A Bayesian Inference Based Computational Tool for Parametric and Nonparametric Medical Diagnosis
Abstract
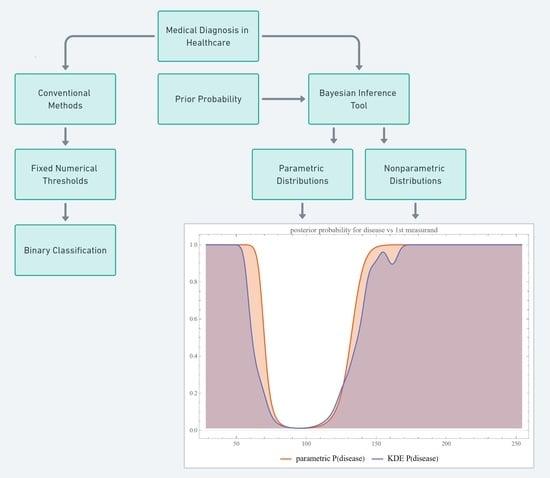
1. Introduction
2. Methods
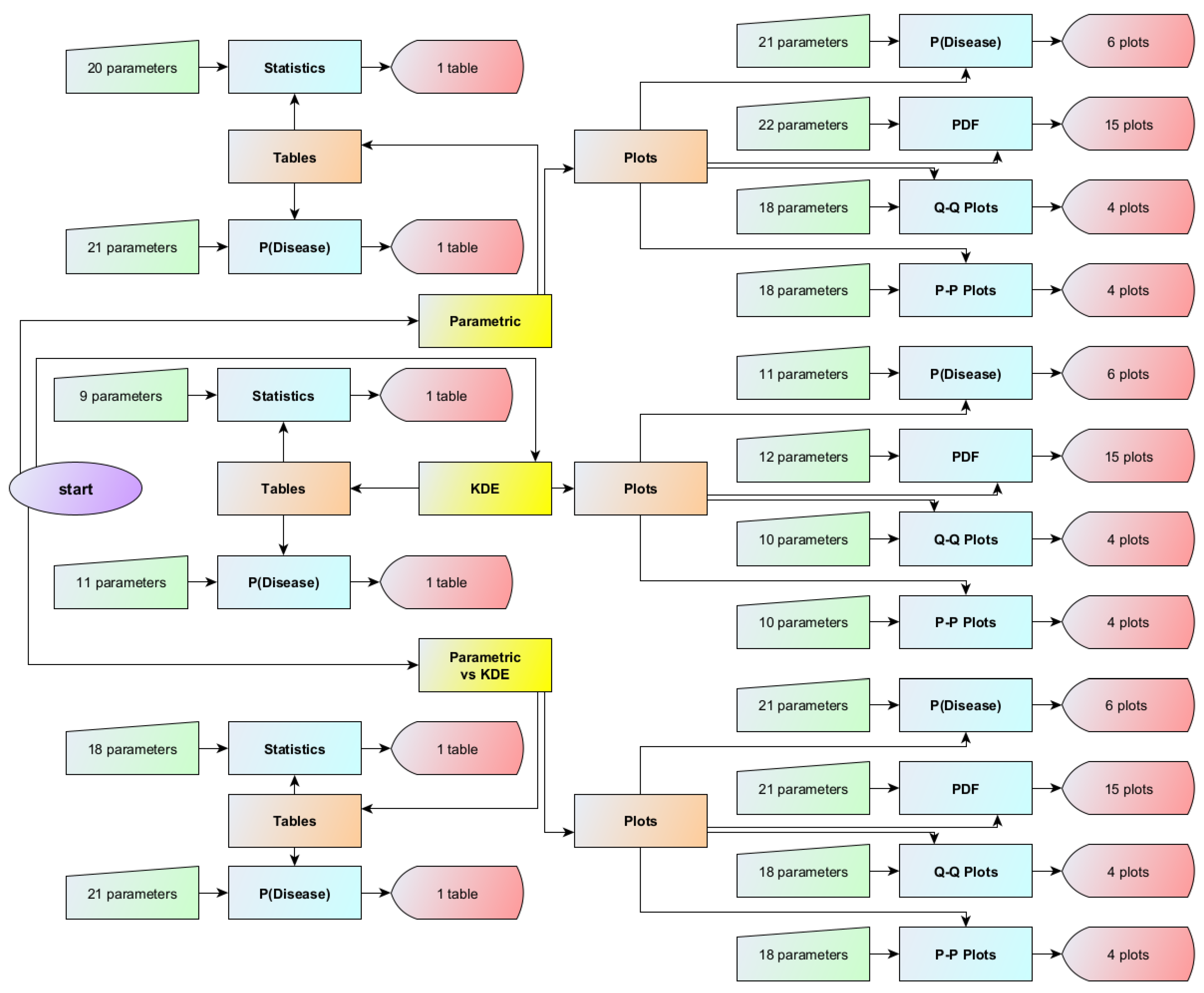
2.1. The Program
Datasets
2.2. Computational Methods
2.2.1. Bayesian Diagnostic Approach
2.2.2. Parametric Distributions
- Normal Distribution
- 1.1
- Univariate
- 1.2
- Bivariate
- Lognormal Distribution
- 2.1
- Univariate
- 2.2
- Bivariate
- Gamma Distribution
- 3.1
- Univariate
- 3.2
- Bivariate
- Copula Distributions
2.2.3. Nonparametric Distributions
Histograms
Kernel Density Estimators (KDEs)
2.3. Interface of the Program
2.3.1. Input Parameters
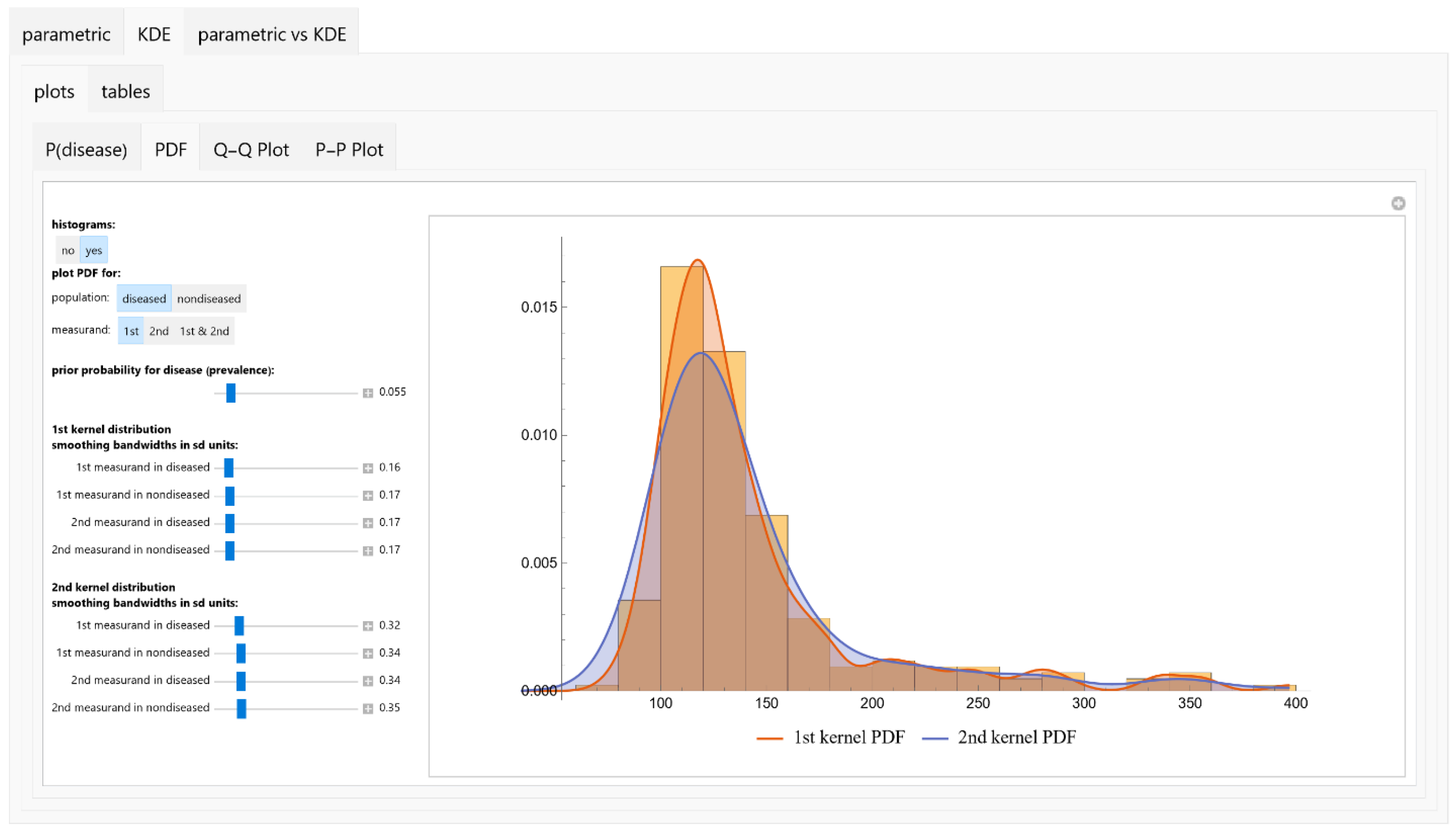
Prior Probability
Parametric Distributions
- Distribution Selection: The user selects the type of distribution from a predefined list:
- 1.1
- Normal Distribution.
- 1.2
- Lognormal Distribution.
- 1.3
- Gamma Distribution.
- Statistical Parameters: For each chosen distribution, the user defines the mean μ and standard deviation σ of the measurand in the respective population.
- Correlation Coefficients: The user specifies the correlation coefficients ρ between the measurands of the first and second diagnostic tests for both diseased and nondiseased populations.
KDEs
- Bandwidth Parameter: For each KDE, the user defines the bandwidth parameter h.
- Correlation Coefficients: As with parametric distributions, the user defines the correlation coefficients ρ between the measurands of the two diagnostic tests.
2.3.2. Output Specifications
Visualizations
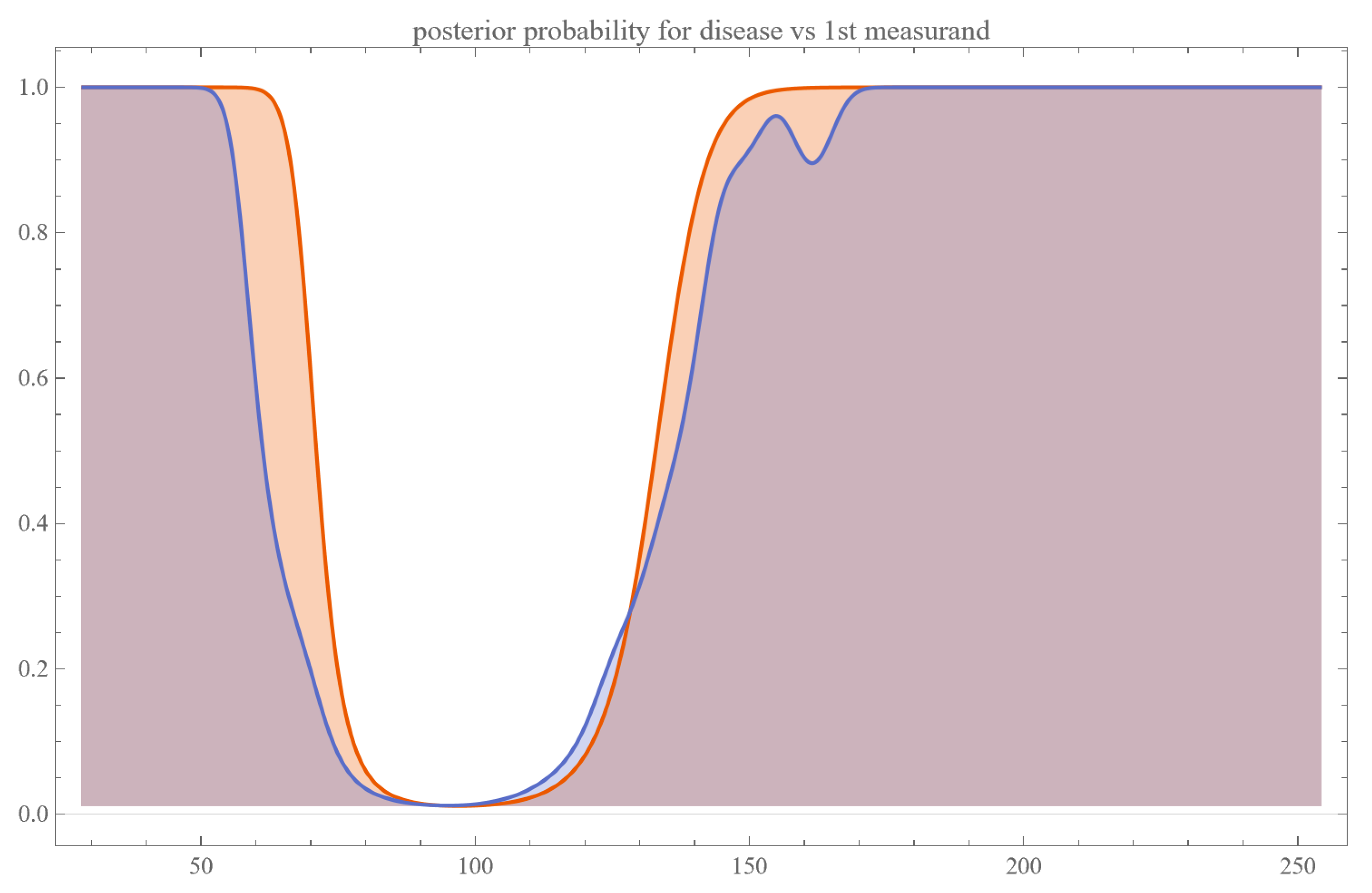
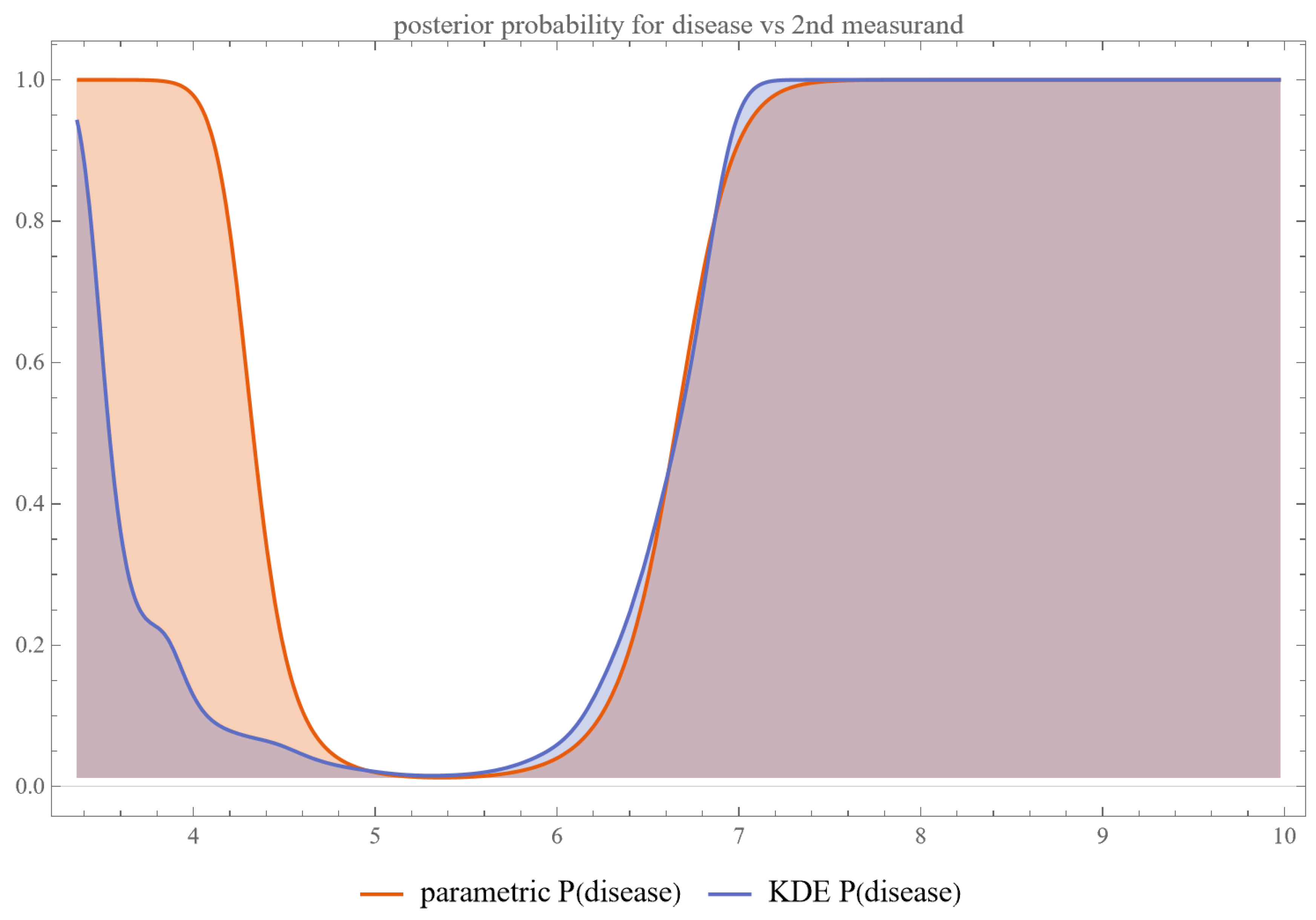
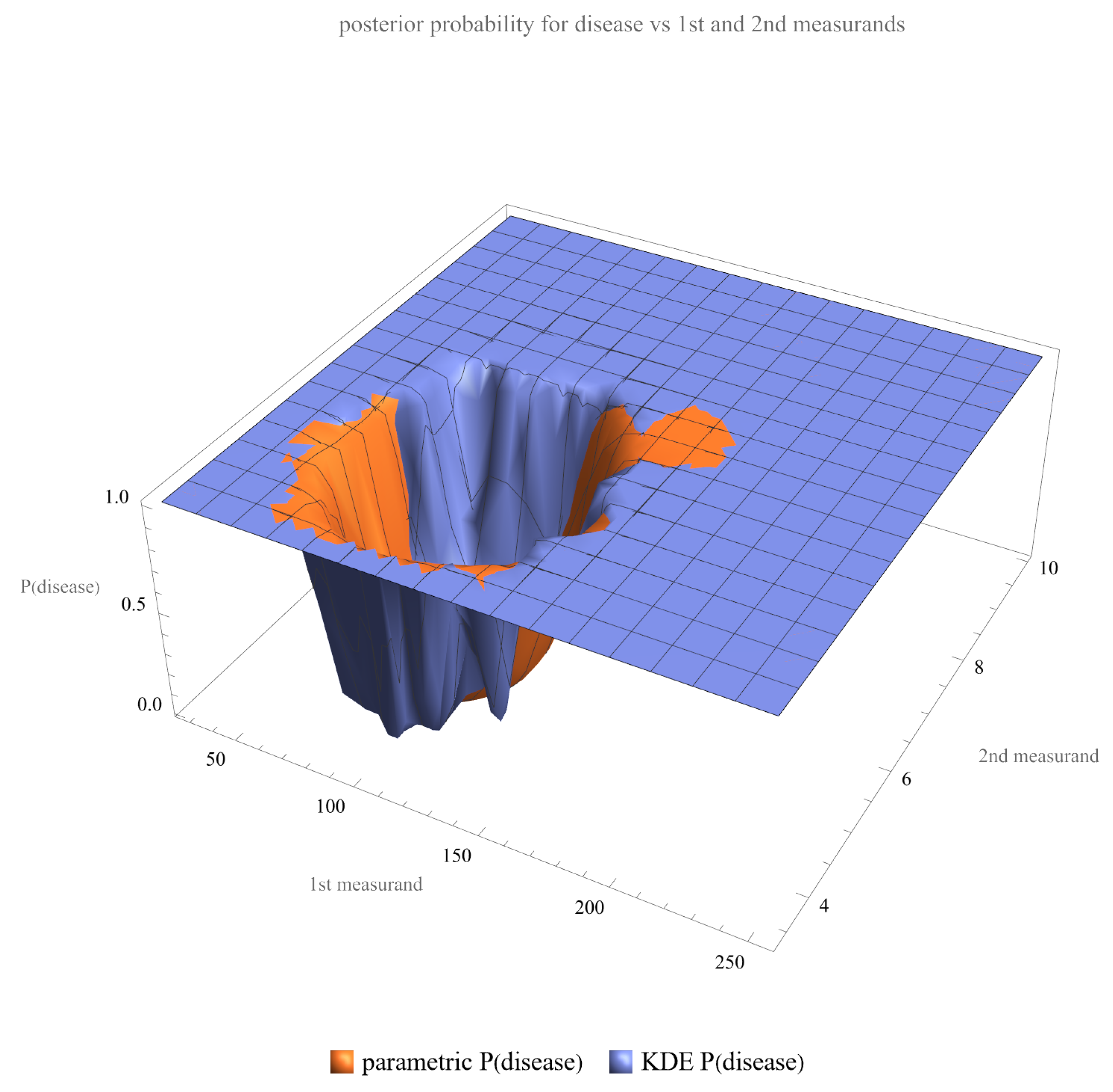
- Posterior Probability of Disease: Plots are generated to show the posterior probability of disease for each measurand and their combination.
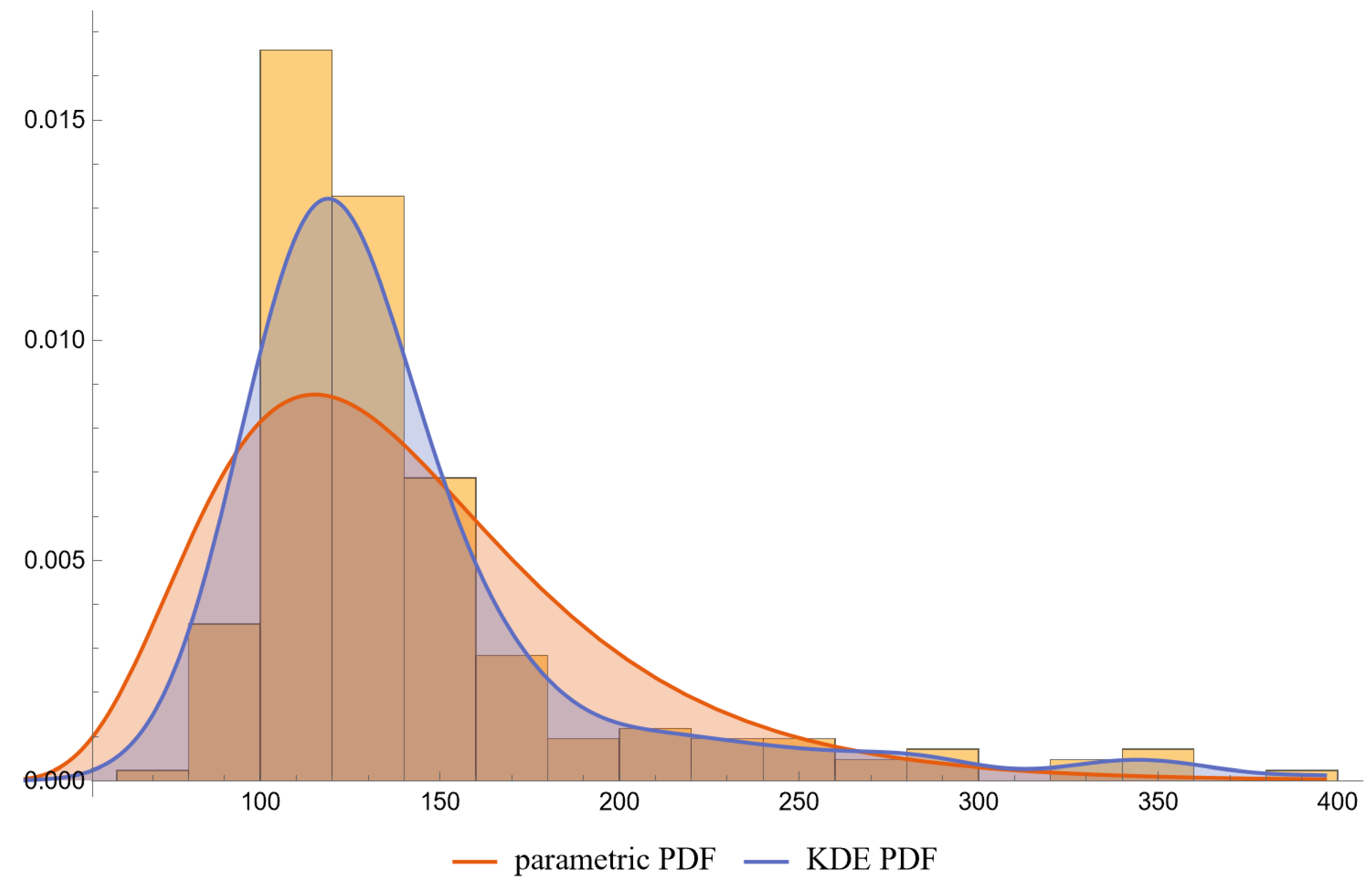
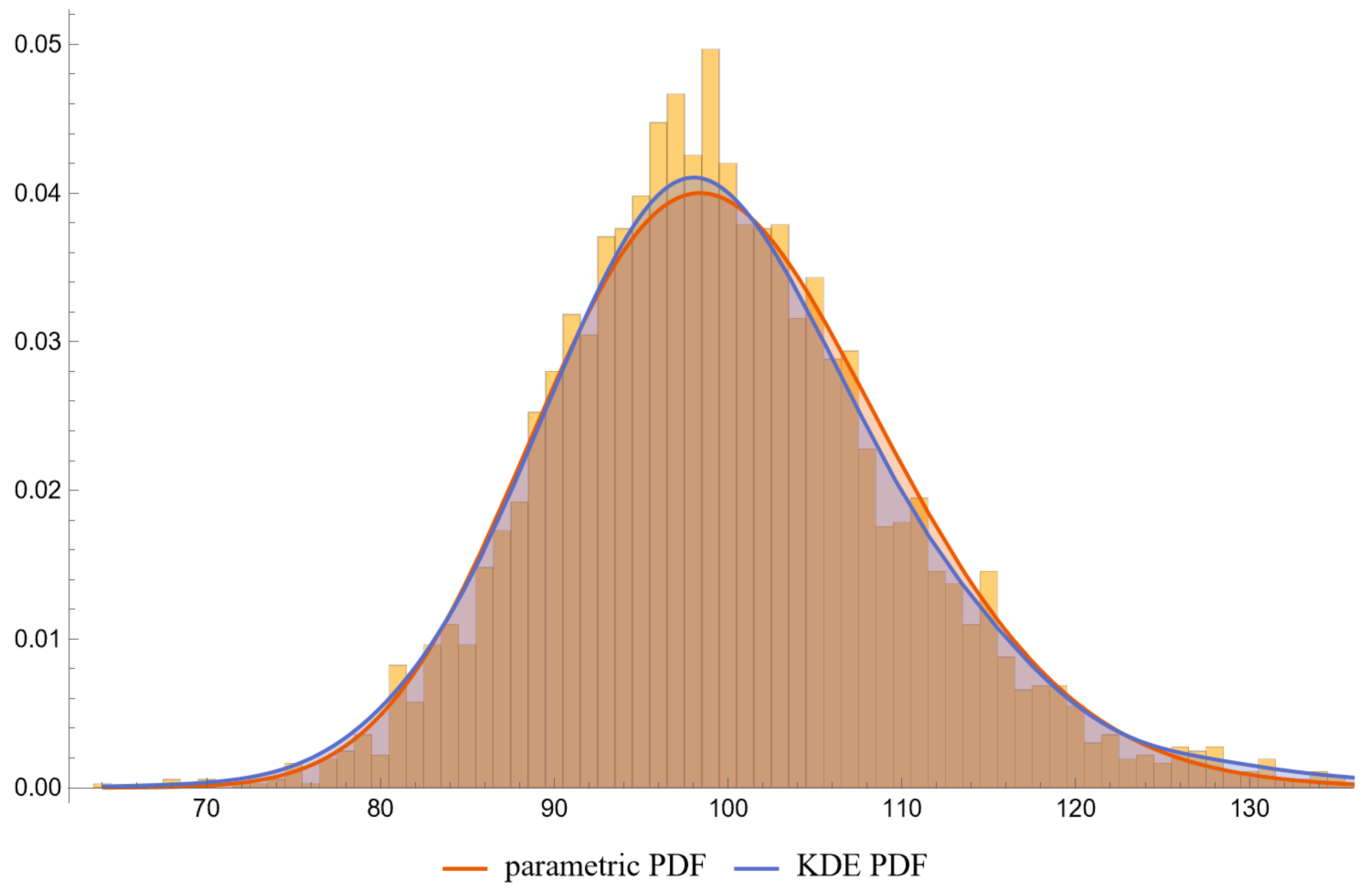
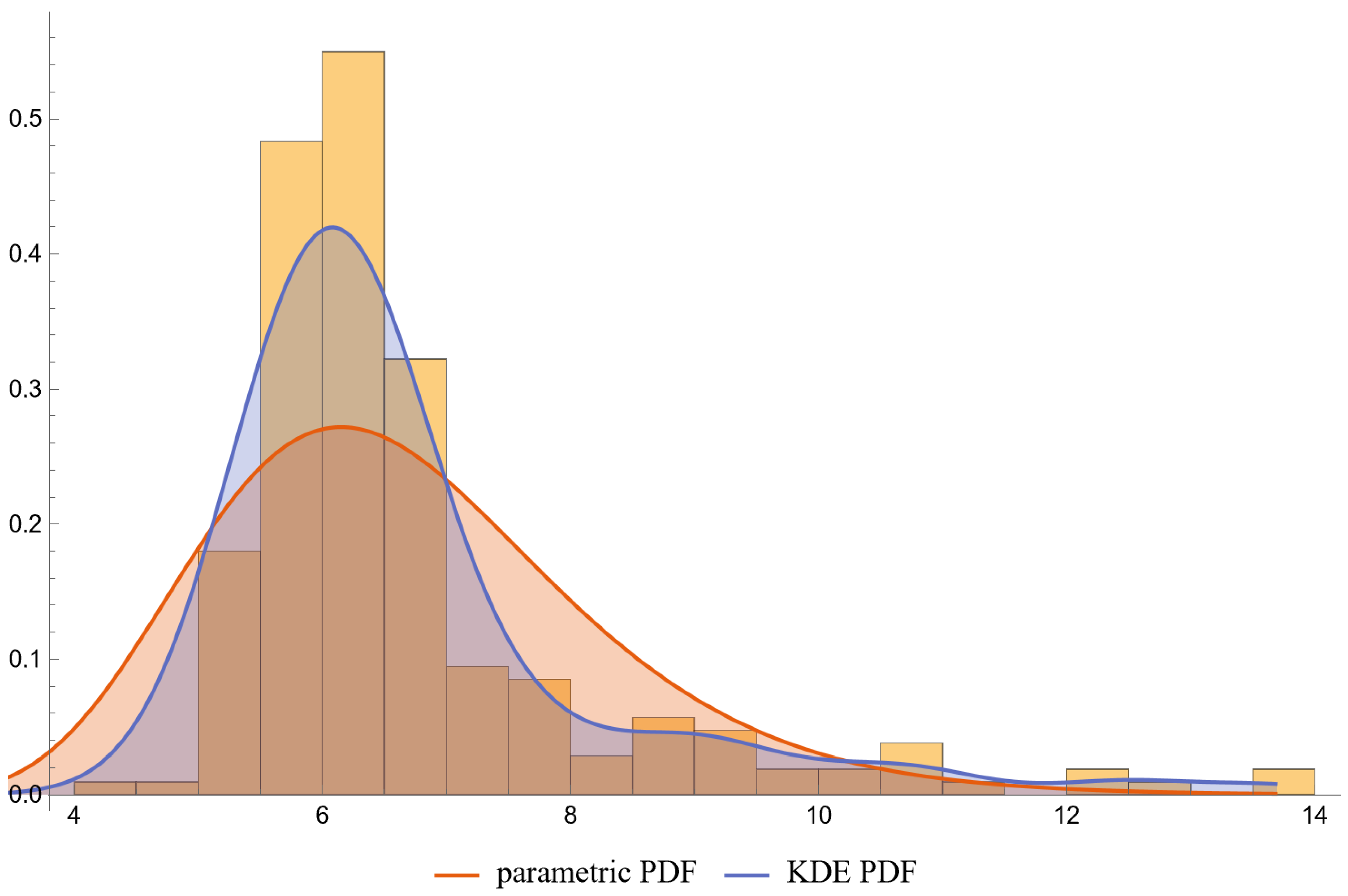
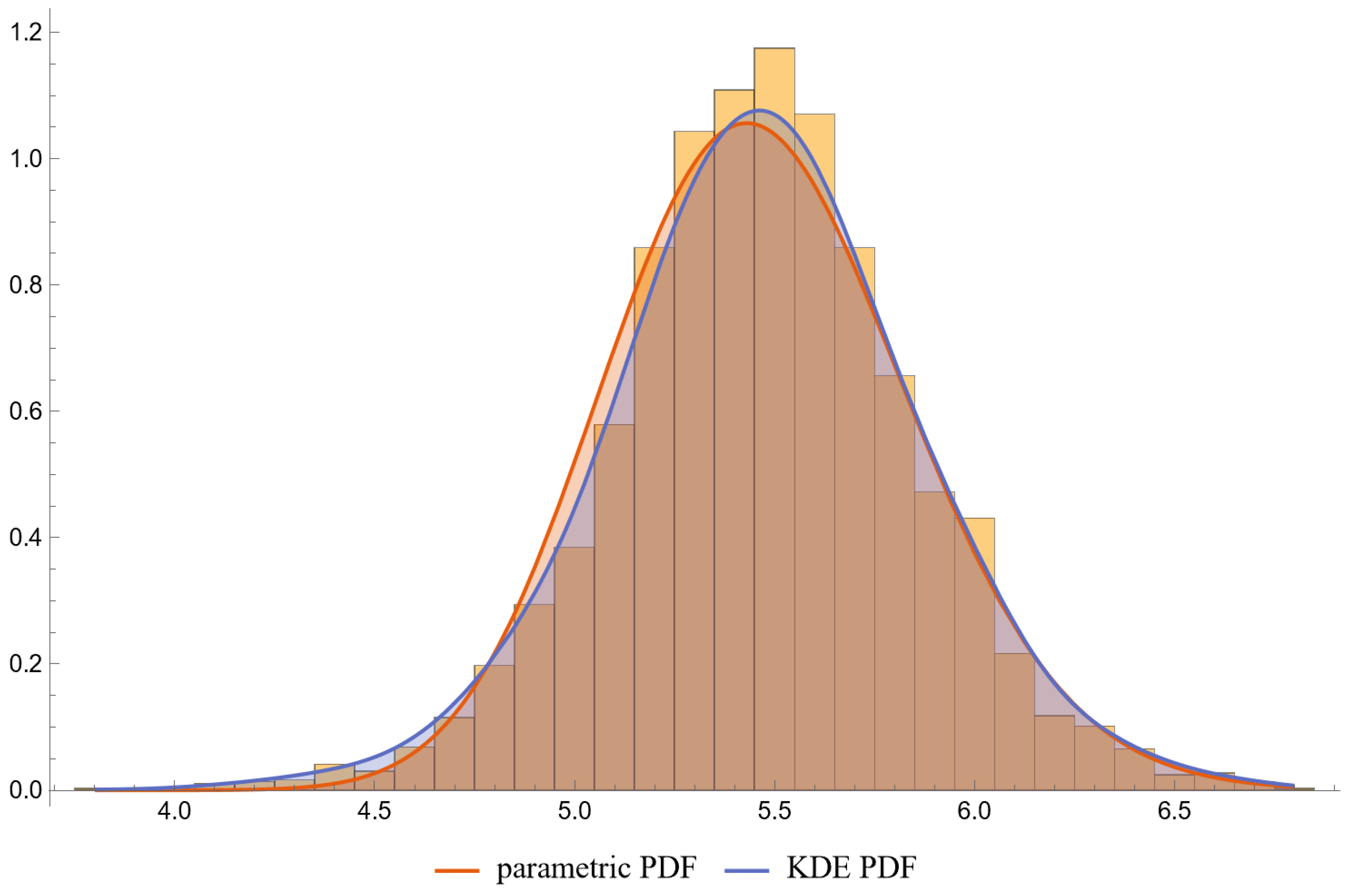
- PDFs: Univariate PDFs for each measurand and the bivariate PDF of their combination are plotted. An option to overlay histograms on these plots is also provided.
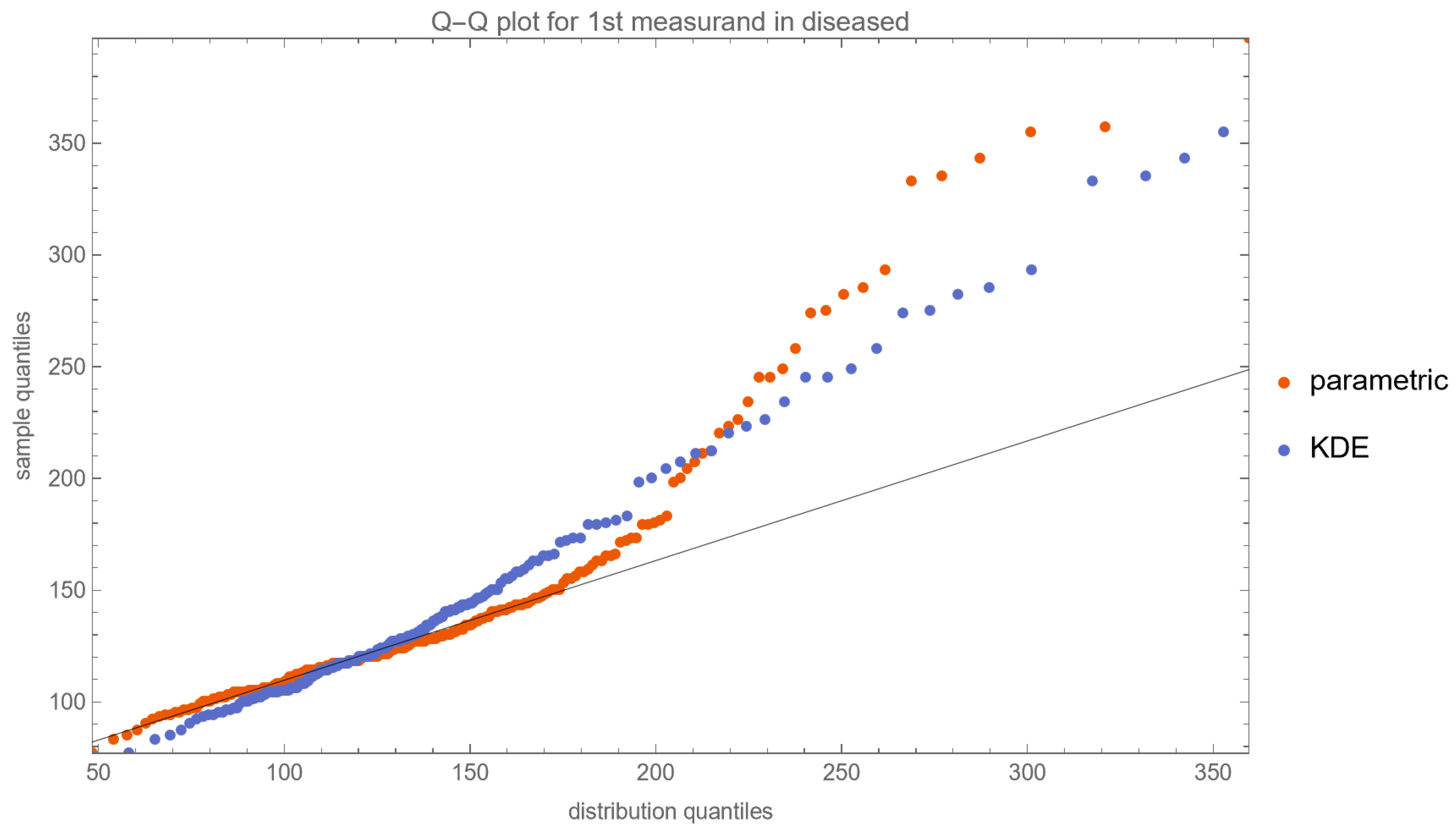
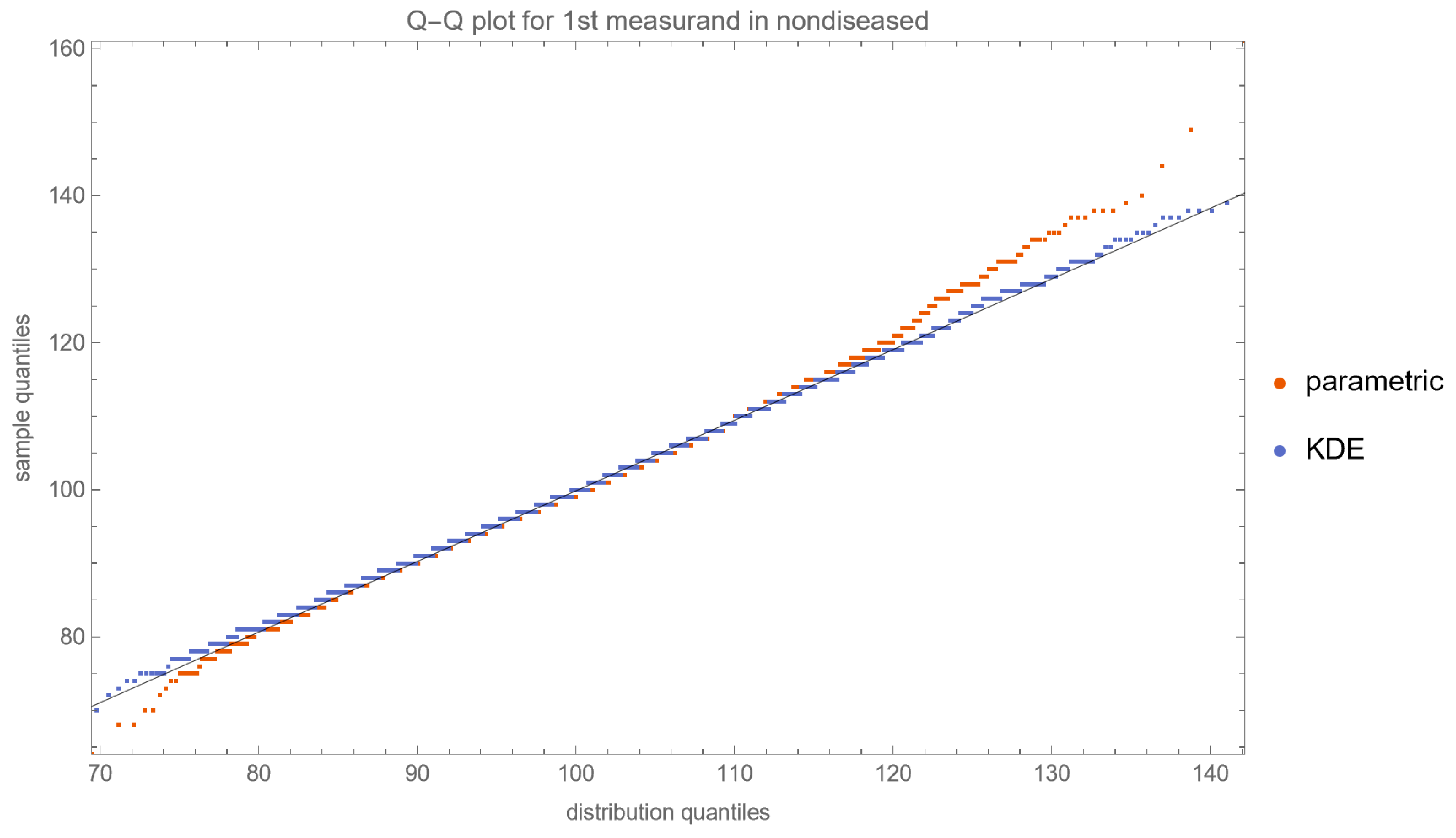
- Quantile–Quantile (Q–Q) Plots: These plots are produced for each measurand to examine its distributional characteristics [21].
- Probability–Probability (P–P) Plots: Similar to Q–Q plots, P–P plots are generated for further assessment of the distribution of each measurand [21].
Tables
- Population Statistics: The program tabulates key statistical metrics such as mean, median, standard deviation, skewness, kurtosis, and prior probability for each user-defined distribution and dataset. For each bivariate distribution of the two measurands in diseased and nondiseased populations, the correlation coefficients are calculated and displayed.
- Posterior Disease Probabilities: For a user-defined pair of test measurement values, the program computes and presents the posterior probabilities for disease for each measurand and their combination.
2.3.3. Illustrative Application
- Age 40–60 years (n = 11,782);
- Valid FPG, HbA1c, and OGTT measurements (n = 4015);
- A negative response to NHANES question DIQ010 regarding a diabetes diagnosis [25] (n = 3854);
- Non-pregnancy status (n = 3854).
3. Results
4. Discussion
4.1. Reevaluation of Traditional Diagnostic Methods
4.2. Challenges and Considerations in Bayesian Analysis for Disease Diagnosis
4.2.1. Ramifications of Incomplete Information
- Over-dependence on Prior Probabilities: The scarcity of empirically derived distributions amplifies reliance on prior probabilities, thereby inducing distortions in the calculation of posterior probabilities. This could result in suboptimal clinical judgments and potentially inaccurate diagnoses [32].
- Elevated Uncertainty: Insufficient data contributes to broader confidence intervals in the computed posterior probabilities, which, in turn, could exacerbate clinical indecisiveness [33].
- Risk of Bias: The introduction of systemic bias due to unrepresentative datasets could compromise the fidelity of Bayesian calculations [7].
- Imperative for Collaborative Research: More coordinated research is needed, including multi-center studies, meta-analyses, and open-access databases—to accumulate and disseminate data essential for effective Bayesian diagnosis [34].
4.2.2. Parametric Versus Nonparametric Bayesian Models
4.2.3. Multimodal Versus Double Sigmoidal Bayesian Probability of Disease Curve
Multimodal Curve
- (a)
- Complex Pathophysiology: Multiple etiological pathways may influence the same measurand in divergent ranges, adding layers of complexity to diagnostic processes [13].
- (b)
- Diagnostic Confounders: External variables affecting the measurand could compromise its efficacy as a standalone diagnostic criterion [38].
- (c)
- Population Subgroups: The existence of demographically or genetically distinct subgroups within the studied population could also account for the observed multimodality [39].
- (d)
- Statistical Artifacts: Demographically or genetically distinct subgroups may be a factor contributing to observed multimodal distributions [39].
Double Sigmoidal Curve
- (a)
- Two Zones of Risk: Such a curve suggests that the risk of the disease is heightened both at low and high extremes of the measurand but reduced in a middle “safe zone.”
- (b)
- Multifactorial Etiology: This might reflect a situation where both deficiency and excess of a particular biological factor contribute to disease risk. For example, both low and elevated levels of hormones may pose challenges to physiological homeostasis.
- (a)
- Threshold Decision-making: Unlike a single sigmoid curve, where one threshold may be adequate for diagnosis, the double sigmoid may necessitate multiple thresholds, defining a “safe zone” for the measurand.
- (b)
- Treatment Strategies: Clinicians must be cautious when intervening based on such a measurand, as moving the measurand too far in either direction could heighten risk.
- (c)
- Population Stratification: This curve shape might imply that different sub-populations or disease subtypes could be better distinguished with additional tests or measurements.
4.3. Shortcomings of This Study
- The OGTT was used as a reference diagnostic method for diabetes mellitus. The diagnostic threshold for 2-h PG was established in relation to the risk of diabetic retinopathy, a microvascular complication of diabetes mellitus [40]. However, glucose tolerance is influenced by complex interactions of factors, both physiological and environmental, which pose significant implications for clinical diagnosis and research. The considerations that could affect glucose tolerance and, therefore, the interpretation of the 2-h PG measurement, include the following:
- (a)
- (b)
- Diurnal Variability: Glucose tolerance is subject to diurnal variation, which could affect the 2-h PG test outcomes. Insulin sensitivity is generally higher in the morning than in the evening [43].
- (c)
- Physical Activity: Exercise improves insulin sensitivity and therefore could affect glucose tolerance tests. The timing and intensity of physical activity could have a direct influence on the 2-h PG results [44].
- (d)
- Dietary Patterns: Short-term and long-term dietary habits, including the macronutrient composition of the diet, may alter the body’s glucose and insulin response [45].
- (e)
- Stress and Emotional States: The acute stress response includes a transient rise in glucose levels as a result of catecholamine release, potentially affecting the 2-h PG test [46].
- (f)
- Medications: Certain medications like corticosteroids, antipsychotics, and diuretics affect glucose metabolism, thereby influencing 2-h PG test outcomes [47].
- (g)
- Genetic Factors: Genetic predispositions influence glucose tolerance, and not accounting for this introduce variability in the 2-h PG test [48].
- The lognormal distributions and the KDE, as parameterized in Table 2, fitted only approximately to the NHANES datasets of FPG and HbA1c measurements. It is well known that biological measurands, such as FPG and HbA1c, may not follow textbook statistical distributions like normal or lognormal distributions. Numerous papers have noted the skewness or kurtosis in the distribution of metabolic variables, urging the use of flexible statistical models [49,50].
Related Statistical Software
5. Conclusions and Future Directions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Correction Statement
Abbreviations
| probability density function | |
| CDF | cumulative distribution function |
| KDE | kernel density estimator |
| OGTT | oral glucose tolerance test |
| PG | plasma glucose |
| 2-h PG | plasma glucose, measured two hours after oral administration of 75 g of glucose, during an OGTT |
| FPG | fasting plasma glucose |
| HbA1c | glycated hemoglobin A1c |
| NHANES | National Health and Nutrition Examination Survey |
Appendix A
Appendix A.1. Formalisms and Notation
Appendix A.1.1. Tuples
Appendix A.1.2. Parameters
- v: prevalence of disease
- μ, m: mean
- σ, s: standard deviation
- ρ, r: correlation coefficient
- k: shape parameter
- : scale parameter
- h: nonparametric kernel density bandwidth
Appendix A.1.3. Functions
- : the inverse of the function
- : determinant of the matrix H
- : probability of the event A
- : conditional probability of the event A given the event B
- : covariance of two jointly distributed random variables and
- : expected value of a random variable
- : natural logarithm
- : likelihood function of the parameter θ given the observed value z of the random variable Z
- : likelihood function of the parameter θ given the observed values z of the random variable Z.
- : loglikelihood function of the parameter θ given the observed value z of the random variable Z
- : loglikelihood function of the parameter θ given the of observed values z of the random variable Z.
- : probability mass function of a discrete variable X
- : the k th q-quantile of a random variable
- : error function
- : complementary error function
- : gamma function
- : incomplete gamma function
- : regularized incomplete gamma function
- : generalized incomplete gamma function
- : regularized generalized incomplete gamma function
- : kernel function
- : univariate PDF
- : univariate PDF given the multivariate parameter θ
- : bivariate PDF
- : bivariate PDF given the multivariate parameter θ
- : univariate CDF
- : univariate CDF given the multivariate parameter θ
- : bivariate CDF
- : bivariate CDF given the multivariate parameter θ
Appendix A.2. Bayes Theorem
- represents the posterior probability of having the disease given the test results .
- denotes the likelihood of obtaining the test results given the presence of the disease.
- denotes the likelihood of obtaining the test results given the absence of the disease.
- is the prior probability or prevalence of the disease.
- signifies the overall probability of the test results .
Appendix A.3. Parametric Distributions
Appendix A.3.1. Normal Distribution
- (a)
- Univariate
- (b)
- Bivariate
Appendix A.3.2. Lognormal Distribution
- (a)
- Univariate
- (b)
- Bivariate
Appendix A.3.3. Gamma Distribution
- (a)
- Univariate
- (b)
- Bivariate
Appendix A.4. Copulas
Appendix A.4.1. X: Normally Distributed—Y: Lognormally Distributed
Appendix A.4.2. X: Lognormally Distributed—Y: Normally Distributed
Appendix A.4.3. X: Normally Distributed—Y: Gamma Distributed
Appendix A.4.4. X: Gamma Distributed– Y: Normally Distributed
Appendix A.4.5. X: Lognormally Distributed—Y: Gamma Distributed
Appendix A.4.6. X: Gamma Distributed—Y: Lognormally Distributed
Appendix A.5. Nonparametric Distributions
Appendix A.5.1. Histograms
Appendix A.5.2. KDEs
- n is the number of the observed values of the variable.
- h is the bandwidth, a positive scalar that determines the width and smoothness of the kernel.
- is the kernel function.
- (a)
- Univariate KDE
- (b)
- Bivariate KDE
Appendix B
Appendix B.1. Software Availability and Requirements
- B.1.1. Program name: Bayesian Diagnosis
- B.1.2. Project home page: https://www.hcsl.com/Tools/BayesianDiagnosis/ (accessed on 28 September 2023)
- B.1.3. Operating systems: Microsoft Windows, Linux, Apple iOS
- B.1.4. Other software requirements:
- For running the program: Wolfram Player®, freely available at: https://www.wolfram.com/player/ (accessed on 31 August 2023) or Wolfram Mathematica®.
- For editing the datasets: Wolfram Mathematica®.
- B.1.5. System requirements: Intel® i9™ or equivalent CPU and 32 GB of RAM
- B.1.6.6. License: Attribution—Noncommercial—ShareAlike 4.0 International Creative Commons License
Appendix C
Appendix C.1. A Note about the Program
Appendix C.1.1. About the Program Controls
Appendix C.1.2. Range of input parameters
- v: 0.010–0.500
- μ: 0.01–10,000.00
- : 0.01–3000.00
- ρ: −1.000–1.000
- h: 0.01–2.00
- x: 0.01–10,000.00
- y: 0.01–100,00.00
Appendix C.1.3. Datasets
References
- Weiner, E.; Simpson, J.A.; Oxford University Press. The Oxford English Dictionary; Clarendon Press: Oxford, UK, 1989. [Google Scholar]
- Zweig, M.H.; Campbell, G. Receiver-Operating Characteristic (ROC) Plots: A Fundamental Evaluation Tool in Clinical Medicine. Clin. Chem. 1993, 39, 561–577. [Google Scholar] [CrossRef] [PubMed]
- Chatzimichail, T.; Hatjimihail, A.T. A Software Tool for Calculating the Uncertainty of Diagnostic Accuracy Measures. Diagnostics 2021, 11, 406. [Google Scholar] [CrossRef]
- Djulbegovic, B.; van den Ende, J.; Hamm, R.M.; Mayrhofer, T.; Hozo, I.; Pauker, S.G.; International Threshold Working Group (ITWG). When Is Rational to Order a Diagnostic Test, or Prescribe Treatment: The Threshold Model as an Explanation of Practice Variation. Eur. J. Clin. Investig. 2015, 45, 485–493. [Google Scholar] [CrossRef] [PubMed]
- Choi, Y.-K.; Johnson, W.O.; Thurmond, M.C. Diagnosis Using Predictive Probabilities without Cut-Offs. Stat. Med. 2006, 25, 699–717. [Google Scholar] [CrossRef] [PubMed]
- Viana, M.A.G.; Ramakrishnan, V. Bayesian Estimates of Predictive Value and Related Parameters of a Diagnostic Test. Can. J. Stat. Rev. Can. Stat. 1992, 20, 311–321. [Google Scholar] [CrossRef]
- Gelman, A.; Carlin, J.B.; Stern, H.S.; Dunson, D.B.; Vehtari, A.; Rubin, D.B. Bayesian Data Analysis; CRC Press: Boca Raton, FL, USA, 2013. [Google Scholar]
- Van de Schoot, R.; Depaoli, S.; King, R.; Kramer, B.; Märtens, K.; Tadesse, M.G.; Vannucci, M.; Gelman, A.; Veen, D.; Willemsen, J.; et al. Bayesian Statistics and Modelling. Nature Reviews Methods Primers 2021, 1, 1–26. [Google Scholar] [CrossRef]
- Bours, M.J.L. Bayes’ Rule in Diagnosis. J. Clin. Epidemiol. 2021, 131, 158–160. [Google Scholar] [CrossRef]
- Carlin, B.P.; Louis, T.A. Bayesian Methods for Data Analysis; CRC Press: Boca Raton, FL, USA, 2008. [Google Scholar]
- Martin, G.M.; Frazier, D.T.; Maneesoonthorn, W.; Loaiza-Maya, R.; Huber, F.; Koop, G.; Maheu, J.; Nibbering, D.; Panagiotelis, A. Bayesian Forecasting in Economics and Finance: A Modern Review. Int. J. Forecast. 2023. [Google Scholar] [CrossRef]
- Liu, J.; Liu, S.J.; Wong, D.S.H. Process Fault Diagnosis Based on Bayesian Inference. In Computer Aided Chemical Engineering; Elsevier, Amsterdam, The Netherlands, Kraslawski, A., Turunen, I., Eds.; 2013; Volume 32, pp. 751–756. [Google Scholar]
- Dawid, A.P. Present Position and Potential Developments: Some Personal Views: Statistical Theory: The Prequential Approach. J. R. Stat. Soc. Ser. A 1984, 147, 278–292. [Google Scholar] [CrossRef]
- Lehmann, E.L.; Romano, J.P. Testing Statistical Hypotheses; Springer: New York, NY, USA, 2008. [Google Scholar]
- Box, G.E.P.; Cox, D.R. An Analysis of Transformations. J. R. Stat. Society. Ser. B Stat. Methodol. 1964, 26, 211–243. [Google Scholar] [CrossRef]
- D’Agostino, R.; Pearson, E.S. Tests for Departure from Normality. Empirical Results for the Distributions of b2 and √b1. Biometrika 1973, 60, 613–622. [Google Scholar] [CrossRef]
- Velanovich, V. Bayesian Analysis in the Diagnostic Process. Am. J. Med. Qual. Off. J. Am. Coll. Med. Qual. 1994, 9, 158–161. [Google Scholar] [CrossRef]
- Wilkes, E.H. A Practical Guide to Bayesian Statistics in Laboratory Medicine. Clin. Chem. 2022, 68, 893–905. [Google Scholar] [CrossRef]
- Geisser, S.; Johnson, W.O. Modes of Parametric Statistical Inference; John Wiley & Sons: Hoboken, NJ, USA, 2006. [Google Scholar]
- Spiegelhalter, D.J.; Abrams, K.R.; Myles, J.P. Bayesian Approaches to Clinical Trials and Health-Care Evaluation; John Wiley & Sons Australia, Limited: Milton, QLD, Australia, 2004. [Google Scholar]
- Wilk, M.B.; Gnanadesikan, R. Probability Plotting Methods for the Analysis of Data. Biometrika 1968, 55, 1–17. [Google Scholar] [CrossRef] [PubMed]
- ElSayed, N.A.; Aleppo, G.; Aroda, V.R.; Bannuru, R.R.; Brown, F.M.; Bruemmer, D.; Collins, B.S.; Gaglia, J.L.; Hilliard, M.E.; Isaacs, D.; et al. Classification and Diagnosis of Diabetes: Standards of Care in Diabetes-2023. Diabetes Care 2023, 46 (Suppl. S1), S19–S40. [Google Scholar] [CrossRef] [PubMed]
- Sun, H.; Saeedi, P.; Karuranga, S.; Pinkepank, M.; Ogurtsova, K.; Duncan, B.B.; Stein, C.; Basit, A.; Chan, J.C.N.; Mbanya, J.C.; et al. IDF Diabetes Atlas: Global, Regional and Country-Level Diabetes Prevalence Estimates for 2021 and Projections for 2045. Diabetes Res. Clin. Pract. 2022, 183, 109119. [Google Scholar] [CrossRef]
- Centers for Disease Control and Prevention. National Center for Health Statistics, 2005–2016 National Health and Nutrition Examination Survey Data. 2005–2016. Available online: https://wwwn.cdc.gov/nchs/nhanes/default.aspx (accessed on 4 September 2023).
- Centers for Disease Control and Prevention. National Health and Nutrition Examination Survey Questionnaire. 2005–2016. Available online: https://wwwn.cdc.gov/nchs/nhanes/Search/variablelist.aspx?Component=Questionnaire (accessed on 4 September 2023).
- Menke, A.; Rust, K.F.; Savage, P.J.; Cowie, C.C. Hemoglobin A1c, Fasting Plasma Glucose, and 2-Hour Plasma Glucose Distributions in U.S. Population Subgroups: NHANES 2005–2010. Ann. Epidemiol. 2014, 24, 83–89. [Google Scholar] [CrossRef]
- Silverman, B.W. Density Estimation for Statistics and Data Analysis; CRC Press: Boca Raton, FL, USA, 1986. [Google Scholar]
- Obermeyer, Z.; Emanuel, E.J. Predicting the Future–Big Data, Machine Learning, and Clinical Medicine. N. Engl. J. Med. 2016, 375, 1216–1219. [Google Scholar] [CrossRef] [PubMed]
- Topol, E.J. Individualized Medicine from Prewomb to Tomb. Cell 2014, 157, 241–253. [Google Scholar] [CrossRef]
- Tucker, L.A. Limited Agreement between Classifications of Diabetes and Prediabetes Resulting from the OGTT, Hemoglobin A1c, and Fasting Glucose Tests in 7412 U.S. Adults. J. Clin. Med. Res. 2020, 9, 2207. [Google Scholar] [CrossRef]
- Smith, A.F.M.; Gelfand, A.E. Bayesian Statistics without Tears: A Sampling-Resampling Perspective. Am. Stat. 1992, 46, 84–88. [Google Scholar]
- O’Hagan, A.; Buck, C.E.; Daneshkhah, A.; Eiser, J.R.; Garthwaite, P.H.; Jenkinson, D.J.; Oakley, J.E.; Rakow, T. Uncertain Judgements: Eliciting Experts’ Probabilities; John Wiley & Sons: Hoboken, NJ, USA, 2006. [Google Scholar]
- Berger, J.O. Statistical Decision Theory and Bayesian Analysis; Springer Science & Business Media: New York, NY, USA, 1985. [Google Scholar]
- McGrayne, S.B. The Theory That Would Not Die: How Bayes’ Rule Cracked the Enigma Code, Hunted Down Russian Submarines, & Emerged Triumphant from Two Centuries of C; Yale University Press: New Haven, CT, USA, 2011. [Google Scholar]
- Box, G.E.P.; Tiao, G.C. Bayesian Inference in Statistical Analysis; John Wiley & Sons, Incorporated: Hoboken, NJ, USA, 2011. [Google Scholar]
- Tamrakar, S.; Choubey, S.B.; Choubey, A. Computational Intelligence in Medical Decision Making and Diagnosis: Techniques and Applications; CRC Press: Boca Raton, FL, USA, 2023. [Google Scholar]
- Wasserman, L. All of Nonparametric Statistics; Springer Science & Business Media: New York, NY, USA, 2006. [Google Scholar]
- Pearl, J.A. Probabilistic Calculus of Actions. In Uncertainty Proceedings 1994; Lopez de Mantaras, R., Poole, D., Eds.; Morgan Kaufmann: San Francisco, CA, USA, 1994; pp. 454–462. [Google Scholar]
- Heckerman, D.; Geiger, D.; Chickering, D.M. Learning Bayesian Networks: The Combination of Knowledge and Statistical Data. Mach. Learn. 1995, 20, 197–243. [Google Scholar] [CrossRef]
- American Diabetes Association. Classification and Diagnosis of Diabetes: Standards of Medical Care in Diabetes—2021. Diabetes Care 2021, 44 (Suppl. S1), S15–S33. [Google Scholar] [CrossRef]
- Meneilly, G.S.; Elliott, T. Metabolic Alterations in Middle-Aged and Elderly Obese Patients with Type 2 Diabetes. Diabetes Care 1999, 22, 112–118. [Google Scholar] [CrossRef] [PubMed]
- Geer, E.B.; Shen, W. Gender Differences in Insulin Resistance, Body Composition, and Energy Balance. Gend. Med. 2009, 6 (Suppl. S1), 60–75. [Google Scholar] [CrossRef] [PubMed]
- Van Cauter, E.; Polonsky, K.S.; Scheen, A.J. Roles of Circadian Rhythmicity and Sleep in Human Glucose Regulation. Endocr. Rev. 1997, 18, 716–738. [Google Scholar]
- Colberg, S.R.; Sigal, R.J.; Fernhall, B.; Regensteiner, J.G.; Blissmer, B.J.; Rubin, R.R.; Chasan-Taber, L.; Albright, A.L.; Braun, B. Exercise and Type 2 Diabetes: The American College of Sports Medicine and the American Diabetes Association: Joint Position Statement. Diabetes Care 2010, 33, e147–e167. [Google Scholar] [CrossRef]
- Salmerón, J.; Manson, J.E.; Stampfer, M.J.; Colditz, G.A.; Wing, A.L.; Willett, W.C. Dietary Fiber, Glycemic Load, and Risk of Non-Insulin-Dependent Diabetes Mellitus in Women. JAMA J. Am. Med. Assoc. 1997, 277, 472–477. [Google Scholar] [CrossRef]
- Surwit, R.S.; van Tilburg, M.A.L.; Zucker, N.; McCaskill, C.C.; Parekh, P.; Feinglos, M.N.; Edwards, C.L.; Williams, P.; Lane, J.D. Stress Management Improves Long-Term Glycemic Control in Type 2 Diabetes. Diabetes Care 2002, 25, 30–34. [Google Scholar] [CrossRef]
- Pandit, M.K.; Burke, J.; Gustafson, A.B.; Minocha, A.; Peiris, A.N. Drug-Induced Disorders of Glucose Tolerance. Ann. Intern. Med. 1993, 118, 529–539. [Google Scholar] [CrossRef]
- Dupuis, J.; Langenberg, C.; Prokopenko, I.; Saxena, R.; Soranzo, N.; Jackson, A.U.; Wheeler, E.; Glazer, N.L.; Bouatia-Naji, N.; Gloyn, A.L.; et al. New Genetic Loci Implicated in Fasting Glucose Homeostasis and Their Impact on Type 2 Diabetes Risk. Nat. Genet. 2010, 42, 105–116. [Google Scholar] [CrossRef]
- Haeckel, R.; Wosniok, W.; Arzideh, F. A Plea for Intra-Laboratory Reference Limits. Part 1. General Considerations and Concepts for Determination. Clin. Chem. Lab. Med. CCLM/FESCC 2007, 45, 1033–1042. [Google Scholar] [CrossRef] [PubMed]
- Arzideh, F.; Wosniok, W.; Gurr, E.; Hinsch, W.; Schumann, G.; Weinstock, N.; Haeckel, R. A Plea for Intra-Laboratory Reference Limits. Part 2. A Bimodal Retrospective Concept for Determining Reference Limits from Intra-Laboratory Databases Demonstrated by Catalytic Activity Concentrations of Enzymes. Clin. Chem. Lab. Med. CCLM/FESCC 2007, 45, 1043–1057. [Google Scholar] [CrossRef] [PubMed]
- Centers for Disease Control and Prevention. National Center for Health Statistics NHANES—NCHS Research Ethics Review Board Approval. 2023. Available online: https://www.cdc.gov/nchs/nhanes/irba98.htm (accessed on 4 September 2023).
- Forbes, C.; Evans, M.; Hastings, N.; Peacock, B. Statistical Distributions; John Wiley & Sons: Hoboken, NJ, USA, 2011. [Google Scholar]
- Gramacki, A. Nonparametric Kernel Density Estimation and Its Computational Aspects; Springer: Berlin/Heidelberg, Germany, 2017. [Google Scholar]
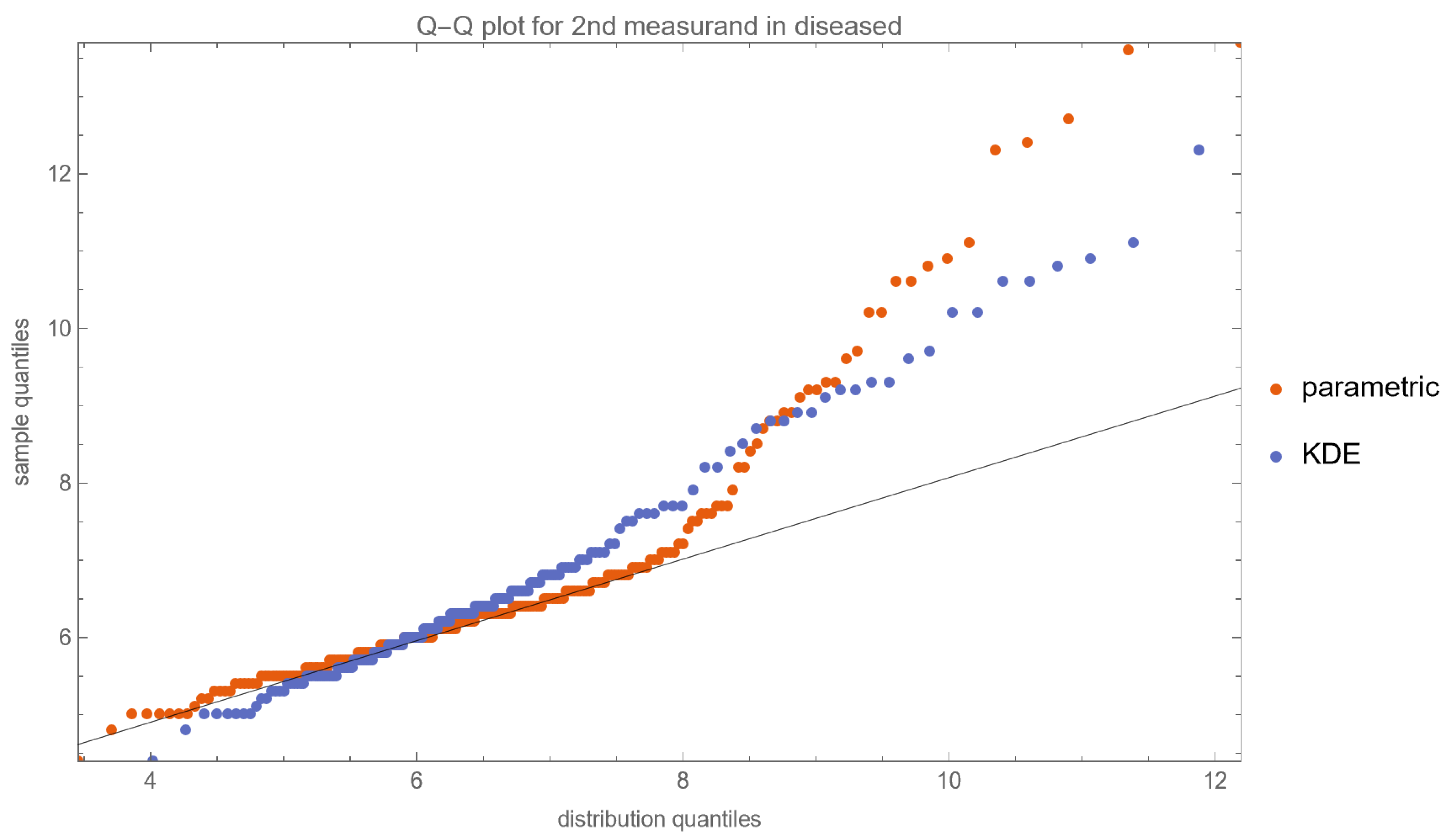
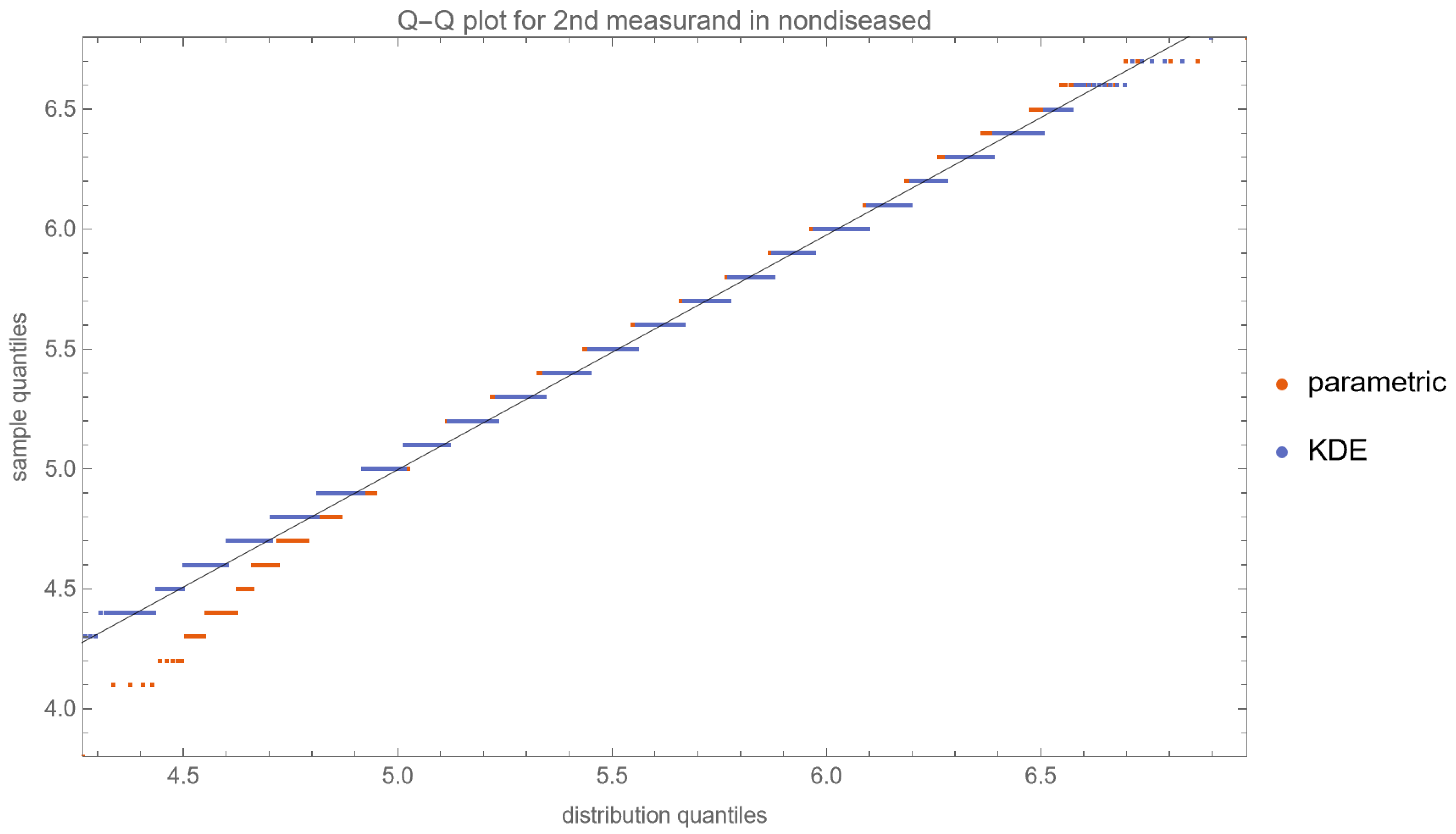
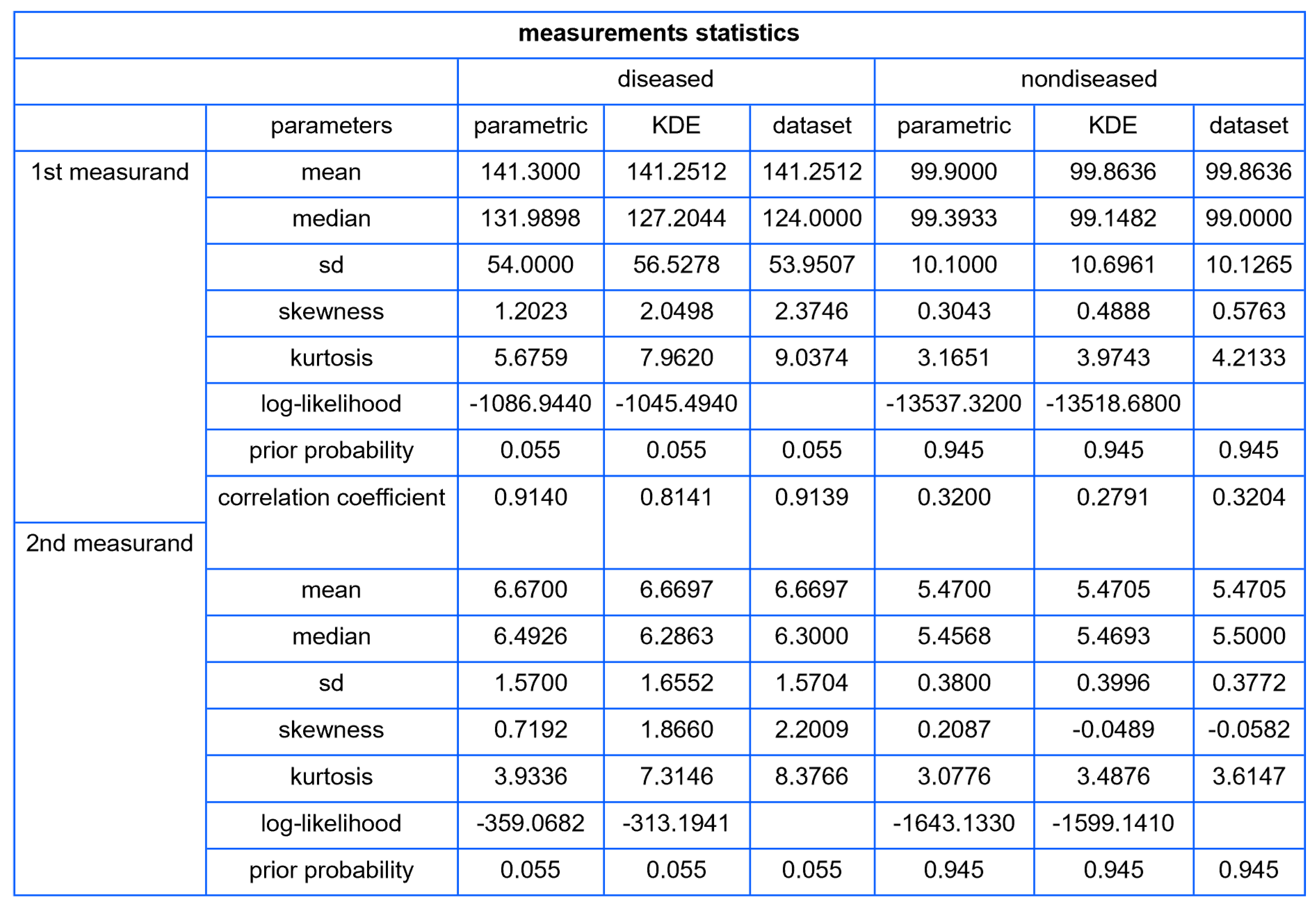
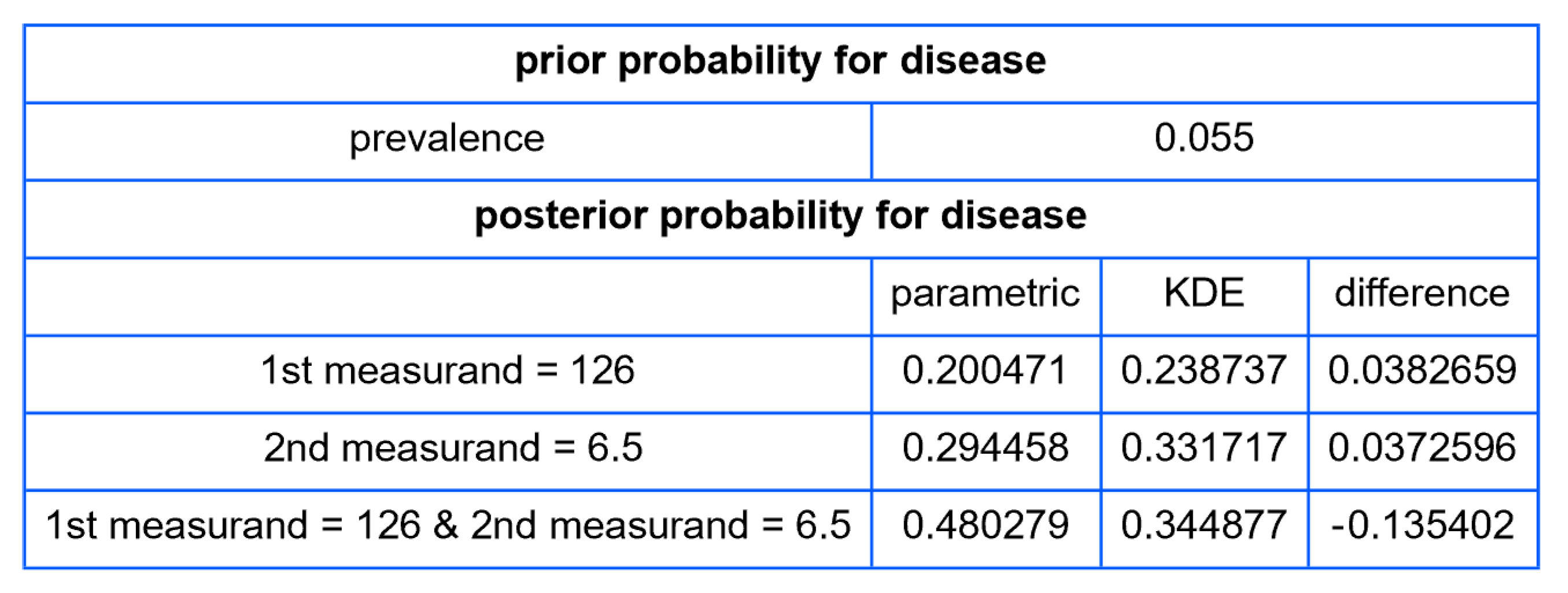
| Diabetic Patients | Nondiabetic Patients | |||
|---|---|---|---|---|
| n | 687 | 10,519 | ||
| Measurand (Units) | FPG (mg/dL) | HbA1c (%) | FPG (mg/dL) | HbA1c (%) |
| Mean | 141.3 | 6.67 | 99.9 | 5.47 |
| Median | 124.0 | 6.30 | 99.0 | 5.50 |
| Standard Deviation | 54.0 | 1.57 | 10.1 | 0.38 |
| Skewness | 2.375 | 2.201 | 0.576 | −0.058 |
| Kurtosis | 9.037 | 8.377 | 4.213 | 3.615 |
| Correlation Coefficient | 0.914 | 0.320 | ||
| Diabetic Patients | Nondiabetic Patients | |||
|---|---|---|---|---|
| Measurand (Units) | FPG (mg/dL) | HbA1c (%) | FPG (mg/dL) | HbA1c (%) |
| Parametric Distribution | Lognormal | Lognormal | Lognormal | Lognormal |
| Parametric Distribution Mean | 141.3 | 6.67 | 99.9 | 5.47 |
| Parametric Distribution SD | 54.0 | 1.57 | 10.1 | 0.38 |
| KDE Smoothing Bandwidth (SD units) | 0.32 | 0.34 | 0.34 | 0.35 |
| Correlation Coefficient | 0.914 | 0.320 | ||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chatzimichail, T.; Hatjimihail, A.T. A Bayesian Inference Based Computational Tool for Parametric and Nonparametric Medical Diagnosis. Diagnostics 2023, 13, 3135. https://doi.org/10.3390/diagnostics13193135
Chatzimichail T, Hatjimihail AT. A Bayesian Inference Based Computational Tool for Parametric and Nonparametric Medical Diagnosis. Diagnostics. 2023; 13(19):3135. https://doi.org/10.3390/diagnostics13193135
Chicago/Turabian StyleChatzimichail, Theodora, and Aristides T. Hatjimihail. 2023. "A Bayesian Inference Based Computational Tool for Parametric and Nonparametric Medical Diagnosis" Diagnostics 13, no. 19: 3135. https://doi.org/10.3390/diagnostics13193135
APA StyleChatzimichail, T., & Hatjimihail, A. T. (2023). A Bayesian Inference Based Computational Tool for Parametric and Nonparametric Medical Diagnosis. Diagnostics, 13(19), 3135. https://doi.org/10.3390/diagnostics13193135