among all the classifiers used in this study. However, to achieve such high performance, this classifier requires relative runtime of 40.68 (i.e., 40.68 times higher than LBP-based DT), which is ~8.72 times larger than that of “EfficieNet-b0”-based SVM.
4. Conclusions
This paper presents a rigorous study on the identification of COVID-19 infection from CXR images based on machine learning approaches. The feature vectors of CXR images have been extracted successfully by utilizing LBP operator and pre-trained CNNs of nineteen different architectures. Then, PRN, SVM, DT, RF, and KNN classifiers have been applied to classify four-class CXR images comprising COVID-19, normal, lung opacity, and viral pneumonia by utilizing the extracted feature vectors of the CXR images. The performances of LBP-based PRN, LBP-based SVM, LBP-based DT, LBP-based RF, LBP-based KNN, CNN-based SVM, and ensemble-CNN-based SVM classifiers have been investigated in detail on the four-class test images and their performances are analyzed in terms of accuracy, precision, recall, specificity, F1 score, and relative runtime. The effects of using six different learning algorithms used to train the LBP-based PRN are analyzed in detail and the results indicate that the Levenberg–Marquardt learning algorithm provides the best classification performance for using LBP-based PRNs in this study. The results also show that the classification performances of LBP-based classifiers are not up to the mark and are significantly lower than that of CNN-based SVM. Among nineteen different single pre-trained CNN-based SVM classifiers, the use of EfficientNet-b0 CNN architecture performs best in our study. The use of such CNN architecture can achieve overall classification performances of 96.39% accuracy, 92.86% precision, 93.04% recall, 97.59% specificity, and 92.95% F1 score with moderately low relative runtime. To further improve the classification performance of CNN-based SVM, we have also utilized ensemble-CNN-based SVM. Such an ensemble configuration consisting of three pre-trained CNNs (i.e., EfficientNet-b0, DenseNet-201, and DarkNet-53) has provided improved classification performances with 97.41% accuracy, 94.91% precision, 94.81% recall, 98.27% specificity, and 94.86% F1 score but required highest runtime to classify CXR images. We believe that the strategy suggested in this paper will provide doctors and physicians with a complementary tool for the diagnosis and prognosis of COVID-19-infected patients. Moreover, the framework so proposed can be integrated into a decision support system that can diagnose COVID-19 based on CXR images, thus considerably minimizing both human and machine error.