Cancer Metastasis Prediction and Genomic Biomarker Identification through Machine Learning and eXplainable Artificial Intelligence in Breast Cancer Research
Abstract
1. Introduction
2. Materials and Methods
2.1. Data Source and Selection Criteria
2.2. Sample Preparation and RNA Isolation
2.3. Microarray Hybridization and Data Normalization
2.4. Biostatistical Data Analysis
2.5. ML and XAI Approach
2.5.1. Data Preprocessing
2.5.2. ML Algorithms Used for BC Metastasis Prediction
2.5.3. Performance Evaluation Metrics
2.5.4. XAI Approach and Feature Importance
SHapley Additive exPlanations (SHAP)
3. Results
4. Discussion
5. Limitations
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Hamood, H. Adverse Conditions in Breast Cancer Survivors: Incidence, Determinants, and Effect on Quality of Life; University of Haifa: Haifa, Israel, 2020. [Google Scholar]
- Ginsburg, O.; Yip, C.H.; Brooks, A.; Cabanes, A.; Caleffi, M.; Dunstan Yataco, J.A.; Gyawali, B.; McCormack, V.; McLaughlin de Anderson, M.; Mehrotra, R. Breast cancer early detection: A phased approach to implementation. Cancer 2020, 126, 2379–2393. [Google Scholar] [CrossRef] [PubMed]
- Mohammadi, M.; Mohammadi, S.; Hadizadeh, H.; Olfati, M.; Moradi, F.; Tanzifi, G.; Ghaderi, S. Brain metastases from breast cancer using magnetic resonance imaging: A systematic review. J. Med. Radiat. Sci. 2023, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Li, J.; He, T.; Ding, J. Bioinformatics identified 17 immune genes as prognostic biomarkers for breast cancer: Application study based on artificial intelligence algorithms. Front. Oncol. 2020, 10, 330. [Google Scholar] [CrossRef] [PubMed]
- Kudela, E.; Samec, M.; Kubatka, P.; Nachajova, M.; Laucekova, Z.; Liskova, A.; Dokus, K.; Biringer, K.; Simova, D.; Gabonova, E. Breast cancer in young women: Status quo and advanced disease management by a predictive, preventive, and personalized approach. Cancers 2019, 11, 1791. [Google Scholar] [CrossRef]
- Abdollahi, J.; Keshandehghan, A.; Gardaneh, M.; Panahi, Y.; Gardaneh, M. Accurate detection of breast cancer metastasis using a hybrid model of artificial intelligence algorithm. Arch. Breast Cancer 2020, 7, 22–28. [Google Scholar] [CrossRef]
- Tarighati, E.; Keivan, H.; Mahani, H. A review of prognostic and predictive biomarkers in breast cancer. Clin. Exp. Med. 2022, 23, 1–16. [Google Scholar] [CrossRef]
- Barzaman, K.; Karami, J.; Zarei, Z.; Hosseinzadeh, A.; Kazemi, M.H.; Moradi-Kalbolandi, S.; Safari, E.; Farahmand, L. Breast cancer: Biology, biomarkers, and treatments. Int. Immunopharmacol. 2020, 84, 106535. [Google Scholar] [CrossRef]
- Rubinger, L.; Gazendam, A.; Ekhtiari, S.; Bhandari, M. Machine learning and artificial intelligence in research and healthcare. Injury 2023, 54, S69–S73. [Google Scholar] [CrossRef]
- Lee, M. Deep Learning Techniques with Genomic Data in Cancer Prognosis: A Comprehensive Review of the 2021–2023 Literature. Biology 2023, 12, 893. [Google Scholar] [CrossRef]
- Vicini, S.; Bortolotto, C.; Rengo, M.; Ballerini, D.; Bellini, D.; Carbone, I.; Preda, L.; Laghi, A.; Coppola, F.; Faggioni, L. A narrative review on current imaging applications of artificial intelligence and radiomics in oncology: Focus on the three most common cancers. Radiol. Medica 2022, 127, 819–836. [Google Scholar] [CrossRef]
- Rajpal, S.; Rajpal, A.; Agarwal, M.; Kumar, V.; Abraham, A.; Khanna, D.; Kumar, N. XAI-CNVMarker: Explainable AI-based copy number variant biomarker discovery for breast cancer subtypes. Biomed. Signal Process. Control 2023, 84, 104979. [Google Scholar] [CrossRef]
- Chakraborty, D.; Ivan, C.; Amero, P.; Khan, M.; Rodriguez-Aguayo, C.; Başağaoğlu, H.; Lopez-Berestein, G. Explainable artificial intelligence reveals novel insight into tumor microenvironment conditions linked with better prognosis in patients with breast cancer. Cancers 2021, 13, 3450. [Google Scholar] [CrossRef] [PubMed]
- Cansel, N.; Hilal Yagin, F.; Akan, M.; Ilkay Aygul, B. Interpretable estimation of suicide risk and severity from complete blood count parameters with explainable artificial intelligence methods. Psychiatr. Danub. 2023, 35, 62–72. [Google Scholar] [CrossRef]
- Idrees, M.; Sohail, A. Explainable machine learning of the breast cancer staging for designing smart biomarker sensors. Sens. Int. 2022, 3, 100202. [Google Scholar] [CrossRef]
- Van’t Veer, L.J.; Dai, H.; Van De Vijver, M.J.; He, Y.D.; Hart, A.A.; Mao, M.; Peterse, H.L.; Van Der Kooy, K.; Marton, M.J.; Witteveen, A.T. Gene expression profiling predicts clinical outcome of breast cancer. Nature 2002, 415, 530–536. [Google Scholar] [CrossRef] [PubMed]
- Ahsan, M.M.; Mahmud, M.P.; Saha, P.K.; Gupta, K.D.; Siddique, Z. Effect of data scaling methods on machine learning algorithms and model performance. Technologies 2021, 9, 52. [Google Scholar] [CrossRef]
- Yu, B.; Chen, C.; Wang, X.; Yu, Z.; Ma, A.; Liu, B. Prediction of protein–protein interactions based on elastic net and deep forest. Expert Syst. Appl. 2021, 176, 114876. [Google Scholar] [CrossRef]
- Shrestha, K.; Alsadoon, O.H.; Alsadoon, A.; Rashid, T.A.; Ali, R.S.; Prasad, P.; Jerew, O.D. A novel solution of an elastic net regularisation for dementia knowledge discovery using deep learning. J. Exp. Theor. Artif. Intell. 2021, 35, 807–829. [Google Scholar] [CrossRef]
- Taha, A.A.; Malebary, S.J. An intelligent approach to credit card fraud detection using an optimized light gradient boosting machine. IEEE Access 2020, 8, 25579–25587. [Google Scholar] [CrossRef]
- McCarty, D.A.; Kim, H.W.; Lee, H.K. Evaluation of light gradient boosted machine learning technique in large scale land use and land cover classification. Environments 2020, 7, 84. [Google Scholar] [CrossRef]
- Yang, B.; Li, W.; Wu, X.; Zhong, W.; Wang, J.; Zhou, Y.; Zhou, Z. Comparison of Ruptured Intracranial Aneurysms Identification Using Different Machine Learning Algorithms and Radiomics. Diagnostics 2023, 13, 2627. [Google Scholar] [CrossRef]
- Kumar, P.S.; Kumari, A.; Mohapatra, S.; Naik, B.; Nayak, J.; Mishra, M. CatBoost ensemble approach for diabetes risk prediction at early stages. In Proceedings of the 2021 1st Odisha International Conference on Electrical Power Engineering, Communication and Computing Technology (ODICON), Bhubaneswar, India, 8–9 January 2021; pp. 1–6. [Google Scholar]
- Wang, L.; Wu, C.; Tang, L.; Zhang, W.; Lacasse, S.; Liu, H.; Gao, L. Efficient reliability analysis of earth dam slope stability using extreme gradient boosting method. Acta Geotech. 2020, 15, 3135–3150. [Google Scholar] [CrossRef]
- Budholiya, K.; Shrivastava, S.K.; Sharma, V. An optimized XGBoost based diagnostic system for effective prediction of heart disease. J. King Saud Univ. -Comput. Inf. Sci. 2022, 34, 4514–4523. [Google Scholar] [CrossRef]
- Hew, K.F.; Hu, X.; Qiao, C.; Tang, Y. What predicts student satisfaction with MOOCs: A gradient boosting trees supervised machine learning and sentiment analysis approach. Comput. Educ. 2020, 145, 103724. [Google Scholar] [CrossRef]
- Sachdeva, S.; Kumar, B. Comparison of gradient boosted decision trees and random forest for groundwater potential mapping in Dholpur (Rajasthan), India. Stoch. Environ. Res. Risk Assess. 2021, 35, 287–306. [Google Scholar] [CrossRef]
- Zhang, Y.; Ni, M.; Zhang, C.; Liang, S.; Fang, S.; Li, R.; Tan, Z. Research and application of AdaBoost algorithm based on SVM. In Proceedings of the 2019 IEEE 8th Joint International Information Technology and Artificial Intelligence Conference (ITAIC), Chongqing, China, 24–26 May 2019; pp. 662–666. [Google Scholar]
- Sevinç, E. An empowered AdaBoost algorithm implementation: A COVID-19 dataset study. Comput. Ind. Eng. 2022, 165, 107912. [Google Scholar] [CrossRef] [PubMed]
- Yacouby, R.; Axman, D. Probabilistic extension of precision, recall, and f1 score for more thorough evaluation of classification models. In Proceedings of the First Workshop on Evaluation and Comparison of NLP Systems, Online, November 2020; pp. 79–91. [Google Scholar]
- Liu, W.; Wang, S.; Ye, Z.; Xu, P.; Xia, X.; Guo, M. Prediction of lung metastases in thyroid cancer using machine learning based on SEER database. Cancer Med. 2022, 11, 2503–2515. [Google Scholar] [CrossRef]
- Antwarg, L.; Miller, R.M.; Shapira, B.; Rokach, L. Explaining anomalies detected by autoencoders using SHAP. arXiv 2019, arXiv:1903.02407. [Google Scholar]
- Nohara, Y.; Matsumoto, K.; Soejima, H.; Nakashima, N. Explanation of machine learning models using shapley additive explanation and application for real data in hospital. Comput. Methods Programs Biomed. 2022, 214, 106584. [Google Scholar] [CrossRef]
- Kim, Y.; Kim, Y. Explainable heat-related mortality with random forest and SHapley Additive exPlanations (SHAP) models. Sustain. Cities Soc. 2022, 79, 103677. [Google Scholar] [CrossRef]
- Zou, H.; Hastie, T. Regularization and variable selection via the elastic net. J. R. Stat. Soc. Ser. B Stat. Methodol. 2005, 67, 301–320. [Google Scholar] [CrossRef]
- Steyerberg, E.W.; Vickers, A.J.; Cook, N.R.; Gerds, T.; Gonen, M.; Obuchowski, N.; Pencina, M.J.; Kattan, M.W. Assessing the performance of prediction models: A framework for some traditional and novel measures. Epidemiology 2010, 21, 128. [Google Scholar] [CrossRef] [PubMed]
- Islam, M.M.; Haque, M.R.; Iqbal, H.; Hasan, M.M.; Hasan, M.; Kabir, M.N. Breast cancer prediction: A comparative study using machine learning techniques. SN Comput. Sci. 2020, 1, 290. [Google Scholar] [CrossRef]
- Wang, J.; Yang, X.; Cai, H.; Tan, W.; Jin, C.; Li, L. Discrimination of breast cancer with microcalcifications on mammography by deep learning. Sci. Rep. 2016, 6, 27327. [Google Scholar] [CrossRef] [PubMed]
- Epping, M.T.; Meijer, L.A.; Krijgsman, O.; Bos, J.L.; Pandolfi, P.P.; Bernards, R. TSPYL5 suppresses p53 levels and function by physical interaction with USP7. Nat. Cell Biol. 2011, 13, 102–108. [Google Scholar] [CrossRef]
- Liu, M.; Ingle, J.N.; Fridley, B.L.; Buzdar, A.U.; Robson, M.E.; Kubo, M.; Wang, L.; Batzler, A.; Jenkins, G.D.; Pietrzak, T.L. TSPYL5 SNPs: Association with plasma estradiol concentrations and aromatase expression. Mol. Endocrinol. 2013, 27, 657–670. [Google Scholar] [CrossRef] [PubMed]
- Taghavi, A.; Akbari, M.E.; Hashemi-Bahremani, M.; Nafissi, N.; Khalilnezhad, A.; Poorhosseini, S.M.; Hashemi-Gorji, F.; Yassaee, V.R. Gene expression profiling of the 8q22-24 position in human breast cancer: TSPYL5, MTDH, ATAD2 and CCNE2 genes are implicated in oncogenesis, while WISP1 and EXT1 genes may predict a risk of metastasis. Oncol. Lett. 2016, 12, 3845–3855. [Google Scholar] [CrossRef]
- Span, P.; Bussink, J.; Manders, P.; Beex, L.; Sweep, C. Carbonic anhydrase-9 expression levels and prognosis in human breast cancer: Association with treatment outcome. Br. J. Cancer 2003, 89, 271–276. [Google Scholar] [CrossRef]
- Liu, Y.; Baglia, M.; Zheng, Y.; Blot, W.; Bao, P.-P.; Cai, H.; Nechuta, S.; Zheng, W.; Cai, Q.; Shu, X.O. ALDH1A1 mRNA expression in association with prognosis of triple-negative breast cancer. Oncotarget 2015, 6, 41360. [Google Scholar] [CrossRef][Green Version]
- Sakuma, S.; D’Angelo, M.A. The roles of the nuclear pore complex in cellular dysfunction, aging and disease. Semin. Cell Dev. Biol. 2017, 68, 72–84. [Google Scholar] [CrossRef]
- Köhler, A.; Hurt, E. Gene regulation by nucleoporins and links to cancer. Mol. Cell 2010, 38, 6–15. [Google Scholar] [CrossRef] [PubMed]
- Amin, R.; Shukla, A.; Zhu, J.J.; Kim, S.; Wang, P.; Tian, S.Z.; Tran, A.D.; Paul, D.; Cappell, S.D.; Burkett, S. Nuclear pore protein NUP210 depletion suppresses metastasis through heterochromatin-mediated disruption of tumor cell mechanical response. Nat. Commun. 2021, 12, 7216. [Google Scholar] [CrossRef] [PubMed]
- Epping, M.; Hart, A.; Glas, A.; Krijgsman, O.; Bernards, R. PRAME expression and clinical outcome of breast cancer. Br. J. Cancer 2008, 99, 398–403. [Google Scholar] [CrossRef] [PubMed]
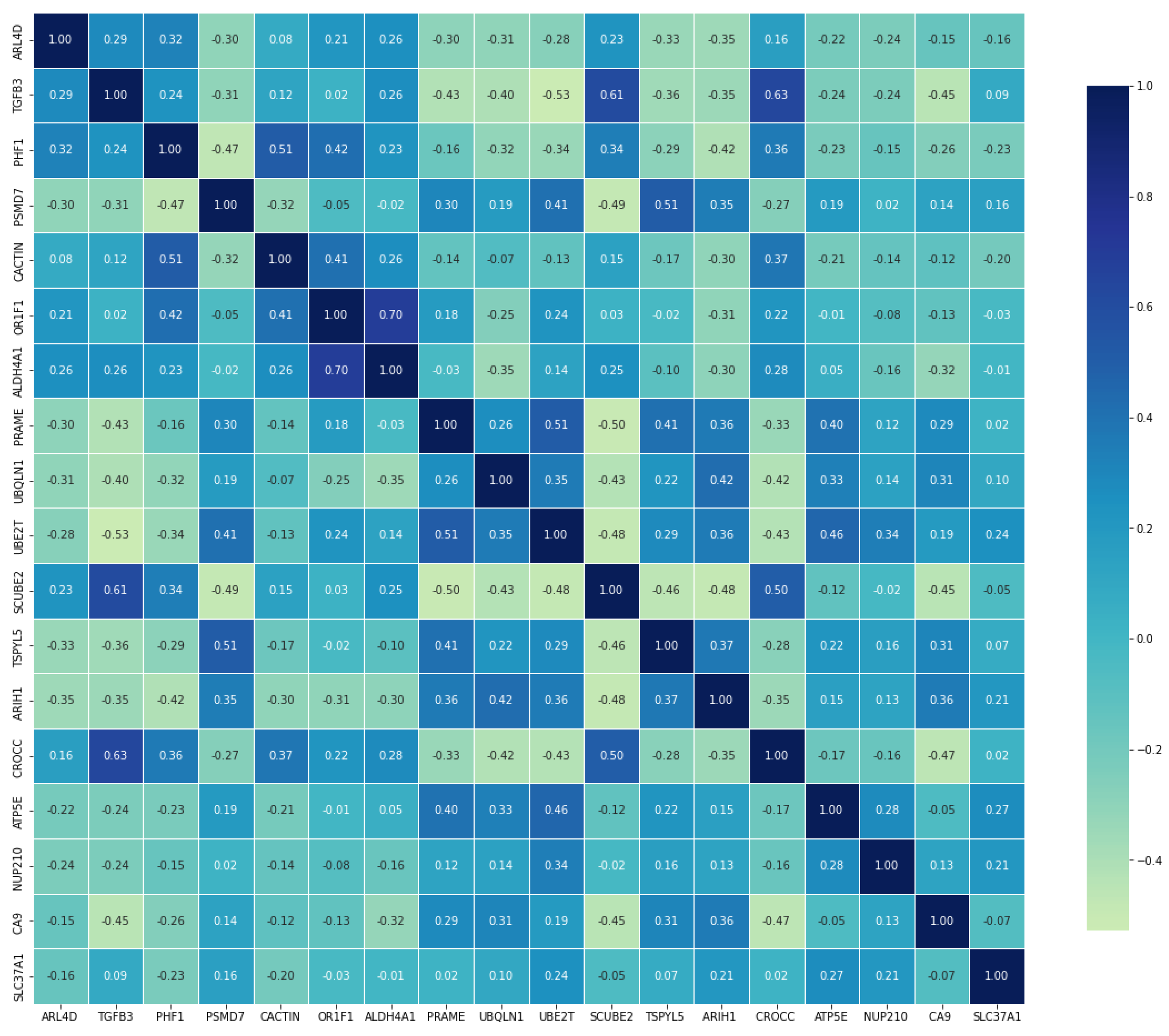
| Gene * | BC Metastasis Status | U-Value | p-Value ** | ES | |
|---|---|---|---|---|---|
| Non-Metastasis | Metastasis | ||||
| ARL4D | 0.401 (1.028) | −0.198 (0.477) | 1093 | <0.001 | 0.143 (medium) |
| TGFB3 | 0.38 (0.994) | −0.248 (0.754) | 1093 | <0.001 | 0.164 (medium) |
| PHF1 | 0.409 (0.916) | −0.221 (0.505) | 788 | <0.001 | 0.173 (medium) |
| PSMD7 | −0.228 (0.67) | 0.458 (0.808) | 887 | <0.001 | 0.188 (medium) |
| CACTIN | 0.248 (0.74) | −0.36 (0.748) | 865 | <0.001 | 0.213 (medium) |
| OR1F1 | 0.404 (1.237) | −0.128 (0.471) | 932.5 | <0.001 | 0.143 (medium) |
| ALDH4A1 | 0.312 (0.713) | −0.412 (0.962) | 982.5 | <0.001 | 0.194 (medium) |
| PRAME | −0.072 (0.226) | 0.744 (1.158) | 1086.5 | <0.001 | 0.135 (medium) |
| UBQLN1 | −0.381 (0.755) | 0.262 (1.06) | 1042 | <0.001 | 0.17 (medium) |
| UBE2T | −0.329 (0.923) | 0.327 (0.635) | 986 | <0.001 | 0.176 (medium) |
| SCUBE2 | 0.245 (0.484) | −0.576 (0.735) | 915 | <0.001 | 0.212 (medium) |
| TSPYL5 | −0.305 (0.599) | 0.51 (0.759) | 961.5 | <0.001 | 0.306 (large) |
| ARIH1 | −0.396 (0.935) | 0.331 (0.792) | 844 | <0.001 | 0.197 (medium) |
| CROCC | 0.455 (0.77) | −0.291 (0.706) | 1022 | <0.001 | 0.191 (medium) |
| ATP5E | −0.401 (0.818) | 0.283 (1.057) | 1013 | <0.001 | 0.196 (medium) |
| NUP210 | −0.094 (0.532) | 0.468 (0.958) | 1075 | 0.001 | 0.117 (medium) |
| CA9 | −0.177 (0.617) | 0.379 (1.171) | 852 | <0.001 | 0.176 (medium) |
| SLC37A1 | −0.121 (0.872) | 0.338 (1.085) | 1051 | 0.002 | 0.0977 (medium) |
| Model | Accuracy | F1 Score | Precision | Recall | AUC | Brier Score |
|---|---|---|---|---|---|---|
| LightGBM | 96 | 96.8 | 100 | 93.8 | 99.3 | 0.024 |
| CatBoost | 84 | 86.7 | 92.9 | 81.2 | 85.1 | 0.057 |
| XGBoost | 92 | 93.8 | 93.8 | 93.8 | 97.9 | 0.026 |
| GBT | 80 | 82.8 | 92.3 | 75 | 94.4 | 0.081 |
| AdaBoost | 88 | 90.9 | 88.2 | 93.8 | 93.1 | 0.077 |
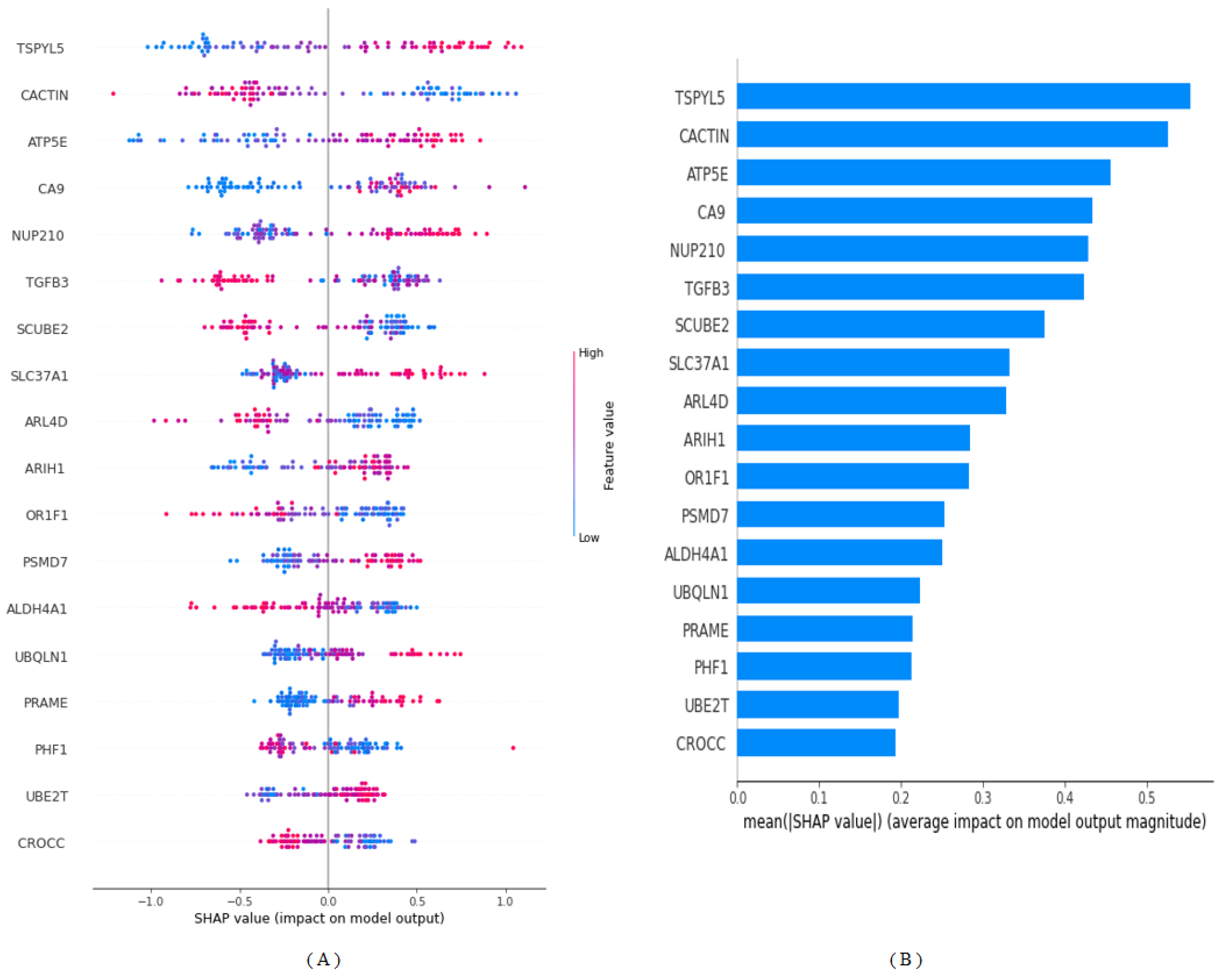
| Gene Name | Importance Score |
|---|---|
| TSPYL5 | 0.092 |
| CACTIN | 0.088 |
| ATP5E | 0.076 |
| CA9 | 0.072 |
| NUP210 | 0.071 |
| TGFB3 | 0.070 |
| SCUBE2 | 0.062 |
| SLC37A1 | 0.055 |
| ARL4D | 0.055 |
| ARIH1 | 0.047 |
| OR1F1 | 0.047 |
| PSMD7 | 0.042 |
| ALDH4A1 | 0.042 |
| UBQLN1 | 0.037 |
| PRAME | 0.035 |
| PHF1 | 0.035 |
| UBE2T | 0.032 |
| CROCC | 0.032 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yagin, B.; Yagin, F.H.; Colak, C.; Inceoglu, F.; Kadry, S.; Kim, J. Cancer Metastasis Prediction and Genomic Biomarker Identification through Machine Learning and eXplainable Artificial Intelligence in Breast Cancer Research. Diagnostics 2023, 13, 3314. https://doi.org/10.3390/diagnostics13213314
Yagin B, Yagin FH, Colak C, Inceoglu F, Kadry S, Kim J. Cancer Metastasis Prediction and Genomic Biomarker Identification through Machine Learning and eXplainable Artificial Intelligence in Breast Cancer Research. Diagnostics. 2023; 13(21):3314. https://doi.org/10.3390/diagnostics13213314
Chicago/Turabian StyleYagin, Burak, Fatma Hilal Yagin, Cemil Colak, Feyza Inceoglu, Seifedine Kadry, and Jungeun Kim. 2023. "Cancer Metastasis Prediction and Genomic Biomarker Identification through Machine Learning and eXplainable Artificial Intelligence in Breast Cancer Research" Diagnostics 13, no. 21: 3314. https://doi.org/10.3390/diagnostics13213314
APA StyleYagin, B., Yagin, F. H., Colak, C., Inceoglu, F., Kadry, S., & Kim, J. (2023). Cancer Metastasis Prediction and Genomic Biomarker Identification through Machine Learning and eXplainable Artificial Intelligence in Breast Cancer Research. Diagnostics, 13(21), 3314. https://doi.org/10.3390/diagnostics13213314









