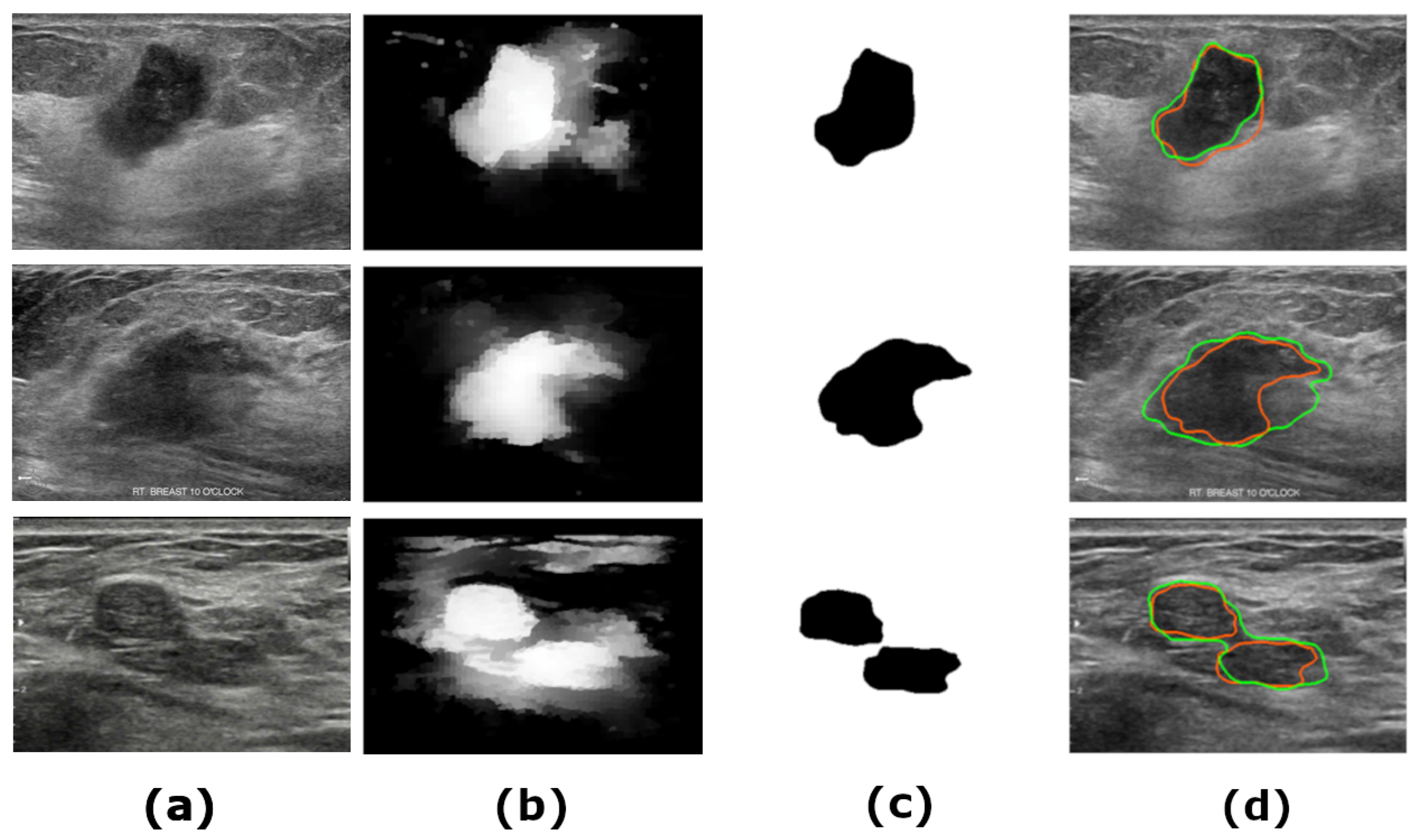
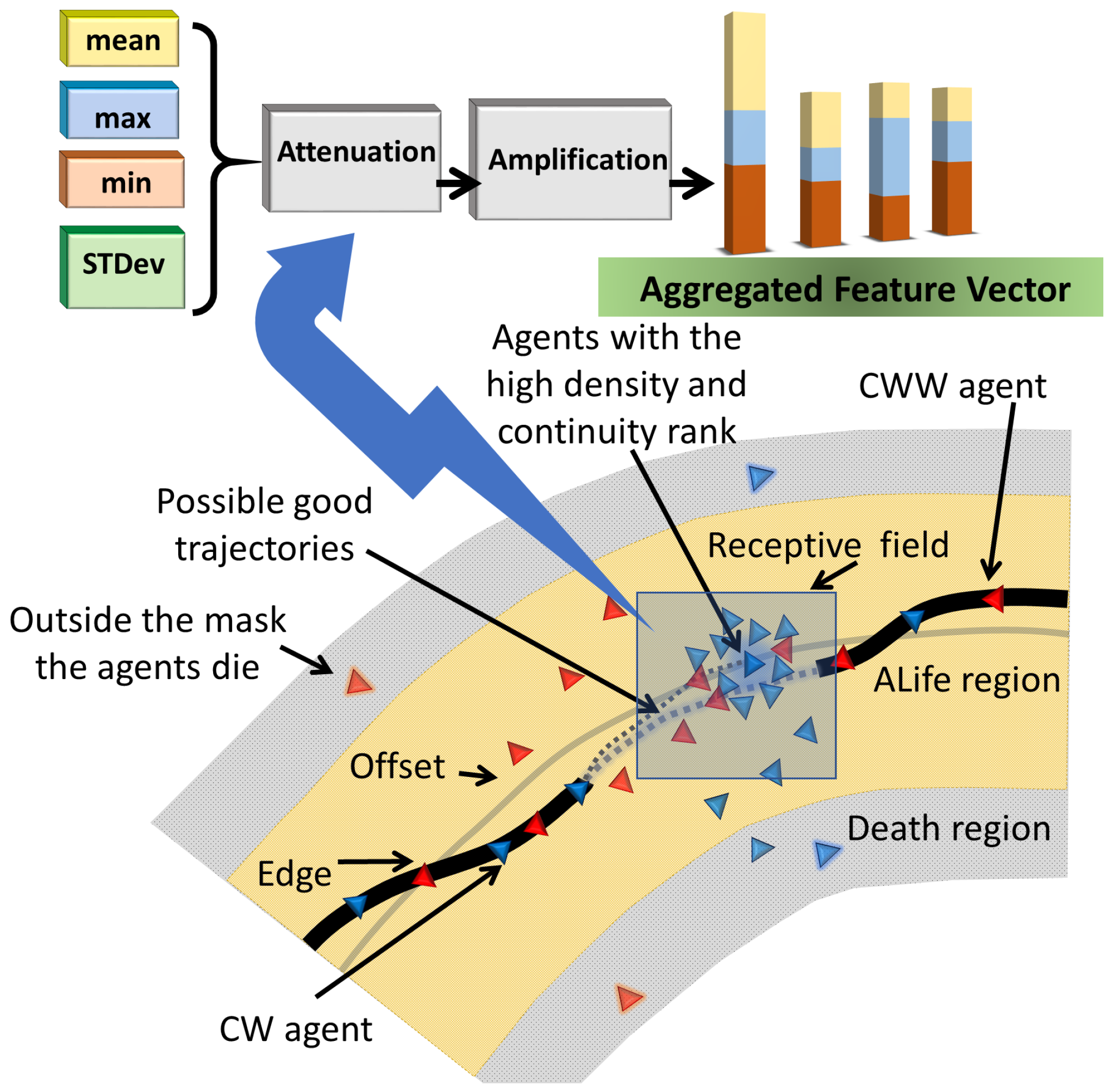
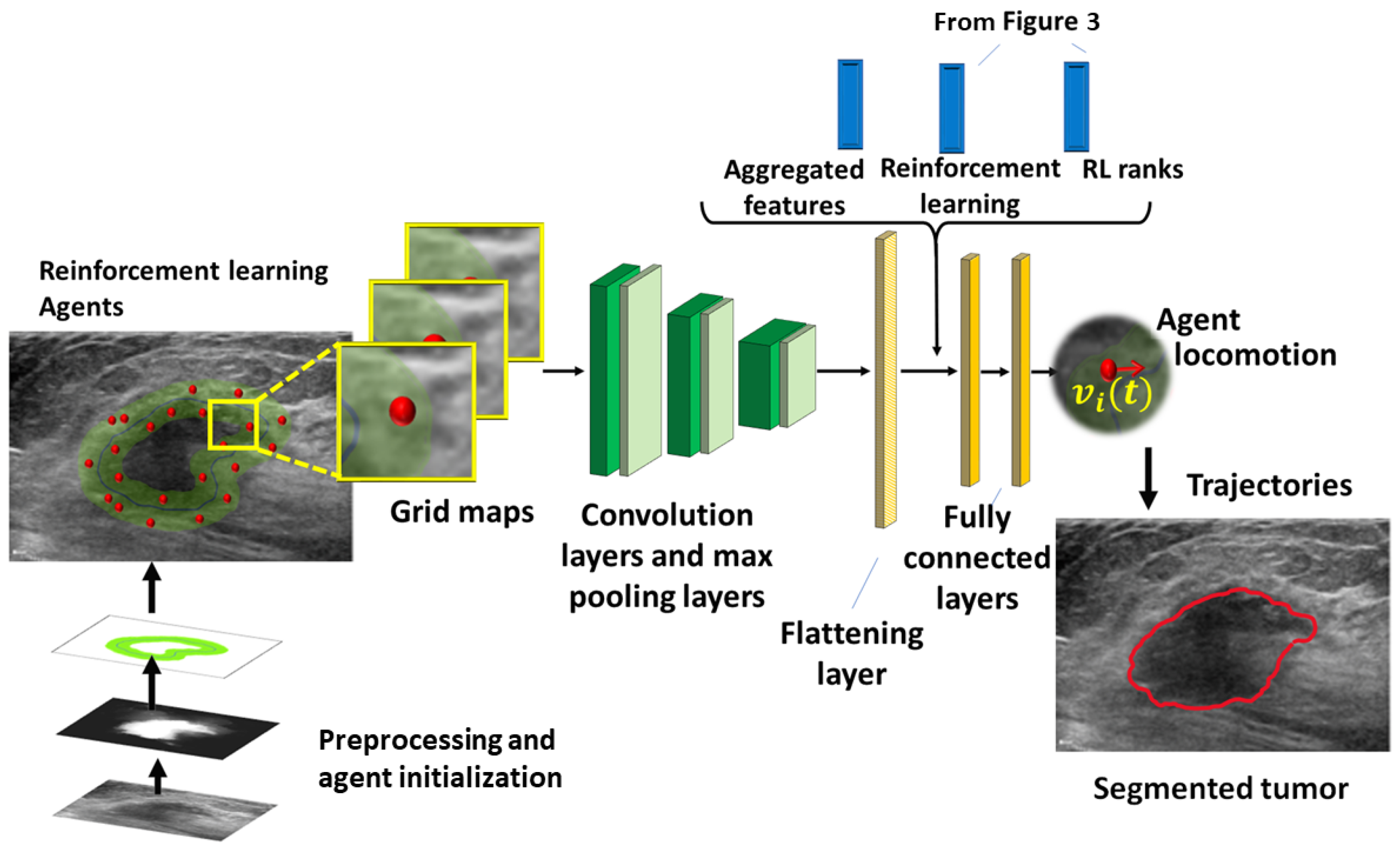
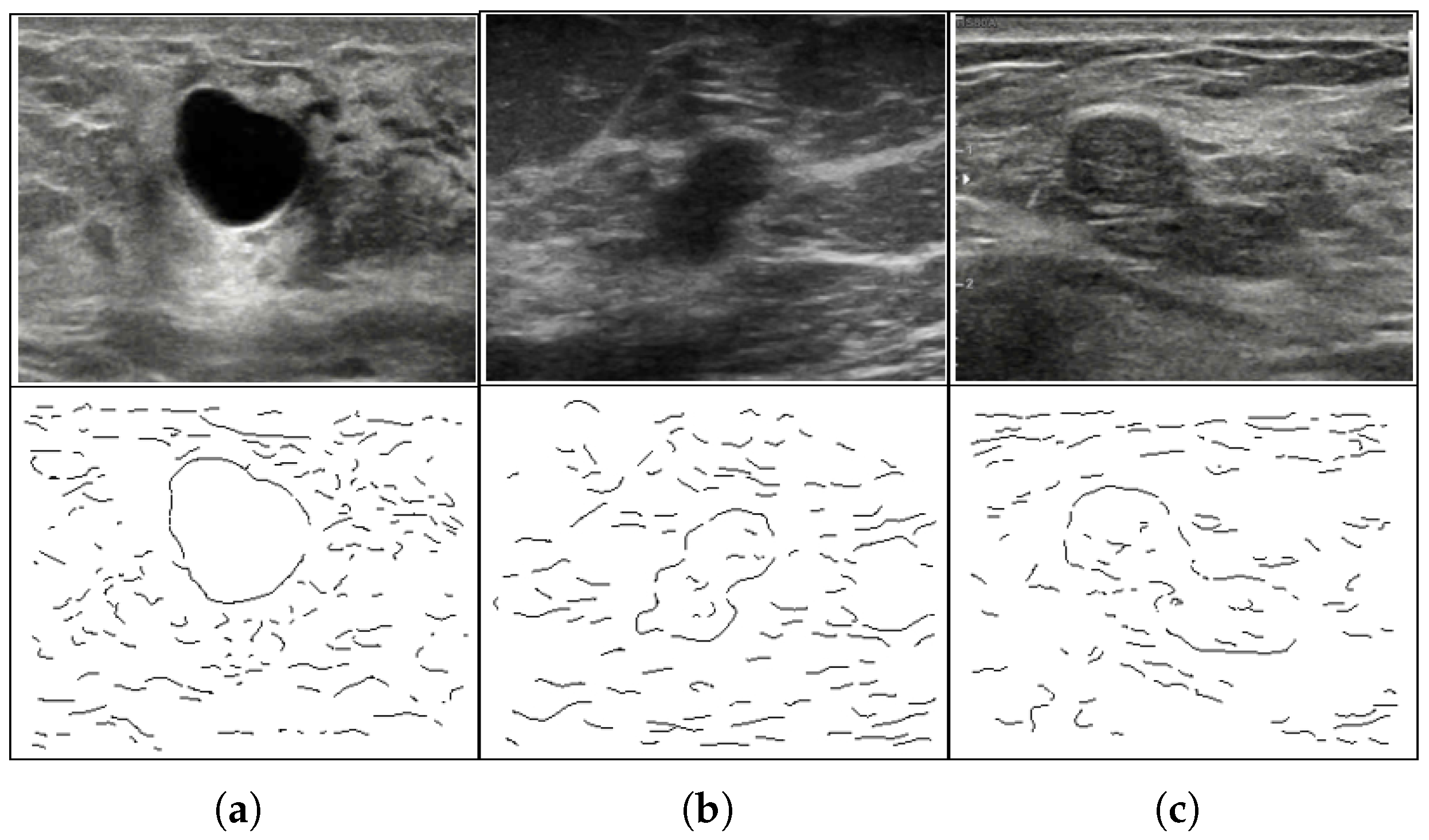
1. Introduction
Worldwide, breast cancer is the most common cancer in women [
1]. If diagnosed early, the survival rates are significantly improved. In Asia, US examination is included in routine cancer screening due to the high prevalence of females with dense breast tissue [
2]. Accurate segmentation assists in evaluating the type and stage of the tumor as well as its possible progression. However, US images are often hard to interpret. They require expertise and experience. Detection and segmentation of breast tumors is a difficult task even for skilled clinicians. They also present a challenge for CAD (computer-assisted diagnostic) systems. The US machine receives an echo of the emitted sound waves. The image of this echo (the US image) is characterized by speckle noise, irregular tumor shapes, uneven textures, poorly defined boundaries, poor contrast, and multiple acoustic shadows. The recent advances in convolutional neural networks (CNNs) and deep learning (DL) tools show much promise for the segmentation of the US images. Nevertheless, the learned features are often not interpretable. The training requires a large amount of ground truth data (usually thousands of images). Moreover, the DL trained on a specific set often fails when a different US machine or a different protocol is used [
3]. Zhang et al. [
4] notes that the “application of these DL models in clinically realistic environments can result in poor generalization and decreased accuracy, mainly due to the domain shift across different hospitals, scanner vendors, imaging protocols, and patient populations…the error rate of a DL was 5.5% on images from the same vendor, but increased to 46.6% for images from another vendor”. The nature of CNNs is to capture the local contextual information. However, they often miss tumors or produce over or under-segmented boundaries. One of the reasons is that the global structure of the object is not captured.
The proposed model is motivated by the autonomous vehicles of Braitenberg’s [
5] and Reynolds’ bird flocks [
6] proposed in the 1980s. Baitenberg’s seminal work in 1984 used reactive robots or vehicles with relatively simple sensor–motor connections aimed to simulate neuro-mechanisms in biological brains. In this paper, Braitenberg’s work is considered a metaphor for artificial life, suggesting that the complex behavior may be a result of a relatively simple design. The ideas are modified here. The robots do not know their final goal. They follow the rewards defined by a certain reward system. Additionally, they can be corrected by the neural network (NN), which observes the population and knows their final destination. Therefore, the NN is a metaphor for a “Universal God”, “Galactic Force”, etc. The idea is supported by artificial life (Alife) [
7], based on tracing agents [
8] and fusion images. It has been shown that, under certain conditions, Alife outperforms the state-of-the-art. Furthermore, Alife requires smaller training sets. The drawback of [
7] is the requirement of the additional set of elasticity images, which are not always available. Further, the above ALife is based on fixed rules and requires extensive training performed manually or by a genetic algorithm. Therefore, an extension of this model is offered. The new model does not require the elasticity images and achieves the same or better accuracy. It uses a relatively small training set, and is capable of handling previously unseen data.
On this level, the basic ideas are to: (1) generate a preliminary binary mask and offset trajectories to guide the agents, (2) train the agents using the convolution network, and (3) include the RL to replace the requirement of the image fusion. The model combines multi-agent reinforcement learning, neighbor message passing, and deep learning to generate trajectories of the agents, approximating the boundary of the lesion. Message passing is based on principal neighborhood aggregation [
9]. Multi-agent deep reinforcement learning (MADRL) employs the Gestalt laws (GL) [
10] of shape perception to construct the reward system.
Summarizing the new elements above, the novelty of the proposed model is as follows.
- -
A new hybrid of the DL, Alife, and RL, has been offered. The method combines the strengths of the DL neural networks with the efficiency and simplicity of ALife and the agents’ communication and learning by RL.
- -
A reward system based on the GL.
- -
An original classification of the images based on the properties of the edge maps included in the training algorithm.
- -
Verification by previously unseen data.
We also consider it valuable that the proposed bio-inspired model does not use the standard genetic framework which often slows down the optimization. The agents are identical, they do not evolve, do not mate, and do not have leaders. This makes it possible to achieve fast computational times, e.g., 5–20 s per image 500 × 500 (
Section 7). The agents communicate and try to maximize their rewards. Hence, the model is able to generate complicated patterns, fit irregular structures, and segment the image efficiently. The proposed MADRL algorithm has been tested on two US datasets against 13 state-of-the-art algorithms. They include active contours (AC), edge linking, level set methods (LS), superpixels, machine learning, deep learning algorithms, and their modifications. The numerical results reveal that the MADRL outperforms its competitors in terms of accuracy when applied to high-complexity images. Note that the paper performs cross-field testing, i.e., compares the segmentation methods based on different principles. The most popular and original algorithms from six classes have been selected. First of all, consider the deformable models, i.e., AC and LS. They have been massively used for edge-based segmentation and extensively used for segmentation of the US images of the breast and thyroids. The latest survey [
11] considers the deformable models of the most frequently used approach for segmentation of the US images. The adaptive diffusion flow [
12] is one of the most remarkable versions of the AC. It establishes a joint framework between the classical gradient vector flow and the image restoration. The recent AC energy-based pressure approach [
13] introduces an “energy balance” between the inner region and the outer region. Finally, the hybrid AC-LS method [
14] based on the local variations of the grey level shows excellent results on various synthetic and real medical images. According to a survey in [
7], these three algorithms outperform 29 top models from this class (
https://sites.google.com/view/edge-driven-marl-siit-biomed/additional-references, accessed on 28 June 2023). The pioneering LS work is Osher, 1988 [
15]. The propagating fronts, with velocity depending on the front curvature, became revolutionary in the image processing of the 2000s. The distance-regularized edge-based DRLSE [
16] is one of the most popular. Introducing the third dimension solves the problem of the multiple ACs and their possible merge. We refer the reader to various modifications of the LS given in the excellent review [
17]. The DRLSE, the saliency-driven region/edge-based LS [
18] and a correntropy-based LS [
19] have been shown to outperform eight top algorithms from this class (
https://sites.google.com/view/edge-driven-marl-siit-biomed/additional-references, accessed on 28 June 2023).
Edge linking methods connect pieces of the object boundary of the edge map into a closed contour subject. The algorithm [
20] is based on edge tracing and a Bayesian contour extrapolation. The ratio contour method [
21] encodes the GL laws of proximity and continuity combined with a saliency measure based on the relative gap length and average curvature. Some of these ideas have been used in our model. The above models outperform four edge-linking procedures (
https://sites.google.com/view/edge-driven-marl-siit-biomed/additional-references, accessed on 28 June 2023). Note that the early variants apply the graph-based approach, where the graph represents candidate parts of the boundary, and the weights represent the affinity between them. The graph is partitioned to optimize a selected utility function. Further, the edge linking evolved in different directions. Some prominent examples are connected components [
22], pixel following [
23], and the dot completion game [
24].
A recent review [
25] claims that the superpixel models are one of the most important tools for image segmentation. In particular, the simple linear iterative clustering (SLIC) proposed in [
26] is one of the most efficient. We compare the RL with the recent version of SLIC [
3], which integrates the NN and k-NN techniques. Our second superpixel reference is [
27]. This model applies the superpixels to training, whereas segmentation is performed by the LS. The prior knowledge regarding the possible shape of the object is included in the LS functional.
Machine learning (ML) (deep learning and neural networks) is one of the most promising fields of study aimed at processing and recognizing US images [
28,
29]. The ML algorithm adapts itself to generate the required features instead of selecting them manually. The most successful models are CNNs and DL networks [
30,
31].
We consider U-Net [
32] as one of the most prominent and influential models of this type. The U-net is often treated as “the benchmarking DL model for medical image processing” [
33]. A hybrid of the ML and the Chan–Vese algorithm [
34] is proposed by [
35]. It combines the k-NN method and the support vector machine. We also test against a semi-supervised DL generative adversarial network proposed by [
36]. The DL applies the multi-scale framework and combines the ground truth and the probability maps obtained from the un-annotated data. The model outperforms the ASDNet, ZengNet, and DANNet (
https://sites.google.com/view/edge-driven-marl-siit-biomed/additional-references, accessed on 28 June 2023). We also test against the selective kernel [
37]. Currently, the selective kernel is one of the most developed U-Net algorithms, which outperforms many recent modifications of the U-Net. It should be noted that the above is not a complete review of the state-of-art of medical image segmentation. The above models and some of their modifications have been selected as the benchmark for testing. The selection criteria are publications in a reputable journal, references, tests against similar and cross-field models, and availability of the code. Our numerical results show that the proposed DL reinforcement learning outperforms 13 selected models from different classes (cross-field testing). The advantage against the second-best performing model on the images characterized by a high complexity is as high as a 97% (success rate) against 77%. The results are subjected to the selected set of images and the proposed accuracy measures. The developed code is applicable to breast cancer diagnostics (automated breast ultrasound), US-guided biopsies, as well as for projects related to automatic breast scanners. A demo video illustrating the algorithm is at (
https://drive.google.com/file/d/1kqW68mdQ1QmkasA1gnXNFRRaavhvpA-M/view?usp=drive_link, accessed on 30 August 2023).
4. Network Training
The PPO [
51] is used for training by an on-policy gradient algorithm [
60] based on the DRL actor–critic techniques. The PPO produces smooth gradient updates to ensure a stable policy space and reduced hyperparameter tuning. The agent trajectories are standardized and used to construct the surrogate loss
and the least square error (
loss)
. The loss function is minimized using the Adam optimizer [
83]. The policy is updated over
epochs and the value function is updated over
epochs.
where
denotes a stochastic estimate of the expected value over a mini-batch of transitions. The clipping parameter
controls the range of the probability ratio
used for the update.
is a generalized advantage estimate [
84] that smooths the discounted rewards and reduces the variance of the policy gradients to make the training stable. In addition, it indicates how good a particular state is. The estimate is defined by
where
is the smoothing factor,
is the discount factor for the future rewards,
is the time step of the episode, and
i is the index of the agent. The term
implies the advantage of the new state over the previous state. It takes into account the immediate reward
and the expected future value
, discounted by a factor of
.
The value function
is used to update the policy function. The
loss is given by
where
K is the total number of trajectories,
is the value function approximated by the network by the parameters
.
is the estimated discounted return.
Note that the policy and value networks are characterized by the same architecture but their parameters are not shared. The optimal policy is being generated by the networks simultaneously.
The pseudocode of the training algorithm is presented as Algorithm 1, while
Table 1 lists its hyperparameters.
| Algorithm 1 Multi-agent deep reinforcement learning |
- 1:
for iterations do - 2:
Initialize - 3:
for Agent do - 4:
// data collection - 5:
Run the policy for time steps - 6:
Collect observations, rewards and actions , where - 7:
Estimate the advantage - 8:
Break - 9:
end for - 10:
//update the policy function - 11:
for do - 12:
Calculate surrogate loss - 13:
Update with the learning rate by the Adam optimizer based on - 14:
end for - 15:
//update the value function - 16:
for do - 17:
Calculate the surrogate loss - 18:
Update with the learning rate by the Adam optimizer based on - 19:
end for - 20:
end for
|
Training Data
The network has been pre-trained using the transfer learning scheme [
85]. The MADRL has been tested on 1000 US images from
http://www.onlinemedicalimages.com (accessed on 17 June 2023) of Thammasat University Hospital, Thailand, Bangkok (Philips iU22 US machine) and
https://www.ultrasoundcases.info (accessed on 20 July 2023) from The Gelderse Vallei Hospital, Ede, The Netherlands (Hitachi US machine). Three experienced radiologists with Thammasat University Hospital provided the ground truth using the electronic pen on a Microsoft Surface Tablet. The final ground truth is obtained by the majority rule. The image resolution ranges from
to
with a 60:40 ratio for training and evaluation. The training edge maps are categorized according to their complexity. The categories include the complexity of the shape, the length of the gaps relative to the contour, and the maximum gap on the contour (
Section 5). The approach allows us to find a suitable solution in a less difficult environment fast and to progressively advance the complex environments. When stability and convergence are established in each category, the training is completed.
6. Numerical Experiments
MADRL has been tested against 13 state-of-the-art methods discussed in the introduction. We include the active contours: ADF-AC [
12], ALF-AC [
14], EBF-AC [
13], level set methods: DR-LS [
16], CB-LS [
19], SR-LS [
18], edge linking: BS-EL [
20], CR-EL [
21], superpixels: PK-SP [
27], SC-SP [
3], and the machine learning/deep learning algorithms ML-AC [
35], SS-GAN [
36], and S-U-NET [
37].
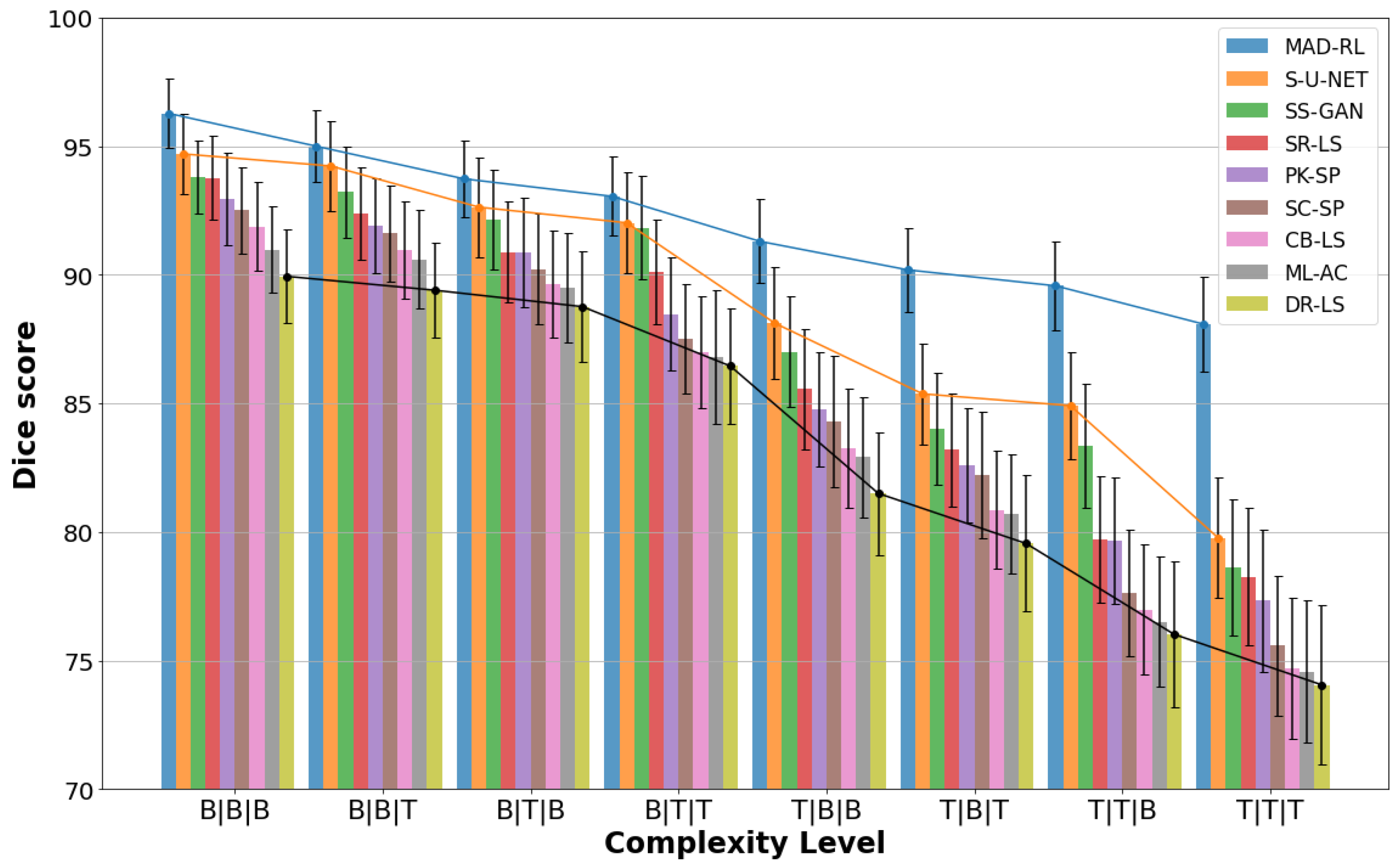
Figure 7 shows samples of the segmented US images by MADRL vs. the benchmark methods for different levels of complexity. The Figure is complemented by
Table 2,
Table 3,
Table 4,
Table 5,
Table 6,
Table 7,
Table 8 and
Table 9 corresponding to the increasing complexity.
Table 2 shows the segmentation results for the simplest complexity B|B|B. Nine methods produce
. However,
and
dropped by 25% and 16%. The methods with 100% segmentation show the maximum
(acceptable). All methods manage
, where
is the lowest. The best
Consider the impact of
and
on the efficiency of the US segmentation.
Table 3 and
Table 4 show the results for B|T|B and B|B|T. Clearly, the maximum size of the gap
has a greater impact on accuracy than their total length. In particular, eight methods for B|B|T have
and three methods fall below 90%. However, in the case of B|T|B only six methods remain in the 100% category (including the proposed MADRL). Moreover, the number of successfully segmented images drops below 90% for the five methods. The lowest
with
.
The results for the complexity B|T|T are given by
Table 5. It combines a large total gap length, a large maximum gap, and a simple shape S=B. Only SR-LS, PK-SP, SC-SP, SS-GAN, S-U-NET, and the proposed MADRL achieve the segmentation rate 100%. Nonetheless, the RL has only a marginal improvement over the S-U-NET. For instance,
and a
, while the
and a
.
Table 6 and
Table 7 shows the significance of the tumor shape. Even T|B|B presents a challenge. The occurrence of a leakage at the boundaries and irregularity of the geometry of the tumor reduce the success rate. The exception is S-U-NET and the proposed MADRL. Note that SR-LS, PK-SP, SS-GAN, and S-U-NET maintain an acceptable success rate above 90%. However, the proposed method outperforms them with
the best
and the best
.
The complexity T|T|B indicates an irregular shape and a considerable edge leakage (
Table 8). This configuration finally breaks S-U-NET although its success rate remains above 90%. The ACs and LSMs fail. SR-LS achieves a modest
. The edge-linking methods BS-EL and CR-EL fail with a success rate below 30%. However, MADRL stands out with
,
and
.
The most difficult type, T|T|T (
Table 9), shows a low success rate for the majority of competing methods. The edge linking methods BS-EL and CR-EL have an unacceptable
. The complexity of the shape combined with the edge leaks leads to the failure of the AC and LS models. The minimum and maximum success rates of the ACs are 13% and 27%, respectively. The LS is slightly better. The minimal success rate
and the maximum
. One of the main disadvantages of the deformable shapes is a strong dependence on the initialization and the inability to handle strong edge leaks. The T|T|T images are irregular, having acoustic shadows and artifacts. As the result, the edge detector generates strong false boundaries and the edge-based AC and LS produce a poor segmentation.
Further, the drawback of the superpixel methods is a potential loss of the true edge if it gets inside a generated superpixel. SC-SP and PK-SP show a success rate of less than 60%. Their . Our main competitor S-U-NET has . We conjecture that this is due to the complexity of the shape, insufficient training data, and the inability of the DL methods to capture the global information. This comment applies to all ML and DL methods presented in the tables. Note that only the S-U-NET achieves a reasonable , which is slightly below the threshold of 80. Further, is also an acceptable result. However, MADRL has a significantly higher , , and .
One of the main reasons for the failure of the DL methods is that the algorithms do not understand the global context. As opposed to that, MADRL combines the DL which allows for the automatic feature extraction with self-trained reinforcement agents capable of communicating the information throughout the entire population. Another reason is a lack of annotated data. Usually, a pure DL model requires one-tenth of thousands of samples. However, the proposed model employs only about 2000 annotated images.
9. Conclusions and Future Work
A new multi-agent reinforcement learning (MADRL) algorithm tailored for breast US segmentation has been proposed and analyzed. The combination of the DL with the tracing RL agents demonstrates significant improvements over the existing state-of-the-art. The method shows an excellent performance across a variety of complex US images, effectively overcoming challenges, such as tumor heterogeneity, boundary leakage, and the global shape context problem. Notably, unlike the CNNs, which learn to extract patterns and features from the data, the MADRL understands the structure of the image through the dynamics of the agents.
The proposed model is motivated by the autonomous vehicles of Braitenberg [
5] aimed to simulate neuro-mechanisms in biological brains. The ideas of Braitenberg have been successfully adapted to the segmentation of US images. The new reward system based on the Gestalt Laws of shape perception has been developed and implemented. The system includes two continuity measures, proximity measure, density, and closure measure. The numerical training shows that 50% of the rewards can be attributed to the continuity and 20% to the density measure. The final solution regarding the locomotion of the agents is generated by the DNN, which observes the entire population and knows their goal. This global knowledge of the image has been achieved using the dynamics of the agents and their ability to exchange information. Another source of information is the classification of the training images into eight categories. Therefore, the system is fed by samples with increasing complexity. This allows the DNN to adapt to the image features using an iterative approach, i.e., start from the simplest category and finalize the training at the hardest level.
The numerical experiments test the proposed MADRL against 13 benchmark methods. The analysis of the results shows that the majority of the competing algorithms perform satisfactorily at the first 3–4 levels of complexity. The complexity of the tumor shape has the most profound impact on the accuracy of the results. When the shape of the tumor changes from simple to complex, 12 benchmark methods from 13 fail for some images. The success rate changes from 45% for conventional edge linking to 96% for a variant of the generative adversary network [
36]. Only S-U-NET [
37] retained the original 100% success rate with the Dice coefficient 88%. The success rate S-U-NET drops below 100% only when the shape of the tumor is complex and the max boundary gap is large and when all three criteria indicate high complexity. In this case,
and
, respectively. However, MADRL outperforms its main competitor, keeping
and
.
The reward function is a variant of the mathematical representation of the Gestalt Laws of shape perception. The numerical experiments with the reward function show that the continuity of the trajectory constitutes the most important reward (over 50%). The density (a variant of proximity) is the second important parameter (around 19%).
The model shows the lowest standard deviation and an excellent statistical significance in terms of the p-values.
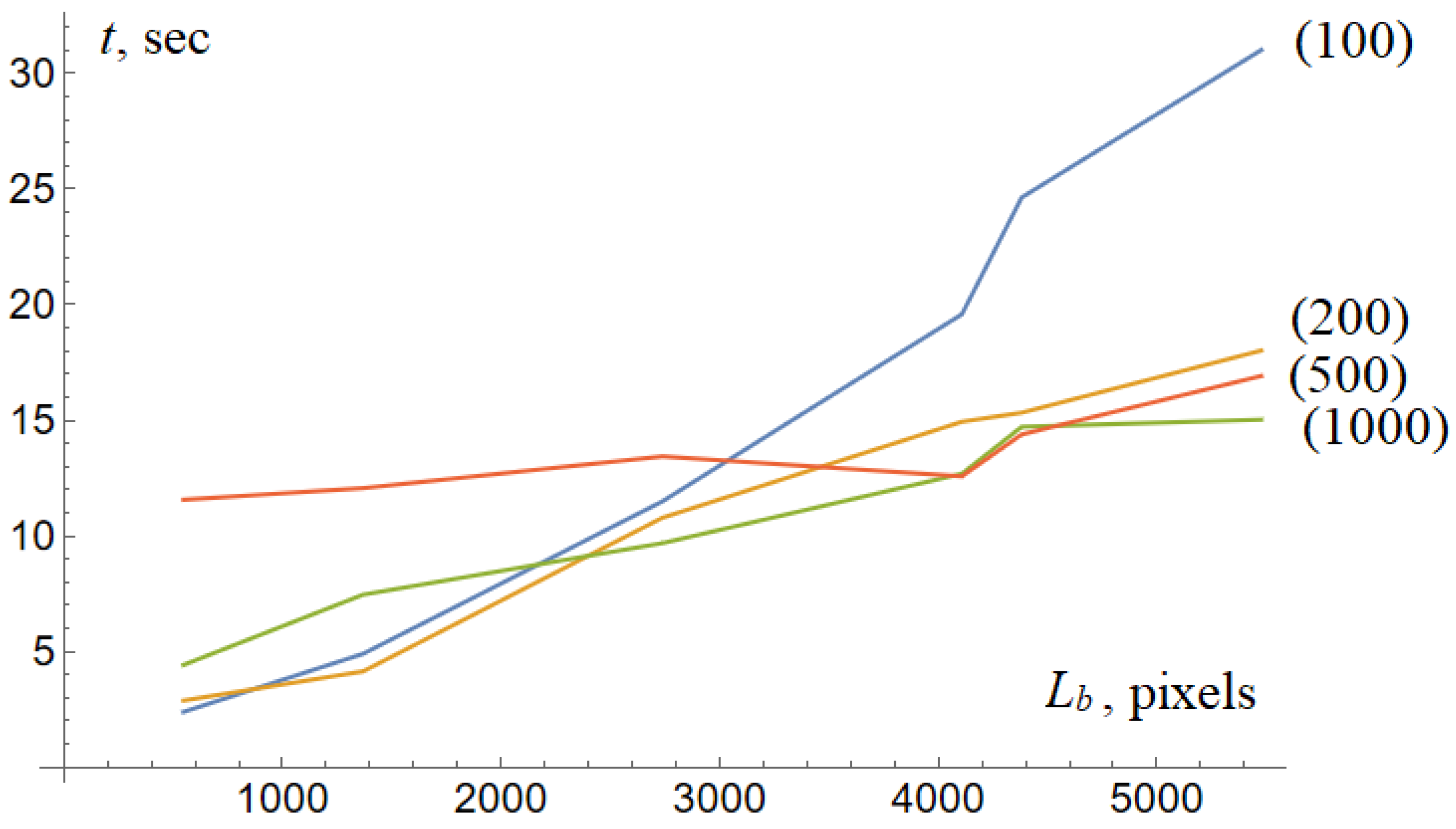
The computational time depends on the size of the tumor. Our preliminary experiments show that the computational time is a linear function of the length of the tumor boundary. In other words, the model shows excellent computational performance. The average time required for the 500 × 500 image ranges from 5 to 20 s. The model processes an image 1000 × 1000 at 30 s.
Note the limitations of this study. While our method has shown significant progress in segmenting US tumors, the results may vary depending on the quality of the images and the complexity of the tumor when the complex images are from an unseen database. The model has many training parameters. Most likely they require substantial re-training and transfer learning if applied to different types of medical imagery, such as MRI, CT-scans, etc. However, the design of a universal segmentation model is outside the scope of this study. One of the specific features of the US images is a sufficiently smooth boundary of the tumor (although globally the shape can be very irregular). This helps the agents accurately approximate the boundary. The model requires modifications if applied to objects with many sharp turns. In this case, the agents “wiggle” at the corners. This requires modifications and specific training.
Our future research focuses on optimizing this approach for real-time segmentation and exploring its potential for classification of the breast tumors. Another goal is to apply it to US imagery of different human organs. The most probable candidates are the US images of thyroids. From the viewpoint of computer science, one of the most interesting questions is the relationship between the number of agents, the length of the boundary, and the complexity of the tumor (eight categories). This open problem requires further research and massive numerical experiments. From the viewpoint of clinical application, the speed remains one of the most important factors. Doctors do not want to wait 20 s. The result must appear immediately on the screen, i.e., 2–3 s. However, the simplicity of the model mentioned above makes it possible to conjecture that this goal is achievable.