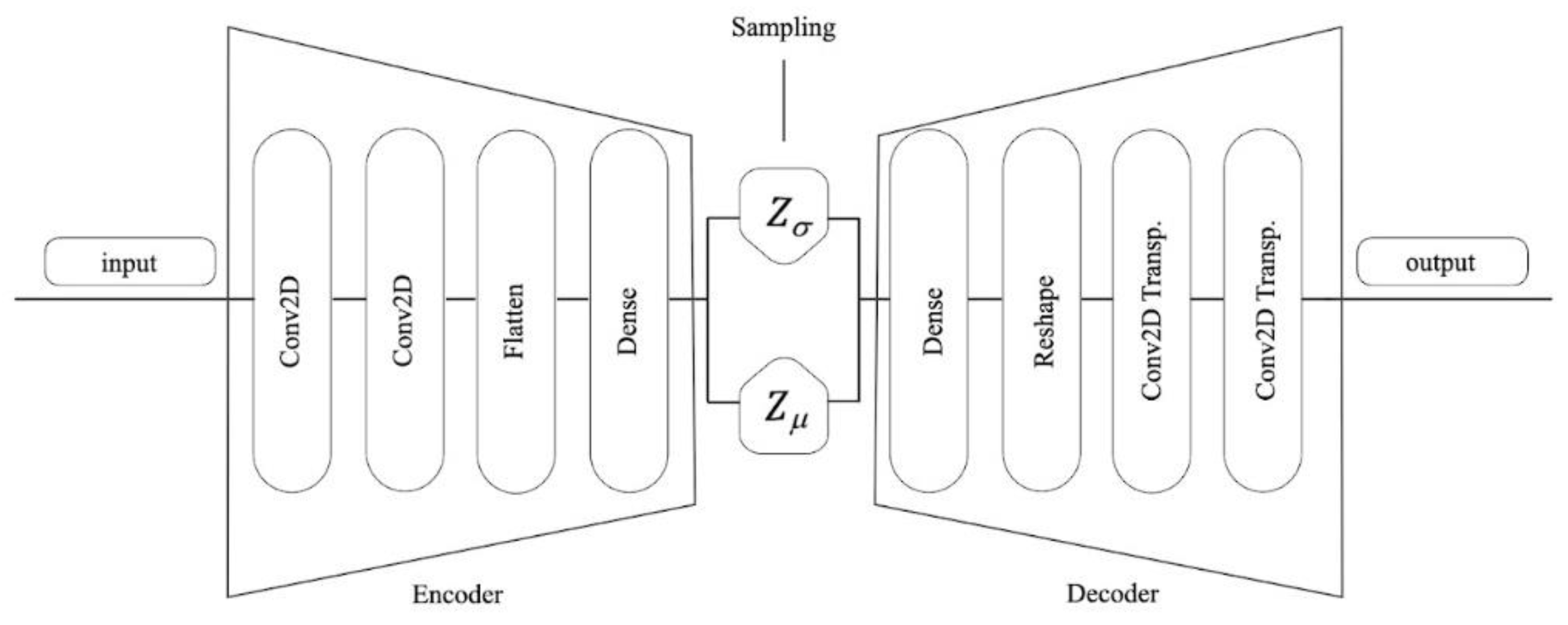
1. Introduction
AD is a degenerative neurological disorder characterized by the slowing of background oscillations or the downward shift of fundamental brain oscillations [
1]. This is the first cause of dementia, and the major sign is fast cognitive deterioration. It was estimated that 35.6 million people lived with dementia worldwide in 2010, with numbers expected to almost double every 20 years, to 65.7 million in 2030 and 115.4 million in 2050. In 2010, 58% of all people with dementia lived in countries with low or middle incomes, with this proportion anticipated to rise to 63% in 2030 and 71% in 2050 [
2]. Cortical neuron loss, axonal disease, and cholinergic deficits are hypothesized to cause these disorders [
3,
4]. The condition is more common in those over the age of 65, and the rate of occurrence rises exponentially with age [
5]. The number of people with neurological disorders is about 50 million and is projected to exceed 100 million by 2050 [
6]. An intermediary stage between Healthy Ageing (HA), sometimes referred to as Healthy Control (HC), and AD has been widely recognized as a stage called MCI. MCI is a term that refers to persons who have many Alzheimer’s disease symptoms but not enough to fulfil the diagnostic criteria for the disease and with more cognitive loss than is typical for their age [
7]. Patients with MCI may gradually regain normalcy. Recent studies estimated that the conversion rate from MCI to AD is approximately 15% per year [
4], whereas this rate is only 1–2% of the global population [
8]. While there is no known cure for AD or MCI, diagnosis and treatment of them in the early stages of the condition can considerably slow the disease’s course and delay the progression of MCI to dementia. As a result, early detection of AD and MCI is critical [
9,
10].
In ref. [
10] after performing pre-processing and dividing the signal into 5-s windows, extracting latent features from 2D gray-scale representations of PSD spectra, and then using a convolutional deep learning network to separate the three classes, AD vs. MCI vs. HC, an accuracy of 83.33% was achieved. Alzheimer’s disease and other neurological disorders such as epilepsy, stroke, and Parkinson’s disease can be diagnosed using a variety of methods. These include Magnetic Resonance Imaging (MRI), Single Photon Emission Computed Tomography (SPECT), and Positron Emission Tomography (PET) [
11]. Neuroimaging technologies that are inexpensive and readily available, such as EEG, have also been examined for the diagnosis of AD. Electroencephalography (EEG) is a non-invasive, low-cost, and portable technique, with a high temporal resolution, that reflects the electrical activity of the brain [
12]. EEG is a reliable tool for the early detection of AD, according to a number of clinical investigations, because the disease has an effect on the complexity, synchronization, and rhythm of the signals, which reduces complexity and causes rhythm to slow. The slowing of the rhythm in AD patients’ EEG signals can be explained by an increase in theta and delta frequency range activity and a decrease in alpha and beta frequency range activity [
13]. Differentiating Alzheimer’s disease from MCI and healthy people has been a challenging issue in research as well. For this reason, the effect of various methods such as MRI, FMRI, EEG, etc. in differentiating these three cases has been studied. In the meta-analysis study that investigated olfactory function to differentiate these three cases, it was shown that olfactory identification was more profoundly impaired in patients with AD than in those with MCI [
13]. In another study, using EEG findings, an algorithm was designed to distinguish Alzheimer’s from MCI, and as a result, an accuracy, sensitivity, and specificity of 96.5%, 96.21%, and 97.96% were achieved [
14].
Recently, novel DA methods have attracted the use of DNNs to map data space from high-dimensional to low-dimensional and realize feature extraction to reconstruct the artificial data. There are two typical deep learning strategies for DA: autoencoder (AE) and generative adversarial networks [
14]. Most recent studies have employed binary classification to distinguish AD and MCI from HC in EEG data. An efficient approach for detecting AD using the EEG data of AD and HC patients was developed by [
13]. They used three different feature sets and an SVM classifier (spectral-, wavelet-, and complexity-based features). Support vector machines (SVMs) are supervised learning models that examine data for regression and classification. They also include associated learning methods. As a consequence, they are able to attain a binary classification accuracy of 96% [
12] performed a time-frequency analysis of EEG signals using Fourier transforms and wavelet transforms. Although they used EEG signals of the AD, MCI, and HC subjects, they performed only binary classification and obtained a maximum accuracy of 92% in dealing with HC and MCI. Few studies have compared AD, MCI, and HC [
14] extracted statistical and spectral features from EEG data for the classification of AD, MCI, and HC. Using random forest classification, they were able to reach an accuracy of 88.79%. Subsequently [
15] proposed a new method to discriminate between AD, MCI, and HC classes using time-frequency domain analysis with continuous wavelet transform (CWT) and bispectral representation (BiS). They fed the extracted features into a multilayer perceptron classifier and obtained an accuracy of 89.24% for a three-class scheme.
Data augmentation (DA) comprises the generation of new samples to augment an existing dataset by transforming the existing samples in a manner that increases the accuracy and stability of the classification or regression. Exposing the classifier to more variable representations of its training samples makes the model more invariant and robust to transformations of the type that it is likely to encounter when attempting to generalize to unseen samples. In recent years, DA techniques have received widespread attention and achieved appreciable performance boosts for DL on EEG signals [
15]. The most important methods for EEG data augmentation include a noise addition (17%) deep learning method. We have two main categories for adding noise to the EEG signals for the purpose of DA: (1) Add various types of noise such as Gaussian, Poisson, salt-and-pepper noise, etc., with different parameters (for instance: mean and standard deviation to the raw signal; (2) Convert EEG signals to sequences of images and add noise to the images. In this paper, adding noise to the raw signal is used.
This study proposes a new approach based on EEG signals for discriminating between AD, MCI, and HC in two-class and three-class classifications. This method is based on PSD features and the SVM classifier. In addition, the dataset in this study has a class imbalance problem (CIP), which is a severely imbalanced distribution of classes. In the case of machine learning, the majority class overwhelms the minority class. The majority class gradient component is substantially longer than the minority class. As a result, the majority class dominates weight upgrades more than the other classes, resulting in a rapid decrease in the majority class error and an increase in the minority class error. Oversampling, down-sampling, and algorithmic-level methods such as the threshold-moving technique and cost-sensitive learning are among the most common CIP handling methods. This research focuses on data augmentation strategies [
16], which are a type of excessive sampling methodology. Data augmentation is the process of creating new samples to supplement current data sets and improve classification or regression accuracy and stability [
17]. It usually generates additional samples from lesser-known classes to match the number of samples in each class, resulting in new data collection. One of the major challenges for machine-learning algorithms that operate under the assumption that data is evenly distributed across classes is class imbalance in imbalanced AD data. For the first time in the AD domain, different approaches to data augmentation—noise addition [
16], VAE-based data production [
14], and hybrid—are used independently and evaluated in this study, which is based on classic methods and deep neural networks.
The following sections of the paper are organized as follows.
Section 2 describes the studied dataset and explains the signal pre-processing, feature extraction, and classification methods used in this paper.
Section 3 displays the results of the study, and
Section 4 contains the discussion. Finally,
Section 5 presents the conclusions and further work.
4. Discussion
Analyzing EEG signals is a quick, inexpensive, and widely available method with a high temporal resolution that enables the research of the dynamic processes involved in the control of the intricate brain functioning system [
23]. As a result, it can be used to detect cognitive impairment in the early stages of major neurocognitive disorders, such as Alzheimer’s disease and MCI [
24]. In order to identify appropriate biomarkers for the early detection of severe neurocognitive disorders such as Alzheimer’s, the study of EEG signal data has therefore grown in importance in recent years [
25]. Another important use of EEG signal data is the easier and more accurate differentiation of neurocognitive disorders from MCI. Due to the difficulty in differentiating between the two (AD and MCI) in the clinic and the similarities in the clinical picture, clinicians in this field, including psychiatrists and neurologists, are occasionally unable to distinguish between them accurately using only an interview and clinical and cognitive evaluation [
26]. Techniques that can aid clinicians in the diagnosis and assist them in making better clinical decision assessments seem to be vital.
According to clinicians in this field, the clinical significance of this distinction and the more precise diagnosis of the two is that the treatment approaches for these two disorders are dissimilar. On the other hand, beginning treatment for Alzheimer’s disease as soon as possible helps to better control the disease and slow the progression of cognitive and behavioral symptoms. It can be a challenge to distinguish between persons who are entirely healthy and have cognitive complaints and those who have MCI since cognitive symptoms can often overlap in patients with different stages of the disease and are on a continuous spectrum.
To improve the accuracy of this distinction as much as possible, a variety of machine learning techniques have been applied in recent studies. Therefore, any technique that aids clinicians in making a more precise clinical diagnosis in this field will have clinical benefits. As a result, the current study’s goal was to apply various machine learning approaches to analyze and interpret the data generated from EEG signals in order to distinguish between Alzheimer’s disease, MCI, and the healthy population. Accuracy and F1scores were the two metrics employed in this study, and the results were interpreted based on these two parameters at intervals of 5, 10, and 15 s. As previously mentioned, three different data augmentation techniques were used in the field of machine learning to evaluate and interpret the data. Finally, they were compared in terms of accuracy in differentiating the three groups. This is due to the obvious difference in the amount of data in the different groups.
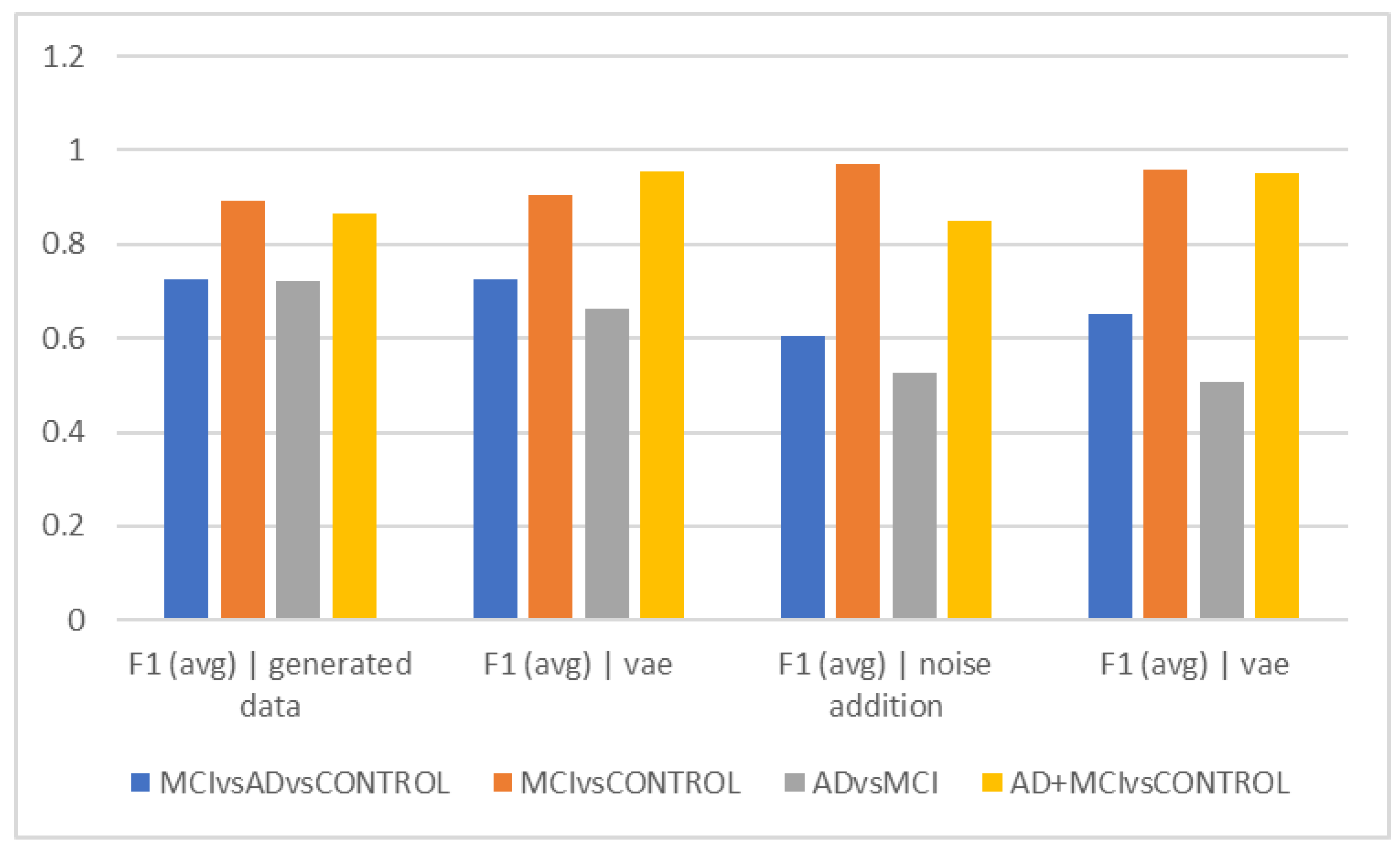
As shown in
Figure 2a, using the noise-addition technique, the HC and MCI could be distinguished with the highest degree of accuracy (acc = 97.22%), while the Alzheimer’s group and MCI could be distinguished with the lowest level of accuracy (acc = 50.68%) using the hybrid technique. Therefore, it seems that the clinical challenge for therapists in differentiating between these two groups also exists in the interpretation of EEG findings. When interpreting the findings, it may be claimed that it appears that the MCI group’s attributes are so distinct from those of healthy individuals that their differentiation is carried out as precisely as possible. However, it has been difficult to distinguish MCI patients from Alzheimer’s patients due to the similarities in their characteristics and EEG findings. In other words, the EEG alterations in these people are more similar to those in Alzheimer’s patients and clearly different from the group of healthy people. The spectrum model of cognitive impairment, which sees MCI as a transitional state between healthy people and those with Alzheimer’s disease, may need to be updated in light of this. This finding can prove that about half of the patients with MCI also receive a diagnosis of Alzheimer’s disease within a period of 4 years [
14,
16,
27,
28]. On the other hand, this issue might be caused by the type of samples used in the dataset, as there might be individuals with more severe MCI who are more similar to individuals with milder types of Alzheimer’s disease in terms of features.
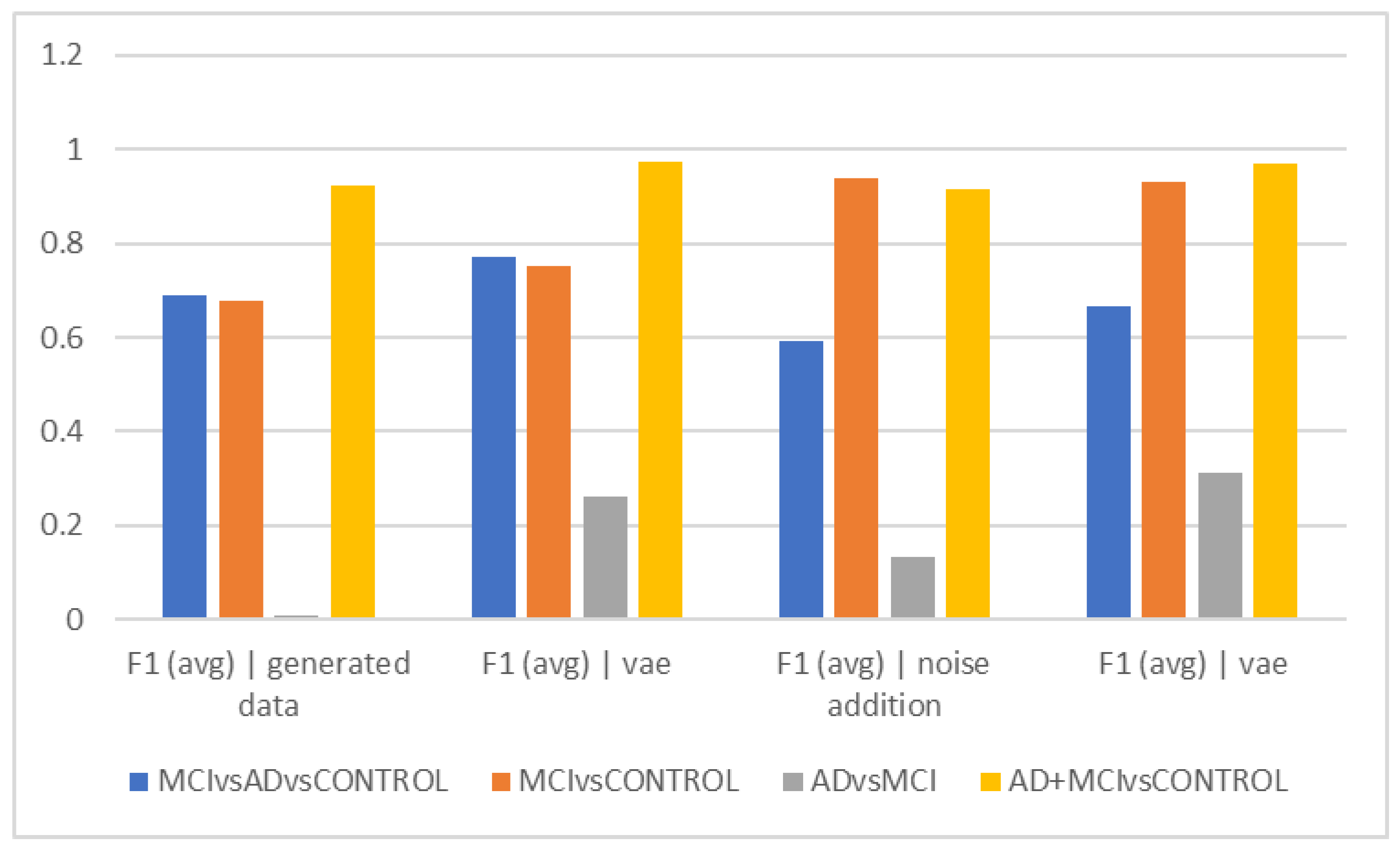
The best accuracy (72.08%) in separating the Alzheimer’s group from MCI was obtained when no data augmentation technique was applied, which can suggest that the data augmentation techniques utilized by machine learning are unable to differentiate between these two groups. The F1Score was used to compare groups in 5-s epochs, and in fact, we also took the data’s distribution into account (
Figure 2d). The ability to distinguish the Alzheimer’s group from MCI was less than in
Figure 2a, despite the general trend of the F1Score in separating groups being similar to
Figure 2a. According to its interpretation, the accuracy of differentiating these two groups decreased when the heterogeneity of the groups and data was taken into consideration using the F1Score. According to F1Score, the biggest problem was in differentiating the Alzheimer’s group from MCI, and when VAEs and hybrid techniques were used, higher scores were obtained in this area.
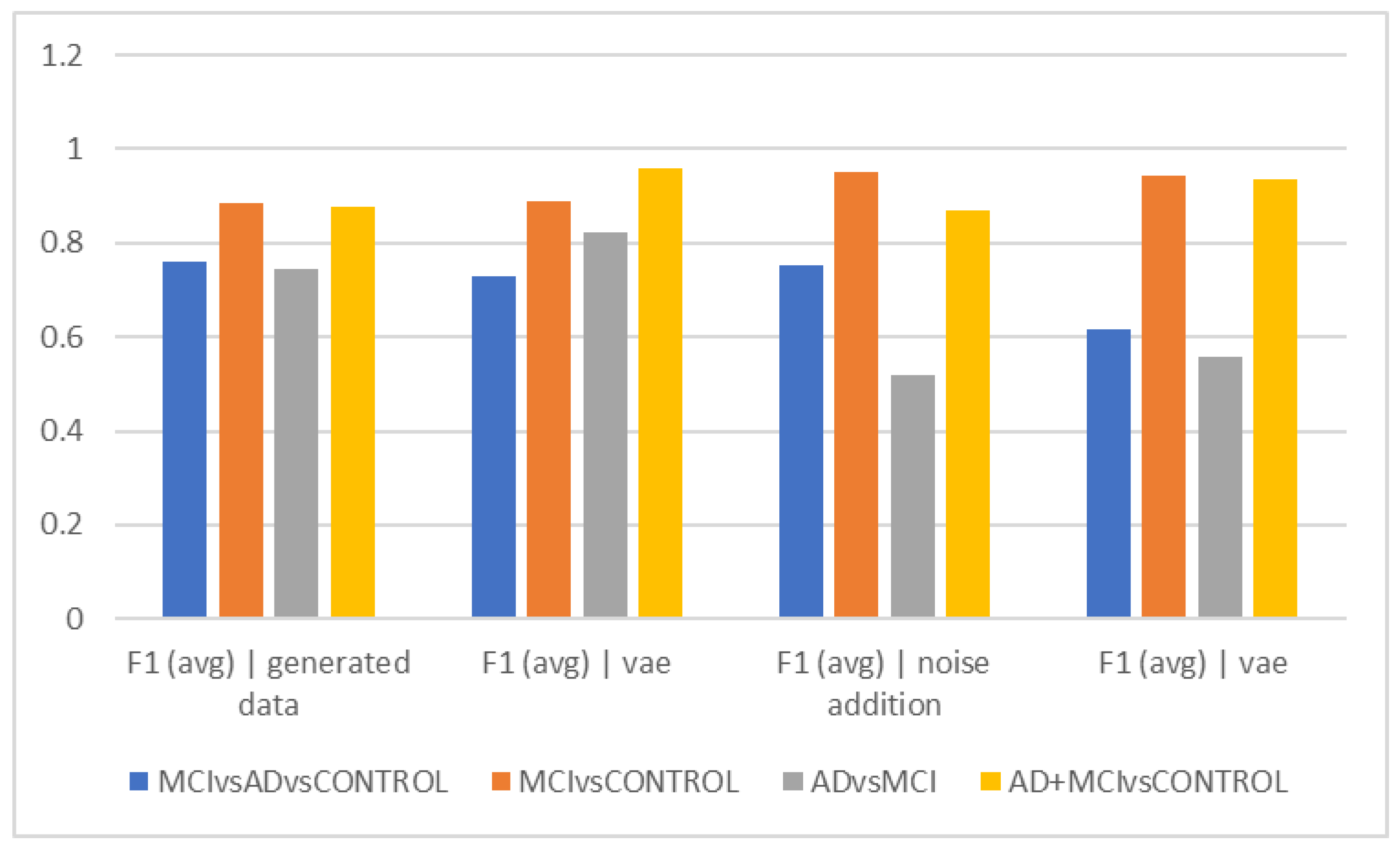
Figure 2b shows that the graph’s overall trend in epochs of 10 s is comparable to
Figure 2a. This indicates that the differentiation of the Alzheimer’s and MCI groups still has the lowest accuracy, and the VAE method worked with higher accuracy in this differentiation. The highest accuracy of differentiation is in differentiating healthy people from the total of Alzheimer’s and MCI people, and the VAEs method still performed the best. The interpretation of the above finding can again points to the greater similarity of the two groups of Alzheimer’s and MCI in the EEG findings and the more obvious difference with healthy people.
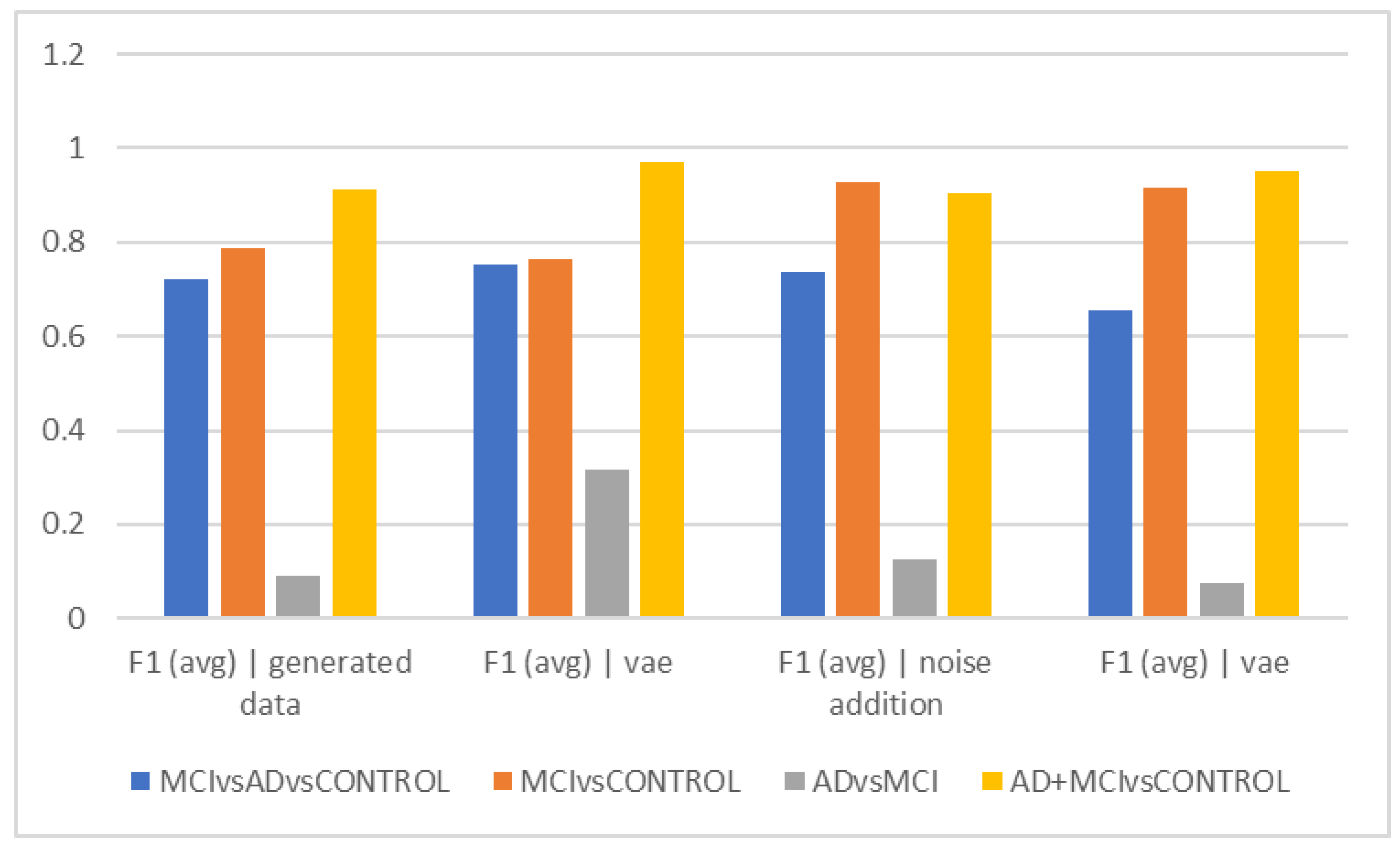
The VAE method is more effective than other ways in differentiating AD and MCI, according to the interpretation of
Figure 2e, which looks at the F1Score at ten-second intervals. However, it is estimated to be less than the accuracy because of the data imbalance in this F1Score difference.
Figure 2c demonstrates that similar to the previous figures, the accuracy trend in 15-s epochs is similar, with the maximum accuracy in differentiating between the normal and MCI groups as well as between healthy individuals and the sum of the other two groups. Still, distinguishing between those with Alzheimer’s disease and those with MCI remains the most difficult and least accurate task; in these cases, approaches that did not require more data and then the VAE method performed better. Based on the F1Score, hybrid and noise-addition techniques performed better in
Figure 2f in differentiating between healthy individuals and MCI. Additionally, the hybrid approach had better effectiveness in separating MCI from Alzheimer’s disease.
It is clear from a thorough examination of all the results that using data augmentation techniques often improves the accuracy of distinguishing between groups. Hence, using these techniques is advised in future research. On the other hand, no appreciable differences between time intervals were found that would affect how well different data augmentation techniques differentiate between classes. The hybrid technique shows low accuracy in almost all trials when separating MCI from Alzheimer’s. One of the potential causes of the low accuracy of the model is that there are not many actual MCI patients in the data set and the model has not been trained on a wide range of MCI patients. Alzheimer’s patients are described as having moderate severity in the dataset’s description, which suggests that they may function similarly to and closely resemble people with more severe MCI. As a result of the variety of MCI patients, it is suggested to incorporate as many cases of this type as possible in the dataset. By doing so, the effect of this variety on the stages of the MCI samples can be reduced, and the model can more accurately distinguish between MCI and Alzheimer’s disease by taking into account MCI cases with varying severity. Based on our current knowledge in the field of classical machine learning projects that have used feature extraction (pca) and classical machine learning classifiers (svm), there is no similar study that has used data augmentation methods to improve accuracy. Therefore, it is not possible to compare the results of our study with previous works in this field.
The biggest difficulty in this study is differentiating MCI from Alzheimer’s. On the other hand, it is a challenge for clinicians to differentiate between the two. The VAE method has typically been able to achieve this distinction more effectively than alternative techniques. Therefore, the method of increasing data based on VAEs may be introduced as an effective parameter in increasing the accuracy of differentiating MCI from Alzheimer’s. The ability to distinguish between individuals with minor cognitive issues such as MCI, more serious ones such as Alzheimer’s, and healthy people is another benefit of adopting these techniques. The significance of this distinction for therapists is to prevent classifying healthy people as sick while also ensuring that those who have cognitive issues of any degree are identified quickly and are given the appropriate treatment. Therefore, the effectiveness of these approaches to distinguish between individuals with MCI and Alzheimer’s disease and the general population was also examined in this study. The VAE approach has been more successful in this differentiation when looking broadly at several time frames. Therefore, this approach is suggested when attempting to separate healthy individuals from those who have cognitive issues of any kind. Due to the lack of serious cognitive symptoms in the early stages of MCI, it may be challenging for clinicians to distinguish patients with MCI from healthy individuals. Noise addition and hybrid methods have consistently outperformed other ways of separating a group of healthy individuals from MCI, and in difficult cases in this field, these two strategies appear to be more effective than others. The originality of this work is that it used data augmentation techniques to improve classification accuracy, which have not been used in previous classical machine learning studies that use PCA feature extraction and traditional SVM machine learning classifiers. Approaches such as GAN and VAE have been used to augment synthetic EEG data in methods that use deep learning models to distinguish between classes.
One of the study’s limitations is the imbalanced dataset and lack of variation, particularly in the MCI group, where the abundance of features causes an imbalanced distribution of data for analysis. Future research should take into account the fact that disease symptoms, etiology, and kind of treatment might vary widely, which may have had an impact on the interpreted EEG data. Additionally, disease symptoms may change with age, which leads to changes in EEG features. It is proposed that more emphasis be given to methods that address this issue in future research, given that the primary challenge is to reliably distinguish the Alzheimer’s group from MCI (which, of course, is a challenge for therapists in the clinic as well). This issue can be resolved by collecting datasets more frequently and with a wider variety, as well as by using machine learning techniques for data augmentation (such as the VAE method this paper proposes).