Clinical Evidence of Circulating Tumor DNA Application in Aggressive Breast Cancer
Abstract
1. Introduction
2. General Overview of Cell-Free DNA
3. The Importance of Testing for Circulating Tumor DNA
4. Analytical and Clinical Validity of Cfdna
5. Gene Analysis
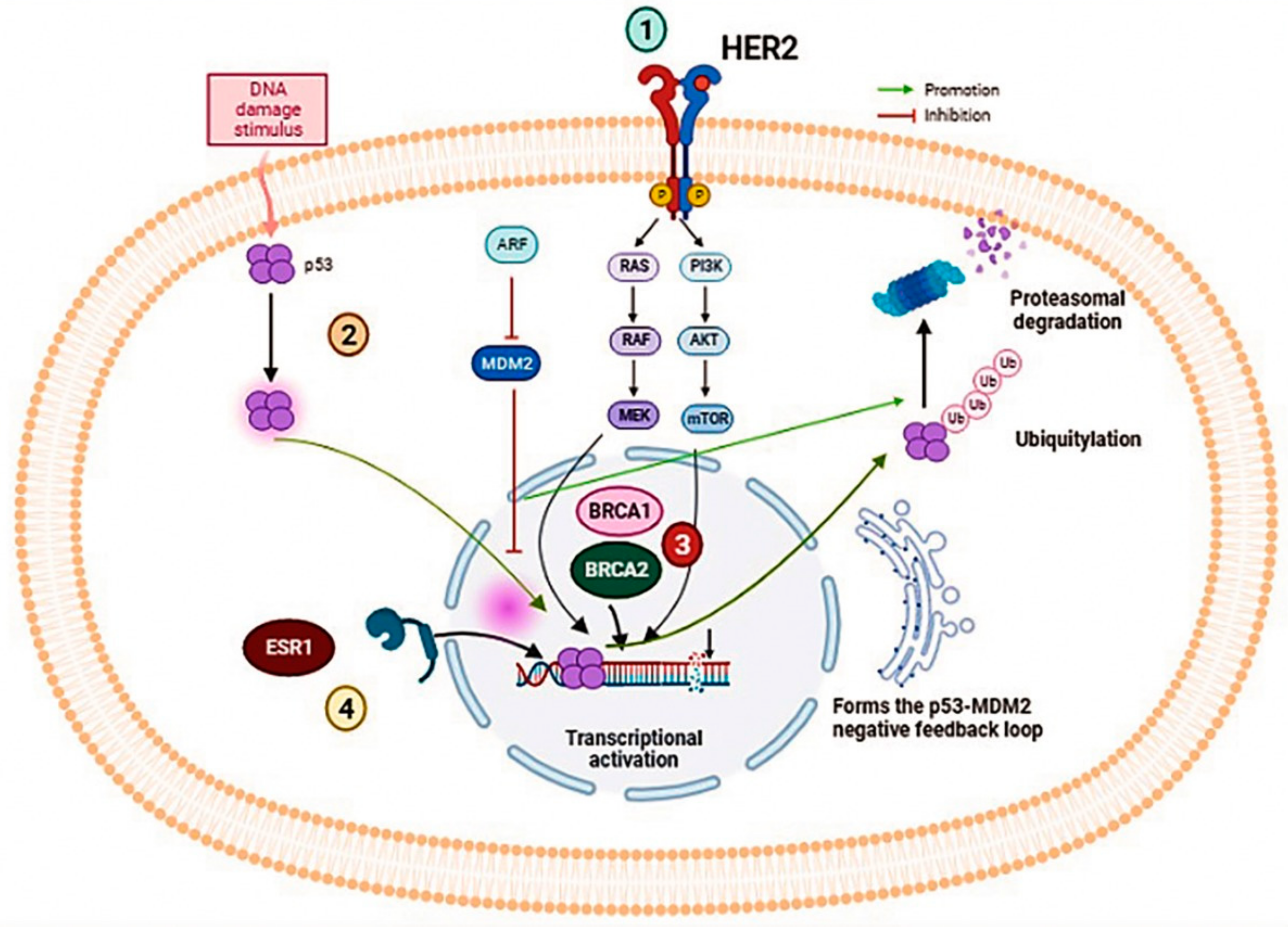
5.1. BRCA1/2 Genes
5.2. ESR1 Gene
5.3. PIK3CA Gene
5.4. TP53 Gene
5.5. ERBB2 Gene
6. Conclusions
7. Perspectives
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Elbasheer, M.M.A.; Alkhidir, A.G.A.; Mohammed, S.M.A.; Abbas, A.A.H.; Mohamed, A.O.; Bereir, I.M.; Abdalazeez, H.R.; Noma, M. Spatial distribution of breast cancer in Sudan 2010–2016. PLoS ONE 2019, 14, e0211085. [Google Scholar] [CrossRef] [PubMed]
- Beckmann, M.W.; Niederacher, D.; Schnürch, H.G.; Gusterson, B.A.; Bender, H.G. Multistep carcinogenesis of breast cancer and tumour heterogeneity. J. Mol. Med. Berl. Ger. 1997, 75, 429–439. [Google Scholar] [CrossRef] [PubMed]
- Tsang, J.Y.S.; Tse, G.M. Molecular Classification of Breast Cancer. Adv. Anat. Pathol. 2020, 27, 27–35. [Google Scholar] [CrossRef] [PubMed]
- Barzaman, K.; Karami, J.; Zarei, Z.; Hosseinzadeh, A.; Kazemi, M.H.; Moradi-Kalbolandi, S.; Safari, E.; Farahmand, L. Breast cancer: Biology, biomarkers, and treatments. Int. Immunopharmacol. 2020, 84, 106535. [Google Scholar] [CrossRef]
- Alimirzaie, S.; Bagherzadeh, M.; Akbari, M.R. Liquid biopsy in breast cancer: A comprehensive review. Clin. Genet. 2019, 95, 643–660. [Google Scholar] [CrossRef]
- Asleh, K.; Riaz, N.; Nielsen, T.O. Heterogeneity of triple negative breast cancer: Current advances in subtyping and treatment implications. J. Exp. Clin. Cancer Res. 2022, 41, 265. [Google Scholar] [CrossRef]
- Ma, L.; Liang, Z.; Zhou, H.; Qu, L. Applications of RNA Indexes for Precision Oncology in Breast Cancer. Genomics Proteomics Bioinformatics 2018, 16, 108–119. [Google Scholar] [CrossRef]
- Shang, M.; Chang, C.; Pei, Y.; Guan, Y.; Chang, J.; Li, H. Potential Management of Circulating Tumor DNA as a Biomarker in Triple-Negative Breast Cancer. J. Cancer 2018, 9, 4627–4634. [Google Scholar] [CrossRef]
- Ignatiadis, M.; Sledge, G.W.; Jeffrey, S.S. Liquid biopsy enters the clinic-implementation issues and future challenges. Nat. Rev. Clin. Oncol. 2021, 18, 297–312. [Google Scholar] [CrossRef]
- Mandel, P.; Metais, P. Nuclear Acids In Human Blood Plasma. Comptes Rendus Seances Soc. Biol. Fil. 1948, 142, 241–243. [Google Scholar]
- Leon, S.A.; Shapiro, B.; Sklaroff, D.M.; Yaros, M.J. Free DNA in the serum of cancer patients and the effect of therapy. Cancer Res. 1977, 37, 646–650. [Google Scholar]
- Lo, Y.M.; Corbetta, N.; Chamberlain, P.F.; Rai, V.; Sargent, I.L.; Redman, C.W.; Wainscoat, J.S. Presence of fetal DNA in maternal plasma and serum. Lancet 1997, 350, 485–487. [Google Scholar] [CrossRef]
- Chiu, R.W.K.; Akolekar, R.; Zheng, Y.W.L.; Leung, T.Y.; Sun, H.; Chan, K.C.A.; Lun, F.M.F.; Go, A.T.J.I.; Lau, E.T.; To, W.W.K.; et al. Non-invasive prenatal assessment of trisomy 21 by multiplexed maternal plasma DNA sequencing: Large scale validity study. BMJ 2011, 342, c7401. [Google Scholar] [CrossRef]
- Castelli, J.; Cabel, L.; Bidard, F.-C.; Duvergé, L.; Bachet, J.-B. ADN tumoral circulant: Principes, applications actuelles en radiothérapie et développement futur. Cancer/Radiothérapie 2018, 22, 653–659. [Google Scholar] [CrossRef]
- Cervena, K.; Vodicka, P.; Vymetalkova, V. Diagnostic and prognostic impact of cell-free DNA in human cancers: Systematic review. Mutat. Res. Rev. Mutat. Res. 2019, 781, 100–129. [Google Scholar] [CrossRef]
- Bettegowda, C.; Sausen, M.; Leary, R.J.; Kinde, I.; Wang, Y.; Agrawal, N.; Bartlett, B.R.; Wang, H.; Luber, B.; Alani, R.M.; et al. Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci. Transl. Med. 2014, 6, 224ra24. [Google Scholar] [CrossRef]
- De Vlaminck, I.; Martin, L.; Kertesz, M.; Patel, K.; Kowarsky, M.; Strehl, C.; Cohen, G.; Luikart, H.; Neff, N.F.; Okamoto, J.; et al. Noninvasive monitoring of infection and rejection after lung transplantation. Proc. Natl. Acad. Sci. USA 2015, 112, 13336–13341. [Google Scholar] [CrossRef]
- Rainer, T.H.; Wong, L.K.S.; Lam, W.; Yuen, E.; Lam, N.Y.L.; Metreweli, C.; Lo, Y.M.D. Prognostic use of circulating plasma nucleic acid concentrations in patients with acute stroke. Clin. Chem. 2003, 49, 562–569. [Google Scholar] [CrossRef]
- Chang, C.P.-Y.; Chia, R.-H.; Wu, T.-L.; Tsao, K.-C.; Sun, C.-F.; Wu, J.T. Elevated cell-free serum DNA detected in patients with myocardial infarction. Clin. Chim. Acta Int. J. Clin. Chem. 2003, 327, 95–101. [Google Scholar] [CrossRef]
- Rodrigues Filho, E.M.; Simon, D.; Ikuta, N.; Klovan, C.; Dannebrock, F.A.; Oliveira de Oliveira, C.; Regner, A. Elevated Cell-Free Plasma DNA Level as an Independent Predictor of Mortality in Patients with Severe Traumatic Brain Injury. J. Neurotrauma 2014, 31, 1639–1646. [Google Scholar] [CrossRef]
- Yao, W.; Mei, C.; Nan, X.; Hui, L. Evaluation and comparison of in vitro degradation kinetics of DNA in serum, urine and saliva: A qualitative study. Gene 2016, 590, 142–148. [Google Scholar] [CrossRef] [PubMed]
- Su, Y.-H.; Wang, M.; Brenner, D.E.; Norton, P.A.; Block, T.M. Detection of Mutated K-ras DNA in Urine, Plasma, and Serum of Patients with Colorectal Carcinoma or Adenomatous Polyps. Ann. N. Y. Acad. Sci. 2008, 1137, 197–206. [Google Scholar] [CrossRef] [PubMed]
- Botezatu, I.; Serdyuk, O.; Potapova, G.; Shelepov, V.; Alechina, R.; Molyaka, Y.; Ananév, V.; Bazin, I.; Garin, A.; Narimanov, M.; et al. Genetic analysis of DNA excreted in urine: A new approach for detecting specific genomic DNA sequences from cells dying in an organism. Clin. Chem. 2000, 46, 1078–1084. [Google Scholar] [CrossRef] [PubMed]
- Pan, W.; Gu, W.; Nagpal, S.; Gephart, M.H.; Quake, S.R. Brain tumor mutations detected in cerebral spinal fluid. Clin. Chem. 2015, 61, 514–522. [Google Scholar] [CrossRef] [PubMed]
- Sriram, K.B.; Relan, V.; Clarke, B.E.; Duhig, E.E.; Windsor, M.N.; Matar, K.S.; Naidoo, R.; Passmore, L.; McCaul, E.; Courtney, D.; et al. Pleural fluid cell-free DNA integrity index to identify cytologically negative malignant pleural effusions including mesotheliomas. BMC Cancer 2012, 12, 428. [Google Scholar] [CrossRef]
- Thierry, A.R.; El Messaoudi, S.; Gahan, P.B.; Anker, P.; Stroun, M. Origins, structures, and functions of circulating DNA in oncology. Cancer Metastasis Rev. 2016, 35, 347–376. [Google Scholar] [CrossRef]
- Ranucci, R. Cell-Free DNA: Applications in Different Diseases. Methods Mol. Biol. 2019, 1909, 3–12. [Google Scholar] [CrossRef]
- Aucamp, J.; Bronkhorst, A.J.; Badenhorst, C.P.S.; Pretorius, P.J. The diverse origins of circulating cell-free DNA in the human body: A critical re-evaluation of the literature. Biol. Rev. Camb. Philos. Soc. 2018, 93, 1649–1683. [Google Scholar] [CrossRef]
- Lui, Y.Y.N.; Chik, K.-W.; Chiu, R.W.K.; Ho, C.-Y.; Lam, C.W.K.; Lo, Y.M.D. Predominant hematopoietic origin of cell-free DNA in plasma and serum after sex-mismatched bone marrow transplantation. Clin. Chem. 2002, 48, 421–427. [Google Scholar] [CrossRef]
- Swarup, V.; Rajeswari, M.R. Circulating (cell-free) nucleic acids—A promising, non-invasive tool for early detection of several human diseases. FEBS Lett. 2007, 581, 795–799. [Google Scholar] [CrossRef]
- Oellerich, M.; Schütz, E.; Beck, J.; Kanzow, P.; Plowman, P.N.; Weiss, G.J.; Walson, P.D. Using circulating cell-free DNA to monitor personalized cancer therapy. Crit. Rev. Clin. Lab. Sci. 2017, 54, 205–218. [Google Scholar] [CrossRef]
- Gould, T.J.; Lysov, Z.; Liaw, P.C. Extracellular DNA and histones: Double-edged swords in immunothrombosis. J. Thromb. Haemost. 2015, 13 (Suppl. S1), S82–S91. [Google Scholar] [CrossRef]
- Liu, Z.; Wang, Z.; Jia, E.; Ouyang, T.; Pan, M.; Lu, J.; Ge, Q.; Bai, Y. Analysis of genome-wide in cell free DNA methylation: Progress and prospect. Analyst 2019, 144, 5912–5922. [Google Scholar] [CrossRef]
- García-Olmo, D.C.; Domínguez, C.; García-Arranz, M.; Anker, P.; Stroun, M.; García-Verdugo, J.M.; García-Olmo, D. Cell-free nucleic acids circulating in the plasma of colorectal cancer patients induce the oncogenic transformation of susceptible cultured cells. Cancer Res. 2010, 70, 560–567. [Google Scholar] [CrossRef]
- Tsaliki, E.; Papageorgiou, E.A.; Spyrou, C.; Koumbaris, G.; Kypri, E.; Kyriakou, S.; Sotiriou, C.; Touvana, E.; Keravnou, A.; Karagrigoriou, A.; et al. MeDIP real-time qPCR of maternal peripheral blood reliably identifies trisomy 21. Prenat. Diagn. 2012, 32, 996–1001. [Google Scholar] [CrossRef]
- Gil, M.M.; Quezada, M.S.; Revello, R.; Akolekar, R.; Nicolaides, K.H. Analysis of cell-free DNA in maternal blood in screening for fetal aneuploidies: Updated meta-analysis. Ultrasound Obstet. Gynecol. Off. J. Int. Soc. Ultrasound Obstet. Gynecol. 2015, 45, 249–266. [Google Scholar] [CrossRef]
- Gielis, E.M.; Ledeganck, K.J.; De Winter, B.Y.; Del Favero, J.; Bosmans, J.-L.; Claas, F.H.J.; Abramowicz, D.; Eikmans, M. Cell-Free DNA: An Upcoming Biomarker in Transplantation. Am. J. Transplant. Off. J. Am. Soc. Transplant. Am. Soc. Transpl. Surg. 2015, 15, 2541–2551. [Google Scholar] [CrossRef]
- Schütz, E.; Fischer, A.; Beck, J.; Harden, M.; Koch, M.; Wuensch, T.; Stockmann, M.; Nashan, B.; Kollmar, O.; Matthaei, J.; et al. Graft-derived cell-free DNA, a noninvasive early rejection and graft damage marker in liver transplantation: A prospective, observational, multicenter cohort study. PLoS Med. 2017, 14, e1002286. [Google Scholar] [CrossRef]
- Fan, Y.; Shi, M.; Chen, S.; Ju, G.; Chen, L.; Lu, H.; Chen, J.; Zheng, S. Analysis of serum cfDNA concentration and integrity before and after surgery in patients with lung cancer. Cell. Mol. Biol. Noisy Gd. Fr. 2019, 65, 56–63. [Google Scholar] [CrossRef]
- Keller, L.; Belloum, Y.; Wikman, H.; Pantel, K. Clinical relevance of blood-based ctDNA analysis: Mutation detection and beyond. Br. J. Cancer 2021, 124, 345–358. [Google Scholar] [CrossRef]
- Franczak, C.; Filhine-Tressarieu, P.; Broséus, J.; Gilson, P.; Merlin, J.-L.; Harlé, A. Clinical Interest of Circulating Tumor DNA in Oncology. Arch. Med. Res. 2018, 49, 297–305. [Google Scholar] [CrossRef] [PubMed]
- Ulrich, B.C.; Paweletz, C.P. Cell-Free DNA in Oncology: Gearing up for Clinic. Ann. Lab. Med. 2018, 38, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Han, X.; Sun, Y. DNA methylation signatures in circulating cell-free DNA as biomarkers for the early detection of cancer. Sci. China Life Sci. 2017, 60, 356–362. [Google Scholar] [CrossRef] [PubMed]
- Dawson, S.-J.; Tsui, D.W.Y.; Murtaza, M.; Biggs, H.; Rueda, O.M.; Chin, S.-F.; Dunning, M.J.; Gale, D.; Forshew, T.; Mahler-Araujo, B.; et al. Analysis of circulating tumor DNA to monitor metastatic breast cancer. N. Engl. J. Med. 2013, 368, 1199–1209. [Google Scholar] [CrossRef] [PubMed]
- Diaz, L.A.; Bardelli, A. Liquid biopsies: Genotyping circulating tumor DNA. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 2014, 32, 579–586. [Google Scholar] [CrossRef]
- Oxnard, G.R.; Paweletz, C.P.; Kuang, Y.; Mach, S.L.; O’Connell, A.; Messineo, M.M.; Luke, J.J.; Butaney, M.; Kirschmeier, P.; Jackman, D.M.; et al. Noninvasive detection of response and resistance in EGFR-mutant lung cancer using quantitative next-generation genotyping of cell-free plasma DNA. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2014, 20, 1698–1705. [Google Scholar] [CrossRef]
- Jamal-Hanjani, M.; Wilson, G.A.; Horswell, S.; Mitter, R.; Sakarya, O.; Constantin, T.; Salari, R.; Kirkizlar, E.; Sigurjonsson, S.; Pelham, R.; et al. Detection of ubiquitous and heterogeneous mutations in cell-free DNA from patients with early-stage non-small-cell lung cancer. Ann. Oncol. Off. J. Eur. Soc. Med. Oncol. 2016, 27, 862–867. [Google Scholar] [CrossRef]
- Qiao, L.; Yu, B.; Liang, Y.; Zhang, C.; Wu, X.; Xue, Y.; Shen, C.; He, Q.; Lu, J.; Xiang, J.; et al. Sequencing shorter cfDNA fragments improves the fetal DNA fraction in noninvasive prenatal testing. Am. J. Obstet. Gynecol. 2019, 221, 345.e1–345.e11. [Google Scholar] [CrossRef]
- Streubel, A.; Stenzinger, A.; Stephan-Falkenau, S.; Kollmeier, J.; Misch, D.; Blum, T.G.; Bauer, T.; Landt, O.; Am Ende, A.; Schirmacher, P.; et al. Comparison of different semi-automated cfDNA extraction methods in combination with UMI-based targeted sequencing. Oncotarget 2019, 10, 5690–5702. [Google Scholar] [CrossRef]
- Domínguez-Vigil, I.G.; Moreno-Martínez, A.K.; Wang, J.Y.; Roehrl, M.H.A.; Barrera-Saldaña, H.A. The dawn of the liquid biopsy in the fight against cancer. Oncotarget 2018, 9, 2912–2922. [Google Scholar] [CrossRef]
- Perakis, S.; Speicher, M.R. Emerging concepts in liquid biopsies. BMC Med. 2017, 15, 75. [Google Scholar] [CrossRef]
- Wan, J.C.M.; Massie, C.; Garcia-Corbacho, J.; Mouliere, F.; Brenton, J.D.; Caldas, C.; Pacey, S.; Baird, R.; Rosenfeld, N. Liquid biopsies come of age: Towards implementation of circulating tumour DNA. Nat. Rev. Cancer 2017, 17, 223–238. [Google Scholar] [CrossRef]
- Boldrin, E.; Nardo, G.; Zulato, E.; Bonanno, L.; Polo, V.; Frega, S.; Pavan, A.; Indraccolo, S.; Saggioro, D. Detection of Loss of Heterozygosity in cfDNA of Advanced EGFR- or KRAS-Mutated Non-Small-Cell Lung Cancer Patients. Int. J. Mol. Sci. 2019, 21, 66. [Google Scholar] [CrossRef]
- Fleischhacker, M.; Schmidt, B. Circulating nucleic acids (CNAs) and cancer—A survey. Biochim. Biophys. Acta 2007, 1775, 181–232. [Google Scholar] [CrossRef]
- Luo, H.; Wei, W.; Ye, Z.; Zheng, J.; Xu, R.-H. Liquid Biopsy of Methylation Biomarkers in Cell-Free DNA. Trends Mol. Med. 2021, 27, 482–500. [Google Scholar] [CrossRef]
- Lo, Y.M.; Zhang, J.; Leung, T.N.; Lau, T.K.; Chang, A.M.; Hjelm, N.M. Rapid clearance of fetal DNA from maternal plasma. Am. J. Hum. Genet. 1999, 64, 218–224. [Google Scholar] [CrossRef]
- Reinert, T.; Schøler, L.V.; Thomsen, R.; Tobiasen, H.; Vang, S.; Nordentoft, I.; Lamy, P.; Kannerup, A.-S.; Mortensen, F.V.; Stribolt, K.; et al. Analysis of circulating tumour DNA to monitor disease burden following colorectal cancer surgery. Gut 2016, 65, 625–634. [Google Scholar] [CrossRef]
- Diehl, F.; Schmidt, K.; Choti, M.A.; Romans, K.; Goodman, S.; Li, M.; Thornton, K.; Agrawal, N.; Sokoll, L.; Szabo, S.A.; et al. Circulating mutant DNA to assess tumor dynamics. Nat. Med. 2008, 14, 985–990. [Google Scholar] [CrossRef]
- Alix-Panabières, C.; Pantel, K. Clinical Applications of Circulating Tumor Cells and Circulating Tumor DNA as Liquid Biopsy. Cancer Discov. 2016, 6, 479–491. [Google Scholar] [CrossRef]
- Stroun, M.; Maurice, P.; Vasioukhin, V.; Lyautey, J.; Lederrey, C.; Lefort, F.; Rossier, A.; Chen, X.Q.; Anker, P. The origin and mechanism of circulating DNA. Ann. N. Y. Acad. Sci. 2000, 906, 161–168. [Google Scholar] [CrossRef]
- Stewart, C.M.; Kothari, P.D.; Mouliere, F.; Mair, R.; Somnay, S.; Benayed, R.; Zehir, A.; Weigelt, B.; Dawson, S.-J.; Arcila, M.E.; et al. The value of cell-free DNA for molecular pathology. J. Pathol. 2018, 244, 616–627. [Google Scholar] [CrossRef] [PubMed]
- Elshimali, Y.I.; Khaddour, H.; Sarkissyan, M.; Wu, Y.; Vadgama, J.V. The Clinical Utilization of Circulating Cell Free DNA (CCFDNA) in Blood of Cancer Patients. Int. J. Mol. Sci. 2013, 14, 18925–18958. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Dittmar, R.L.; Xia, S.; Zhang, H.; Du, M.; Huang, C.-C.; Druliner, B.R.; Boardman, L.; Wang, L. Cell-free DNA copy number variations in plasma from colorectal cancer patients. Mol. Oncol. 2017, 11, 1099–1111. [Google Scholar] [CrossRef] [PubMed]
- Heitzer, E.; Ulz, P.; Geigl, J.B. Circulating tumor DNA as a liquid biopsy for cancer. Clin. Chem. 2015, 61, 112–123. [Google Scholar] [CrossRef]
- Chen, S.; Liu, M.; Zhou, Y. Bioinformatics Analysis for Cell-Free Tumor DNA Sequencing Data. Methods Mol. Biol. 2018, 1754, 67–95. [Google Scholar] [CrossRef]
- Miao, Y.; Fan, Y.; Zhang, L.; Ma, T.; Li, R. Clinical value of plasma cfDNA concentration and integrity in breast cancer patients. Cell. Mol. Biol. Noisy Gd. Fr. 2019, 65, 64–72. [Google Scholar] [CrossRef]
- Merker, J.D.; Oxnard, G.R.; Compton, C.; Diehn, M.; Hurley, P.; Lazar, A.J.; Lindeman, N.; Lockwood, C.M.; Rai, A.J.; Schilsky, R.L.; et al. Circulating Tumor DNA Analysis in Patients With Cancer: American Society of Clinical Oncology and College of American Pathologists Joint Review. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 2018, 36, 1631–1641. [Google Scholar] [CrossRef]
- Song, P.; Wu, L.R.; Yan, Y.H.; Zhang, J.X.; Chu, T.; Kwong, L.N.; Patel, A.A.; Zhang, D.Y. Limitations and opportunities of technologies for the analysis of cell-free DNA in cancer diagnostics. Nat. Biomed. Eng. 2022, 6, 232–245. [Google Scholar] [CrossRef]
- Callens, C.; Bidard, F.-C.; Curto-Taribo, A.; Trabelsi-Grati, O.; Melaabi, S.; Delaloge, S.; Hardy-Bessard, A.-C.; Bachelot, T.; Clatot, F.; De La Motte Rouge, T.; et al. Real-Time Detection of ESR1 Mutation in Blood by Droplet Digital PCR in the PADA-1 Trial: Feasibility and Cross-Validation with NGS. Anal. Chem. 2022, 94, 6297–6303. [Google Scholar] [CrossRef]
- Lin, C.; Liu, X.; Zheng, B.; Ke, R.; Tzeng, C.-M. Liquid Biopsy, ctDNA Diagnosis through NGS. Life 2021, 11, 890. [Google Scholar] [CrossRef]
- Smyth, L.M.; Reichel, J.B.; Tang, J.; Patel, J.A.A.; Meng, F.; Selcuklu, D.S.; Houck-Loomis, B.; You, D.; Samoila, A.; Schiavon, G.; et al. Utility of Serial cfDNA NGS for Prospective Genomic Analysis of Patients on a Phase I Basket Study. JCO Precis. Oncol. 2021, 5, 6–16. [Google Scholar] [CrossRef]
- Chandarlapaty, S.; Chen, D.; He, W.; Sung, P.; Samoila, A.; You, D.; Bhatt, T.; Patel, P.; Voi, M.; Gnant, M.; et al. Prevalence of ESR1 Mutations in Cell-Free DNA and Outcomes in Metastatic Breast Cancer: A Secondary Analysis of the BOLERO-2 Clinical Trial. JAMA Oncol. 2016, 2, 1310–1315. [Google Scholar] [CrossRef]
- Takeshita, T.; Yamamoto, Y.; Yamamoto-Ibusuki, M.; Tomiguchi, M.; Sueta, A.; Murakami, K.; Omoto, Y.; Iwase, H. Analysis of ESR1 and PIK3CA mutations in plasma cell-free DNA from ER-positive breast cancer patients. Oncotarget 2017, 8, 52142–52155. [Google Scholar] [CrossRef]
- Vidula, N.; Rich, T.A.; Sartor, O.; Yen, J.; Hardin, A.; Nance, T.; Lilly, M.B.; Nezami, M.A.; Patel, S.P.; Carneiro, B.A.; et al. Routine Plasma-Based Genotyping to Comprehensively Detect Germline, Somatic, and Reversion BRCA Mutations among Patients with Advanced Solid Tumors. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2020, 26, 2546–2555. [Google Scholar] [CrossRef]
- Weigelt, B.; Comino-Méndez, I.; de Bruijn, I.; Tian, L.; Meisel, J.L.; García-Murillas, I.; Fribbens, C.; Cutts, R.; Martelotto, L.G.; Ng, C.K.; et al. Diverse BRCA1 and BRCA2 Reversion Mutations in Circulating Cell-Free DNA of Therapy-Resistant Breast or Ovarian Cancer. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2017, 23, 6708–6720. [Google Scholar] [CrossRef]
- Del Re, M.; Crucitta, S.; Lorenzini, G.; De Angelis, C.; Diodati, L.; Cavallero, D.; Bargagna, I.; Cinacchi, P.; Fratini, B.; Salvadori, B.; et al. PI3K mutations detected in liquid biopsy are associated to reduced sensitivity to CDK4/6 inhibitors in metastatic breast cancer patients. Pharmacol. Res. 2021, 163, 105241. [Google Scholar] [CrossRef]
- Riva, F.; Bidard, F.-C.; Houy, A.; Saliou, A.; Madic, J.; Rampanou, A.; Hego, C.; Milder, M.; Cottu, P.; Sablin, M.-P.; et al. Patient-Specific Circulating Tumor DNA Detection during Neoadjuvant Chemotherapy in Triple-Negative Breast Cancer. Clin. Chem. 2017, 63, 691–699. [Google Scholar] [CrossRef]
- Savli, H.; Sertdemir, N.; Aydin, D.; Dursun, B.; Kurtas, O.; Reka, S.; Sunnetci-Akkoyunlu, D.; Eren-Keskin, S.; Uygun, K.; Ozden, E.; et al. TP53, EGFR and PIK3CA gene variations observed as prominent biomarkers in breast and lung cancer by plasma cell-free DNA genomic testing. J. Biotechnol. 2019, 300, 87–93. [Google Scholar] [CrossRef]
- Hu, Z.-Y.; Xie, N.; Tian, C.; Yang, X.; Liu, L.; Li, J.; Xiao, H.; Wu, H.; Lu, J.; Gao, J.; et al. Identifying Circulating Tumor DNA Mutation Profiles in Metastatic Breast Cancer Patients with Multiline Resistance. EBioMedicine 2018, 32, 111–118. [Google Scholar] [CrossRef]
- Ma, C.X.; Bose, R.; Gao, F.; Freedman, R.A.; Telli, M.L.; Kimmick, G.; Winer, E.; Naughton, M.; Goetz, M.P.; Russell, C.; et al. Neratinib Efficacy and Circulating Tumor DNA Detection of HER2 Mutations in HER2 Nonamplified Metastatic Breast Cancer. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2017, 23, 5687–5695. [Google Scholar] [CrossRef]
- Turner, N.C.; Kingston, B.; Kilburn, L.S.; Kernaghan, S.; Wardley, A.M.; Macpherson, I.R.; Baird, R.D.; Roylance, R.; Stephens, P.; Oikonomidou, O.; et al. Circulating tumour DNA analysis to direct therapy in advanced breast cancer (plasmaMATCH): A multicentre, multicohort, phase 2a, platform trial. Lancet Oncol. 2020, 21, 1296–1308. [Google Scholar] [CrossRef] [PubMed]
- Baretta, Z.; Mocellin, S.; Goldin, E.; Olopade, O.I.; Huo, D. Effect of BRCA germline mutations on breast cancer prognosis. Medicine (Baltimore) 2016, 95, e4975. [Google Scholar] [CrossRef] [PubMed]
- Pascual, T.; Gonzalez-Farre, B.; Teixidó, C.; Oleaga, L.; Oses, G.; Ganau, S.; Chic, N.; Riu, G.; Adamo, B.; Galván, P.; et al. Significant Clinical Activity of Olaparib in a Somatic BRCA1-Mutated Triple-Negative Breast Cancer with Brain Metastasis. JCO Precis. Oncol. 2019, 3, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Claus, E.B.; Risch, N.; Thompson, W.D. Genetic analysis of breast cancer in the cancer and steroid hormone study. Am. J. Hum. Genet. 1991, 48, 232–242. [Google Scholar]
- Wang, Y.; Cortez, D.; Yazdi, P.; Neff, N.; Elledge, S.J.; Qin, J. BASC, a super complex of BRCA1-associated proteins involved in the recognition and repair of aberrant DNA structures. Genes Dev. 2000, 14, 927–939. [Google Scholar] [CrossRef]
- Algebaly, A.S.; Suliman, R.S.; Al-Qahtani, W.S. Comprehensive study for BRCA1 and BRCA2 entire coding regions in breast cancer. Clin. Transl. Oncol. Off. Publ. Fed. Span. Oncol. Soc. Natl. Cancer Inst. Mex. 2021, 23, 74–81. [Google Scholar] [CrossRef]
- Cancer Genome Atlas Network Comprehensive molecular portraits of human breast tumours. Nature 2012, 490, 61–70. [CrossRef]
- Winter, C.; Nilsson, M.P.; Olsson, E.; George, A.M.; Chen, Y.; Kvist, A.; Törngren, T.; Vallon-Christersson, J.; Hegardt, C.; Häkkinen, J.; et al. Targeted sequencing of BRCA1 and BRCA2 across a large unselected breast cancer cohort suggests that one-third of mutations are somatic. Ann. Oncol. 2016, 27, 1532–1538. [Google Scholar] [CrossRef]
- Clatot, F.; Perdrix, A.; Sefrioui, D.; Sarafan-Vasseur, N.; Di Fiore, F. Intérêt clinique de la détection des mutations circulantes d’ESR1 chez les patientes traitées pour un cancer du sein métastatique exprimant les récepteurs hormonaux. Bull. Cancer (Paris) 2018, 105, 46–54. [Google Scholar] [CrossRef]
- Zundelevich, A.; Dadiani, M.; Kahana-Edwin, S.; Itay, A.; Sella, T.; Gadot, M.; Cesarkas, K.; Farage-Barhom, S.; Saar, E.G.; Eyal, E.; et al. ESR1 mutations are frequent in newly diagnosed metastatic and loco-regional recurrence of endocrine-treated breast cancer and carry worse prognosis. Breast Cancer Res. 2020, 22, 16. [Google Scholar] [CrossRef]
- Leiser, M. ESR1 Mutations Predict Survival, Endocrine Therapy Failure in Early Breast Cancer, 28 Avril 2020. Consulté le: 5 décembre 2022. [En ligne]. Available online: https://www.healio.com/news/hematology-oncology/20200428/esr1-mutations-predict-survival-endocrine-therapy-failure-in-early-breast-cancer (accessed on 24 September 2022).
- Zhu, W.; Ren, C.; Wang, Y.; Wen, L.; Zhang, G.; Liao, N. Prevalence of ESR1 Mutation in Chinese ER-Positive Breast Cancer. OncoTargets Ther. 2020, 13, 615–621. [Google Scholar] [CrossRef]
- Lefebvre, C.; Bachelot, T.; Filleron, T.; Pedrero, M.; Campone, M.; Soria, J.-C.; Massard, C.; Lévy, C.; Arnedos, M.; Lacroix-Triki, M.; et al. Mutational Profile of Metastatic Breast Cancers: A Retrospective Analysis. PLoS Med. 2016, 13, e1002201. [Google Scholar] [CrossRef]
- Shibayama, T.; Low, S.-K.; Ono, M.; Kobayashi, T.; Kobayashi, K.; Fukada, I.; Ito, Y.; Ueno, T.; Ohno, S.; Nakamura, Y.; et al. Clinical significance of gene mutation in ctDNA analysis for hormone receptor-positive metastatic breast cancer. Breast Cancer Res. Treat. 2020, 180, 331–341. [Google Scholar] [CrossRef]
- Mosele, F.; Stefanovska, B.; Lusque, A.; Tran Dien, A.; Garberis, I.; Droin, N.; Le Tourneau, C.; Sablin, M.-P.; Lacroix, L.; Enrico, D.; et al. Outcome and molecular landscape of patients with PIK3CA-mutated metastatic breast cancer. Ann. Oncol. Off. J. Eur. Soc. Med. Oncol. 2020, 31, 377–386. [Google Scholar] [CrossRef]
- Saal, L.H.; Holm, K.; Maurer, M.; Memeo, L.; Su, T.; Wang, X.; Yu, J.S.; Malmström, P.-O.; Mansukhani, M.; Enoksson, J.; et al. PIK3CA mutations correlate with hormone receptors, node metastasis, and ERBB2, and are mutually exclusive with PTEN loss in human breast carcinoma. Cancer Res. 2005, 65, 2554–2559. [Google Scholar] [CrossRef]
- Zhao, J.J.; Cheng, H.; Jia, S.; Wang, L.; Gjoerup, O.V.; Mikami, A.; Roberts, T.M. The p110alpha isoform of PI3K is essential for proper growth factor signaling and oncogenic transformation. Proc. Natl. Acad. Sci. USA 2006, 103, 16296–16300. [Google Scholar] [CrossRef]
- Herrera-Abreu, M.T.; Palafox, M.; Asghar, U.; Rivas, M.A.; Cutts, R.J.; Garcia-Murillas, I.; Pearson, A.; Guzman, M.; Rodriguez, O.; Grueso, J.; et al. Early Adaptation and Acquired Resistance to CDK4/6 Inhibition in Estrogen Receptor-Positive Breast Cancer. Cancer Res. 2016, 76, 2301–2313. [Google Scholar] [CrossRef]
- Juric, D.; Krop, I.; Ramanathan, R.K.; Wilson, T.R.; Ware, J.A.; Sanabria Bohorquez, S.M.; Savage, H.M.; Sampath, D.; Salphati, L.; Lin, R.S.; et al. Phase I Dose-Escalation Study of Taselisib, an Oral PI3K Inhibitor, in Patients with Advanced Solid Tumors. Cancer Discov. 2017, 7, 704–715. [Google Scholar] [CrossRef]
- Dent, S.; Cortés, J.; Im, Y.-H.; Diéras, V.; Harbeck, N.; Krop, I.E.; Wilson, T.R.; Cui, N.; Schimmoller, F.; Hsu, J.Y.; et al. Phase III randomized study of taselisib or placebo with fulvestrant in estrogen receptor-positive, PIK3CA-mutant, HER2-negative, advanced breast cancer: The SANDPIPER trial. Ann. Oncol. 2021, 32, 197–207. [Google Scholar] [CrossRef]
- Juric, D.; Janku, F.; Rodón, J.; Burris, H.A.; Mayer, I.A.; Schuler, M.; Seggewiss-Bernhardt, R.; Gil-Martin, M.; Middleton, M.R.; Baselga, J.; et al. Alpelisib Plus Fulvestrant in PIK3CA-Altered and PIK3CA-Wild-Type Estrogen Receptor-Positive Advanced Breast Cancer: A Phase 1b Clinical Trial. JAMA Oncol. 2019, 5, e184475. [Google Scholar] [CrossRef]
- André, F.; Ciruelos, E.; Rubovszky, G.; Campone, M.; Loibl, S.; Rugo, H.S.; Iwata, H.; Conte, P.; Mayer, I.A.; Kaufman, B.; et al. Alpelisib for PIK3CA-Mutated, Hormone Receptor–Positive Advanced Breast Cancer. N. Engl. J. Med. 2019, 380, 1929–1940. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Zhang, J.; Tong, J.H.M.; Chan, A.W.H.; Yu, J.; Kang, W.; To, K.F. Targeting the Oncogenic p53 Mutants in Colorectal Cancer and Other Solid Tumors. Int. J. Mol. Sci. 2019, 20, 5999. [Google Scholar] [CrossRef] [PubMed]
- Duffy, M.J.; Synnott, N.C.; Crown, J. Mutant p53 in breast cancer: Potential as a therapeutic target and biomarker. Breast Cancer Res. Treat. 2018, 170, 213–219. [Google Scholar] [CrossRef] [PubMed]
- Garrido-Castro, A.C.; Lin, N.U.; Polyak, K. Insights into Molecular Classifications of Triple-Negative Breast Cancer: Improving Patient Selection for Treatment. Cancer Discov. 2019, 9, 176–198. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez, B.J.; Córdoba, G.D.; Aranda, A.G.; Álvarez, M.; Vicioso, L.; Pérez, C.L.; Hernando, C.; Bermejo, B.; Parreño, A.J.; Lluch, A.; et al. Detection of TP53 and PIK3CA Mutations in Circulating Tumor DNA Using Next-Generation Sequencing in the Screening Process for Early Breast Cancer Diagnosis. J. Clin. Med. 2019, 8, 1183. [Google Scholar] [CrossRef]
- Madic, J.; Kiialainen, A.; Bidard, F.-C.; Birzele, F.; Ramey, G.; Leroy, Q.; Rio Frio, T.; Vaucher, I.; Raynal, V.; Bernard, V.; et al. Circulating tumor DNA and circulating tumor cells in metastatic triple negative breast cancer patients. Int. J. Cancer 2015, 136, 2158–2165. [Google Scholar] [CrossRef]
- Ma, F.; Zhu, W.; Guan, Y.; Yang, L.; Xia, X.; Chen, S.; Li, Q.; Guan, X.; Yi, Z.; Qian, H.; et al. ctDNA dynamics: A novel indicator to track resistance in metastatic breast cancer treated with anti-HER2 therapy. Oncotarget 2016, 7, 66020–66031. [Google Scholar] [CrossRef]
- Cooke, T.; Reeves, J.; Lanigan, A.; Stanton, P. HER2 as a prognostic and predictive marker for breast cancer. Ann. Oncol. Off. J. Eur. Soc. Med. Oncol. 2001, 12 (Suppl. 1), S23–S28. [Google Scholar] [CrossRef]
- Houssami, N.; Macaskill, P.; Balleine, R.L.; Bilous, M.; Pegram, M.D. HER2 discordance between primary breast cancer and its paired metastasis: Tumor biology or test artefact? Insights through meta-analysis. Breast Cancer Res. Treat. 2011, 129, 659–674. [Google Scholar] [CrossRef]
- Abraham, J.; Montero, A.J.; Jankowitz, R.C.; Salkeni, M.A.; Beumer, J.H.; Kiesel, B.F.; Piette, F.; Adamson, L.M.; Nagy, R.J.; Lanman, R.B.; et al. Safety and Efficacy of T-DM1 Plus Neratinib in Patients With Metastatic HER2-Positive Breast Cancer: NSABP Foundation Trial FB-10. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 2019, 37, 2601–2609. [Google Scholar] [CrossRef]
- Gevensleben, H.; Garcia-Murillas, I.; Graeser, M.K.; Schiavon, G.; Osin, P.; Parton, M.; Smith, I.E.; Ashworth, A.; Turner, N.C. Noninvasive detection of HER2 amplification with plasma DNA digital PCR. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2013, 19, 3276–3284. [Google Scholar] [CrossRef]
- Kleftogiannis, D.; Ho, D.; Liew, J.X.; Poon, P.S.Y.; Gan, A.; Ng, R.C.-H.; Tan, B.K.-T.; Tay, K.H.; Lim, S.H.; Tan, G.S.; et al. Detection of genomic alterations in breast cancer with circulating tumour DNA sequencing. Sci. Rep. 2020, 10, 16774. [Google Scholar] [CrossRef]
| Gene | Patient Cohort | Molecule Testing Technique | Main Findings | Clinical Significance | References |
|---|---|---|---|---|---|
| ESR1 | 541 postmenopausal women with a diagnosis of MBC. | Analyzed ESR1 mutations (Y537S and D538G) on cell-free DNA (cfDNA) using droplet digital polymerase chain reaction (ddPCR). | D538G (21.1%) Y537S (13.3%) 30 had both mutations. | These mutations were associated with shorter overall survival: - wild-type, 32.1 months - D538G, 25.99 months - Y537S, 19.98 months - Both mutations, 15.15 months. | [72] |
| 86 estrogen receptor-positive BC patients.185 plasma samples (151 plasma samples from 69 MBC patients, and 34 plasma samples from 17 primary BC (PBC) patients). | Multiplex droplet digital PCR assays in a snapshot and serially. | cfDNA ESR1 and PIK3CA mutations were found in 28.9% and 24.6% of MBC patients, respectively. | All patients with ESR1 mutations had resistance to prior AI (aromatase inhibitor) therapy. 85% of patients with ESR1 mutations had resistance to prior SERM (Selective estrogen receptor modulators) therapy. | [73] | |
| BRCA1/2 | 828 patients with advanced breast, ovarian, prostate, or pancreatic cancer. (the study was conducted in accordance with the Declaration of Helsinki). | Plasma-based NGS assay. | Of 828 patients, 60 (7.2%) had at least one BRCA1/2 loss-of-function mutation, 42 patients with germline mutations and 18 (14 patients had breast cancer) with somatic mutations only. | NGS analysis of cfDNA identified high rates of therapeutically relevant mutations, including deleterious BRCA1/2 somatic mutations missed by germline testing. | [74] |
| 24 patients with proven BRCA1/2 germline mutations (19 ovarian cancer patients and 5 patients with MBC who received prior treatment with platinum-based chemotherapy and/or PARP inhibitors). | Targeted massively parallel sequencing of tumor DNA from ovarian cancer patients, cfDNA from ovarian and breast cancer patients, and their germline DNA. | Identification of BRCA1 or BRCA2 reversion mutations in the cfDNA of 4 ovarian cancer patients (21%) and 2 breast cancer patients (40%). | cfDNA sequencing can help identify putative BRCA1/2 reversion mutations which may facilitate patient selection for PARP inhibition therapy. | [75] | |
| PIK3CA | Thirty patients with advanced BC (ABC); | PIK3CA mutation analysis was performed using ddPCR. | The presence of a PI3K mutation in liquid biopsy correlates with worse PFS in patients with ABC receiving CDK4/6i. | Integration of PI3K status assessment with other molecular information could improve the management of patients with aggressive breast cancer and better suggest the best therapeutic strategy. | [76] |
| TP53 | 46 patients with nonmetastatic triple-negative breast cancer; | Characterization of TP53 gene mutations in tumor tissue through massively parallel sequencing (MPS). Monitoring of previously characterized mutations based on ctDNA analysis by ddPCR at four time points: pre-NCT, post-cycle, pre-surgery, and post-surgery. | Results show a marked decrease in ctDNA levels and positivity rate during chemotherapy cycles. | The high prevalence of TP53 mutations in TNBC is a potential biomarker for ctDNA monitoring during NCT, and therefore is a tool for TNBC management. | [77] |
| 113 lung and 18 breast cancer patients | NGS analysis of ctDNA: Panel for hot spot regions in 11 genes for lung cancer and 10 genes for breast cancer. | Variations in the TP53 gene were detected at a high frequency in both tumor types, followed by the PIK3CA gene in breast cancer. | Based on NGS and ddPCR techniques, liquid biopsy could be a very effective method for managing terminal cancer cases and monitoring treatment responses. | [78] | |
| 68 patients with metastatic breast cancer (MBC). | cfDNA and gDNA (Genomic DNA) analysis by next-generation sequencing (NGS) | TP53 mutations occurred in 10 (45.45%) TNBC patients, 9 (36.00%) HER2+ patients, and 7 (22.22%) HR+ patients. TP53 represents the gene with the highest number of somatic mutations. | Mutations in TP53 cDNA and PIK3CA genes likely limit survival and promote disease progression. | [79] | |
| ERBB2 | 636 women with HER2 nonamplified MBC. | ctDNA analysis by NGS. | Results of this study indicate the efficacy of neratinib for HER2-mutated nonamplified breast cancer. | This study supports the potential use of ctDNA to identify patients with HER2-mutated breast cancer to establish a new standard of care. | [80] |
| Multicohort, phase 2a, platform trial of ctDNA testing in 18 UK hospitals.1051 patients were registered in the study. | ddPCR and NGS are used to detect ctDNA mutations. Patients were recruited into four parallel treatment cohorts corresponding to the mutations identified in the ctDNA (ESR1; HER2; AKT1 and PTEN). | The findings of this study demonstrate the clinically relevant activity of targeted therapies against rare HER2 and AKT1 mutations. | The results of this research show that ctDNA analysis, with the technologies used in this study, is accurate enough to be routinely adopted into clinical practice. | [81] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
El Hejjioui, B.; Bouguenouch, L.; Melhouf, M.A.; El Mouhi, H.; Bennis, S. Clinical Evidence of Circulating Tumor DNA Application in Aggressive Breast Cancer. Diagnostics 2023, 13, 470. https://doi.org/10.3390/diagnostics13030470
El Hejjioui B, Bouguenouch L, Melhouf MA, El Mouhi H, Bennis S. Clinical Evidence of Circulating Tumor DNA Application in Aggressive Breast Cancer. Diagnostics. 2023; 13(3):470. https://doi.org/10.3390/diagnostics13030470
Chicago/Turabian StyleEl Hejjioui, Brahim, Laila Bouguenouch, Moulay Abdelilah Melhouf, Hind El Mouhi, and Sanae Bennis. 2023. "Clinical Evidence of Circulating Tumor DNA Application in Aggressive Breast Cancer" Diagnostics 13, no. 3: 470. https://doi.org/10.3390/diagnostics13030470
APA StyleEl Hejjioui, B., Bouguenouch, L., Melhouf, M. A., El Mouhi, H., & Bennis, S. (2023). Clinical Evidence of Circulating Tumor DNA Application in Aggressive Breast Cancer. Diagnostics, 13(3), 470. https://doi.org/10.3390/diagnostics13030470






