The Genomic Landscape of Colorectal Cancer in the Saudi Arabian Population Using a Comprehensive Genomic Panel
Abstract
1. Introduction
2. Materials and Methods
Sample Selection
3. Results
3.1. Cohort Demographics and Clinical Management
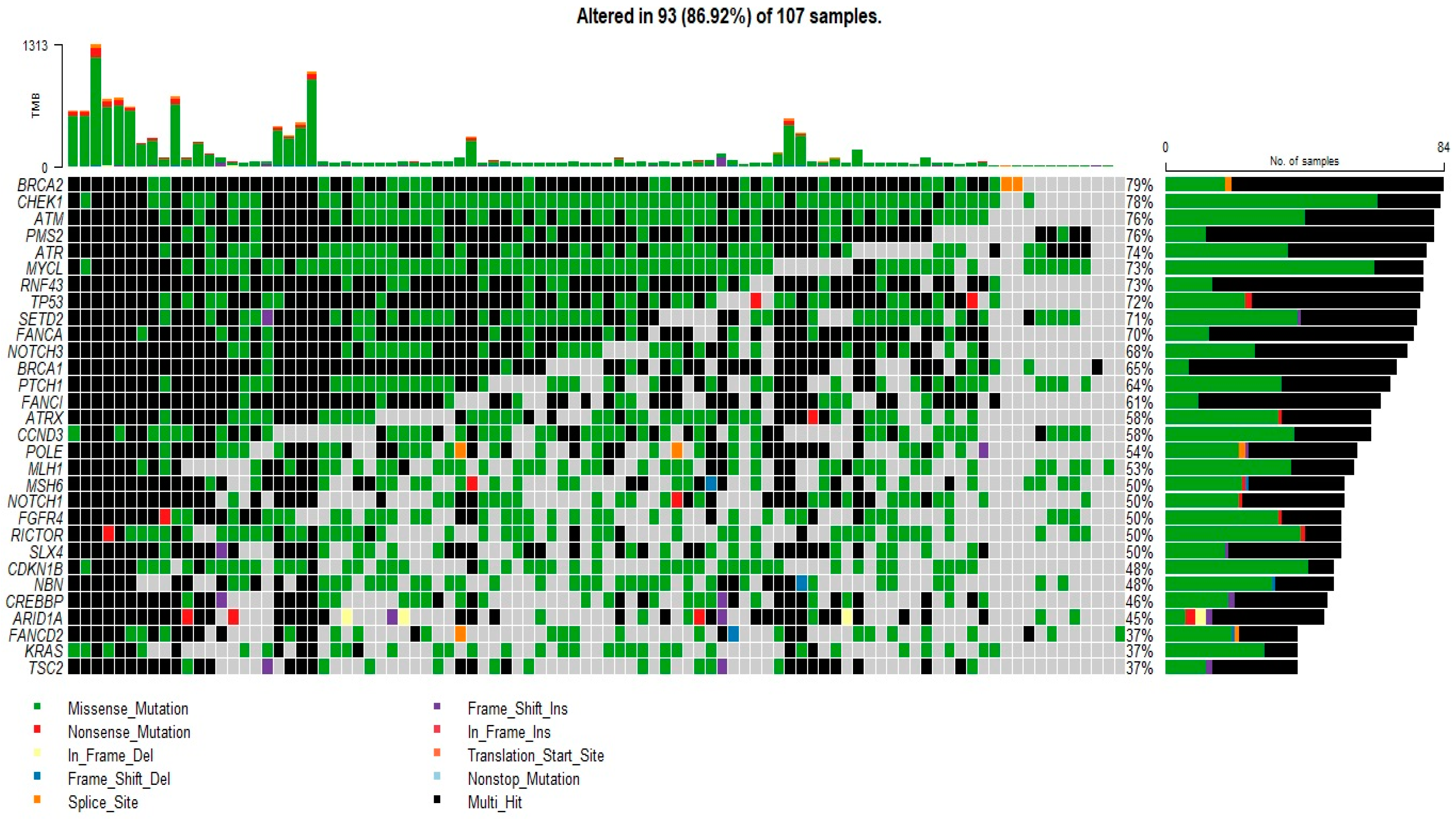
3.2. Mutational Profile Analysis
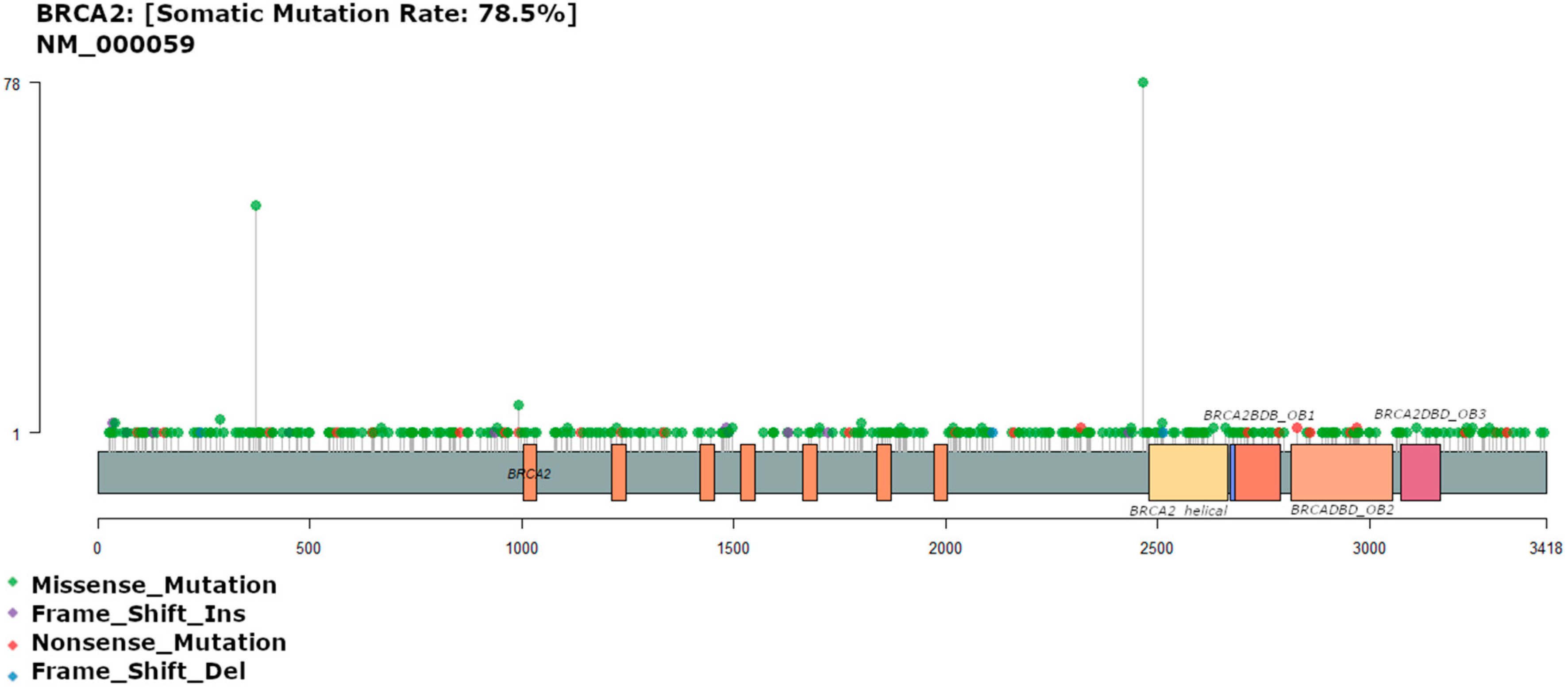
3.3. BRCA2 Mutation
3.4. TP53 Mutations
3.5. KRAS Mutation
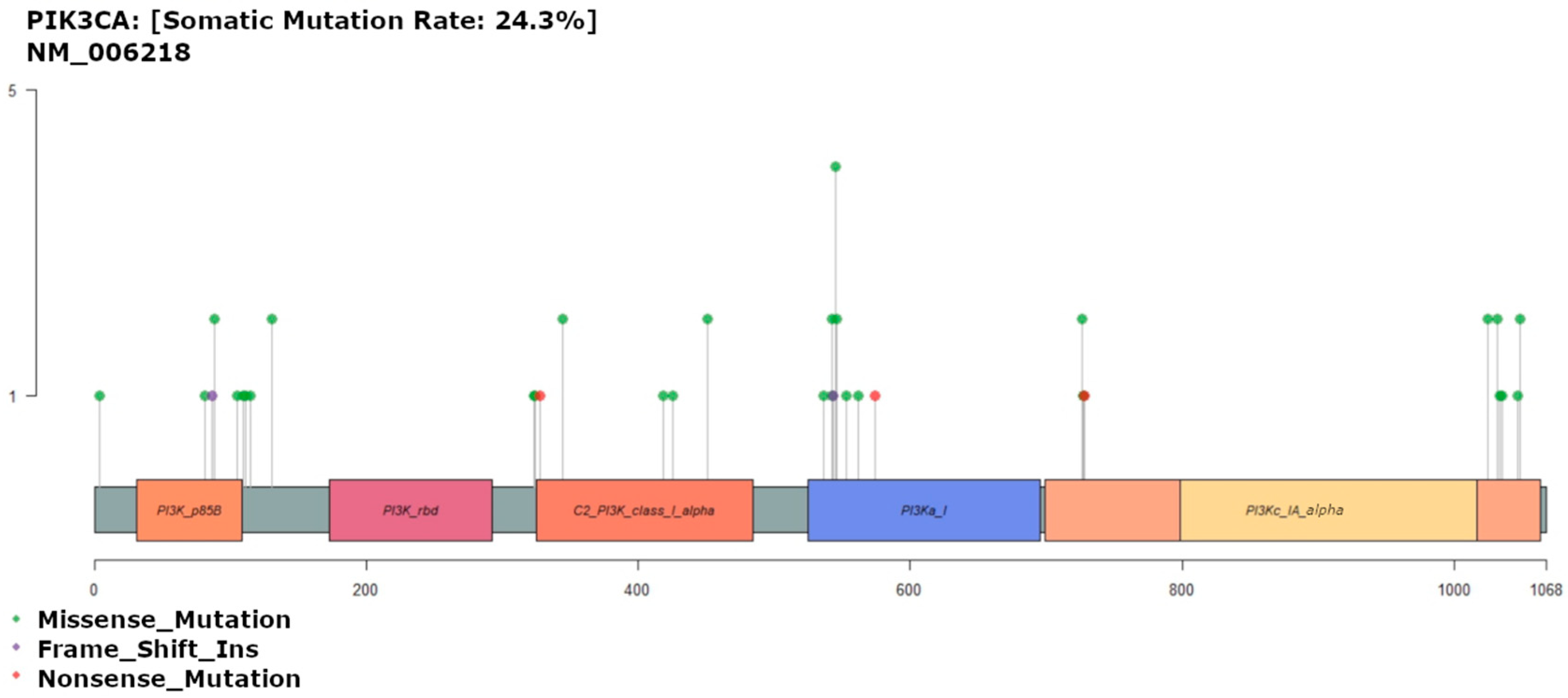
3.6. PIK3CA Mutation
3.7. Survival Curves
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Binalmatrefa, G.; Rodríguez-Moranta, F.; Teule, A.; Medina-Hayas, M. Colorectal cancer: From prevention to personalized medicine. WJG 2014, 20, 6786–6808. [Google Scholar]
- Sanz-Garcia, E.; Grasselli, J.; Argiles, G.; Elez, M.E.; Tabernero, J. Current and advancing treatments for metastatic colorectal cancer. Expert Opin. Biol. Ther. 2016, 16, 93–110. [Google Scholar] [CrossRef]
- Helewa, R.M.; Turner, D.; Wirtzfeld, D.; Park, J.; Hochman, D.; Czaykowski, P.; Singh, H.; Shu, E.; Xue, L.; McKay, A. Does geography influence the treatment and outcomes of colorectal cancer? A population-based analysis. World J. Surg. Oncol. 2013, 11, 14. [Google Scholar] [CrossRef]
- Arafa, M.A.; Farhat, K. Colorectal Cancer in the Arab World—Screening Practices and Future Prospects. Asian Pac. J. Cancer Prev. APJCP 2015, 16, 7425–7430. [Google Scholar] [CrossRef]
- Almatroudi, A. The Incidence Rate of Colorectal Cancer in Saudi Arabia: An Observational Descriptive Epidemiological Analysis. Int. J. Gen. Med. 2020, 13, 977–979. [Google Scholar] [CrossRef]
- Rey, J.M.; Ducros, V.; Pujol, P.; Wang, Q.; Buisine, M.P.; Aissaoui, H.; Maudelonde, T.; Olschwang, S. Improving Mutation Screening in Patients with Colorectal Cancer Predisposition Using Next-Generation. JMD 2017, 19, 589–601. [Google Scholar] [CrossRef][Green Version]
- Alsanea, N.; Almadi, M.A.; Abduljabbar, A.S.; Alhomoud, S.; Alshaban, T.A.; Alsuhaibani, A.; Alzahrani, A.; Batwa, F.; Hassan, A.-H.; Hibbert, D.; et al. National Guidelines for Colorectal Cancer Screening in Saudi Arabia with strength of recommendations and quality of evidence. Ann. Saudi Med. 2015, 35, 189–195. [Google Scholar] [CrossRef]
- Alqahtani, M.; Edwards, C.; Buzzacott, N.; Carpenter, K.; Alsaleh, K.; Alsheikh, A.; Abozeed, W.; Mashhour, M.; Almousa, A.; Housawi, Y.; et al. Screening for Lynch syndrome in young Saudi colorectal cancer patients using microsatellite instability testing and next generation sequencing. Fam. Cancer 2018, 17, 197–203. [Google Scholar] [CrossRef]
- Mosli, M.H.; Al-Ahwal, M.S. Colorectal cancer in the Kingdom of Saudi Arabia: Need for screening. Asian Pac. J. Cancer Prev. 2012, 13, 3809–3813. [Google Scholar]
- Alsanea, N.; Abduljabbar, A.S.; Alhomoud, S.; Ashari, L.H.; Hibbert, D.; Bazarbashi, S. Colorectal cancer in Saudi Arabia: Incidence, survival, demographics and implications for national policies. Ann. Saudi Med. 2015, 35, 196–202. [Google Scholar] [CrossRef]
- Almadi, M.A.; Barkun, A.N. Initial guidelines for colorectal cancer screening in Saudi Arabia: A beginning. Ann. Saudi Med. 2015, 35, 341–342. [Google Scholar] [CrossRef] [PubMed]
- Brandão, C.; Lage, J. Management of Patients with Hereditary Colorectal Cancer Syndromes. Port. J. Gastroenterol. 2015, 22, 204–212. [Google Scholar] [CrossRef] [PubMed]
- AlZaabi, A. Colorectal Cancer in the Arab World. In Cancer in the Arab World; Al-Shamsi, H.O., Abu-Gheida, I.H., Iqbal, F., Al-Awadhi, A., Eds.; Springer: Singapore, 2022; pp. 363–379. [Google Scholar]
- Cummings, C.A.; Peters, E.; Lacroix, L.; Andre, F.; Lackner, M.R. The Role of Next-Generation Sequencing in Enabling Personalized Oncology Therapy. Clin. Transl. Sci. 2016, 9, 283–289. [Google Scholar] [CrossRef] [PubMed]
- Jauhri, M.; Bhatnagar, A.; Gupta, S.; Shokeen, Y.; Minhas, S.; Aggarwal, S. Targeted molecular profiling of rare genetic alterations in colorectal cancer using next-generation sequencing. Med. Oncol. 2016, 33, 106. [Google Scholar] [CrossRef]
- Zhang, L.; Chen, L.; Sah, S.; Latham, G.J.; Patel, R.; Song, Q.; Koeppen, H.; Tam, R.; Schleifman, E.; Mashhedi, H.; et al. Profiling cancer gene mutations in clinical formalin-fixed, paraffin-embedded colorectal tumor specimens using targeted next-generation sequencing. Oncologist 2014, 19, 336–343. [Google Scholar] [CrossRef]
- Harlé, A.; Filhine-Tresarrieu, P.; Husson, M.; Boidot, R.; Rouyer, M.; Dubois, C.; Leroux, A.; Merlin, J.L. Rare RAS Mutations in Metastatic Colorectal Cancer Detected During Routine RAS Genotyping Using Next Generation Sequencing. Target Oncol. 2016, 11, 363–370. [Google Scholar] [CrossRef]
- Timmermann, B.; Kerick, M.; Roehr, C.; Fischer, A.; Isau, M.; Boerno, S.T.; Wunderlich, A.; Barmeyer, C.; Seemann, P.; Koenig, J.; et al. Somatic mutation profiles of MSI and MSS colorectal cancer identified by whole exome next generation sequencing and bioinformatics analysis. PLoS ONE 2010, 5, e15661. [Google Scholar] [CrossRef]
- Dakowicz, D.; Zajkowska, M.; Mroczko, B. Relationship between VEGF Family Members, Their Receptors and Cell Death in the Neoplastic Transformation of Colorectal Cancer. Int. J. Mol. Sci. 2022, 23, 6. [Google Scholar] [CrossRef]
- Moench, R.; Gasser, M.; Nawalaniec, K.; Grimmig, T.; Ajay, A.K.; de Souza, L.C.R.; Cao, M.; Luo, Y.; Hoegger, P.; Ribas, C.M.; et al. Platelet-derived growth factor (PDGF) cross-signaling via non-corresponding receptors indicates bypassed signaling in colorectal cancer. Oncotarget 2022, 13, 1140–1152. [Google Scholar] [CrossRef]
- Ciombor, K.K.; Haraldsdottir, S.; Goldberg, R.M. How Can Next-Generation Sequencing (Genomics) Help Us in Treating Colorectal Cancer? Curr. Color. Cancer Rep. 2014, 10, 372–379. [Google Scholar] [CrossRef][Green Version]
- Zhang, H.; Zheng, X.; Ji, T.; Fu, L.; Bai, D.; Liao, Y.; Zhang, H.; Ding, Y.; Zheng, L. Comparative screening of K-ras mutations in colorectal cancer and lung cancer patients using a novel real-time PCR with ADx-K-ras kit and Sanger DNA sequencing. Cell Biochem. Biophys. 2012, 62, 415–420. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Kothari, N.; Schell, M.J.; Teer, J.K.; Yeatman, T.; Shibata, D.; Kim, R. Comparison of KRAS mutation analysis of colorectal cancer samples by standard testing and next-generation sequencing. J. Clin. Pathol. 2014, 67, 764–767. [Google Scholar] [CrossRef] [PubMed]
- Gao, J.; Wu, H.; Wang, L.; Zhang, H.; Duan, H.; Lu, J.; Liang, Z. Validation of targeted next-generation sequencing for RAS mutation detection in FFPE colorectal cancer tissues: Comparison with Sanger sequencing and ARMS-Scorpion real-time PCR. BMJ Open 2016, 6, e009532. [Google Scholar] [CrossRef] [PubMed]
- Vestergaard, L.K.; Oliveira, D.N.P.; Poulsen, T.S.; Høgdall, C.K.; Høgdall, E.V. Oncomine™ Comprehensive Assay v3 vs. Oncomine™ Comprehensive Assay Plus. Cancers 2021, 13, 5230. [Google Scholar] [CrossRef]
- Di, J.; Yang, H.; Jiang, B.; Wang, Z.; Ji, J.; Su, X. Whole exome sequencing reveals intertumor heterogeneity and distinct genetic origins of sporadic synchronous colorectal cancer. Int. J. Cancer 2018, 142, 927–939. [Google Scholar] [CrossRef] [PubMed]
- Disciglio, V.; Devecchi, A.; Palumbo, O.; Carella, M.; Penso, D.; Milione, M.; Valle, G.; Pierotti, M.A.; Vitellaro, M.; Bertario, L.; et al. Whole exome sequencing and single nucleotide polymorphism array analyses to identify germline alterations in genes associated with testosterone metabolism in a patient with androgen insensitivity syndrome and early-onset colorectal cancer. Chin. J. Cancer 2016, 35, 51. [Google Scholar] [CrossRef]
- Morrow, J.B.; Dallo, F.J.; Julka, M. Community-based colorectal cancer screening trials with multi-ethnic groups: A systematic review. J. Community Health 2010, 35, 592–601. [Google Scholar] [CrossRef]
- Cronan, T.A.; Devos-Comby, L.; Villalta, I.; Gallagher, R. Ethnic differences in colorectal cancer screening. J. Psychosoc. Oncol. 2008, 26, 63–86. [Google Scholar] [CrossRef]
- Baskin, Y.; Calibasi, G.; Amirfallah, A.; Dagdeviren, Y.K.; Canda, A.E.; Sarioglu, S.; Sagol, O.; Ellidokuz, H.; Oztop, I.; Yilmaz, U. KRAS and BRAF mutation frequencies in a series of Turkish colorectal cancer patients. Transl. Cancer Res. 2014, 3, 160–166. [Google Scholar]
- Xue, W.Q.; He, Y.Q.; Zhu, J.H.; Ma, J.Q.; He, J.; Jia, W.H. Association of BRCA2 N372H polymorphism with cancer susceptibility: A comprehensive review and meta-analysis. Sci. Rep. 2014, 4, 6791. [Google Scholar] [CrossRef]
- Li, Q.; Guan, R.; Qiao, Y.; Liu, C.; He, N.; Zhang, X.; Jia, X.; Sun, H.; Yu, J.; Xu, L. Association between the BRCA2 rs144848 polymorphism and cancer susceptibility: A meta-analysis. Oncotarget 2017, 8, 39818–39832. [Google Scholar] [CrossRef] [PubMed]
- Pearlman, R.; Frankel, W.L.; Swanson, B.; Zhao, W.; Yilmaz, A.; Miller, K.; Bacher, J.; Bigley, C.; Nelsen, L.; Goodfellow, P.J.; et al. Prevalence and Spectrum of Germline Cancer Susceptibility Gene Mutations Among Patients With Early-Onset Colorectal Cancer. JAMA Oncol. 2017, 3, 464–471. [Google Scholar] [CrossRef] [PubMed]
- Stoffel, E.M.; Koeppe, E.; Everett, J.; Ulintz, P.; Kiel, M.; Osborne, J.; Williams, L.; Hanson, K.; Gruber, S.B.; Rozek, L.S. Germline Genetic Features of Young Individuals with Colorectal Cancer. Gastroenterology 2018, 154, 897–905. [Google Scholar] [CrossRef] [PubMed]
- Yurgelun, M.B.; Kulke, M.H.; Fuchs, C.S.; Allen, B.A.; Uno, H.; Hornick, J.L.; Ukaegbu, C.I.; Brais, L.K.; McNamara, P.G.; Mayer, R.J.; et al. Cancer Susceptibility Gene Mutations in Individuals with Colorectal Cancer. J. Clin. Oncol. 2017, 35, 1086–1095. [Google Scholar] [CrossRef]
- Asadi, M.; Shanaehbandi, D.; Zarintan, A.; Pedram, N.; Baradaran, B.; Zafari, V.; Shirmohamadi, M.; Hashemzadeh, S. TP53 Gene Pro72Arg (rs1042522) Single Nucleotide Polymorphism as Not a Risk Factor for Colorectal Cancer in the Iranian Azari Population. Asian Pac. J. Cancer Prev. APJCP 2017, 18, 3423–3427. [Google Scholar]
- Ruijs, M.W.; Verhoef, S.; Rookus, M.A.; Pruntel, R.; van der Hout, A.H.; Hogervorst, F.B.; Kluijt, I.; Sijmons, R.H.; Aalfs, C.M.; Wagner, A.; et al. TP53 germline mutation testing in 180 families suspected of Li-Fraumeni syndrome: Mutation detection rate and relative frequency of cancers in different familial phenotypes. J. Med. Gene 2010, 47, 421–428. [Google Scholar] [CrossRef]
- Manoharan, V.; Karunanayake, E.H.; Tennekoon, K.H.; De Silva, S.; Imthikab, A.I.A.; De Silva, K.; Angunawela, P.; Vishwakula, S.; Lunec, J. Pattern of nucleotide variants of TP53 and their correlation with the expression of p53 and its downstream proteins in a Sri Lankan cohort of breast and colorectal cancer patients. BMC Cancer 2020, 20, 72. [Google Scholar] [CrossRef]
- Fortuno, C.; Pesaran, T.; Dolinsky, J.; Yussuf, A.; McGoldrick, K.; Kho, P.F.; James, P.A.; Spurdle, A.B. p53 major hotspot variants are associated with poorer prognostic features in hereditary cancer patients. Cancer Genet. 2019, 235–236, 21–27. [Google Scholar] [CrossRef]
- Nakayama, M.; Oshima, M. Mutant p53 in colon cancer. J. Mol. Cell Biol. 2019, 11, 267–276. [Google Scholar] [CrossRef]
- Younis, N.S.; AlMasoud, E.S.; Al Khawajah, F.; Alghazal, F.J.; AlMofarfesh, H.M.; Al-Khalaf, L.H.; Al Otaibi, M.S.; Alkhamis, S.M.; Al Naser, Z.A.; Al Mousa, Z.H.; et al. Potential genetic biomarker of Saudi Arabian patients with colorectal cancer. Eur. Rev. Med. Pharmacol. Sci. 2022, 26, 3109–3126. [Google Scholar] [CrossRef]
- Margonis, G.A.; Kim, Y.; Spolverato, G.; Ejaz, A.; Gupta, R.; Cosgrove, D.; Anders, R.; Karagkounis, G.; Choti, M.A.; Pawlik, T.M. Association Between Specific Mutations in KRAS Codon 12 and Colorectal Liver Metastasis. JAMA Surg. 2015, 150, 722–729. [Google Scholar] [CrossRef] [PubMed]
- Huang, L.; Guo, Z.; Wang, F.; Fu, L. KRAS mutation: From undruggable to druggable in cancer. Signal Transduct. Target. Ther. 2021, 6, 386. [Google Scholar] [CrossRef] [PubMed]
- Koulouridi, A.; Karagianni, M.; Messaritakis, I.; Sfakianaki, M.; Voutsina, A.; Trypaki, M.; Bachlitzanaki, M.; Koustas, E.; Karamouzis, M.V.; Ntavatzikos, A.; et al. Prognostic Value of KRAS Mutations in Colorectal Cancer Patients. Cancersm 2022, 14, 3320. [Google Scholar] [CrossRef] [PubMed]
- Ranjbar, R.; Mohammadpour, S.; Esfahani, A.T.; Namazian, S.; Yaghob-Taleghani, M.; Baghaei, K.; Tabatabaei, S.A.M.; Pasharavesh, L.; Nazemalhosseini-Mojarad, E. Prevalence and prognostic role of PIK3CA E545K mutation in Iranian colorectal cancer patients. Gastroenterol. Hepatol. Bed Bench 2019, 12, S22–S29. [Google Scholar]
- Zhang, L.; Shay, J.W. Multiple Roles of APC and its Therapeutic Implications in Colorectal Cancer. J. Natl. Cancer Inst. 2017, 109, djw332. [Google Scholar] [CrossRef]
- Almuzzaini, B.; Alghamdi, J.; Alomani, A.; AlGhamdi, S.; Alsharm, A.A.; Alshieban, S.; Sayed, A.; Alhejaily, A.G.; Aljaser, F.S.; Abudawood, M.; et al. Identification of Novel Mutations in Colorectal Cancer Patients Using AmpliSeq Comprehensive Cancer Panel. J. Pers. Med. 2021, 11, 535. [Google Scholar] [CrossRef]
- Canon, J.; Rex, K.; Saiki, A.Y.; Mohr, C.; Cooke, K.; Bagal, D.; Gaida, K.; Holt, T.; Knutson, C.G.; Koppada, N.; et al. The clinical KRAS(G12C) inhibitor AMG 510 drives anti-tumour immunity. Nature 2019, 575, 217–223. [Google Scholar] [CrossRef]
- Cancer Genome Atlas Network. Comprehensive molecular characterization of human colon and rectal cancer. Nature 2012, 487, 330–337. [Google Scholar] [CrossRef]
- Mighton, C.; Lerner-Ellis, J.P. Principles of molecular testing for hereditary cancer. Genes Chromosomes Cancer 2022, 61, 356. [Google Scholar] [CrossRef]
- Vanderwalde, A.; Spetzler, D.; Xiao, N.; Gatalica, Z.; Marshall, J. Microsatellite instability status determined by next-generation sequencing and compared with PD-L1 and tumor mutational burden in 11,348 patients. Cancer Med. 2018, 7, 746–756. [Google Scholar] [CrossRef]
- Shia, J. The diversity of tumours with microsatellite instability: Molecular mechanisms and impact upon microsatellite instability testing and mismatch repair protein immunohistochemistry. Histopathology 2021, 78, 485–497. [Google Scholar] [CrossRef]
- Oh, M.; McBride, A.; Yun, S.; Bhattacharjee, S.; Slack, M.; Martin, J.R.; Jeter, J.; Abraham, I. BRCA1 and BRCA2 Gene Mutations and Colorectal Cancer Risk: Systematic Review and Meta-analysis. JNCI J. Natl. Cancer Inst. 2018, 110, 1178–1189. [Google Scholar] [CrossRef] [PubMed]
- Cullinane, C.M.; Creavin, B.; O’connell, E.P.; Kelly, L.; O’Sullivan, M.J.; Corrigan, M.A.; Redmond, H.P. Risk of colorectal cancer associated with BRCA1 and/or BRCA2 mutation carriers: Systematic review and meta-analysis. Br. J. Surg. 2020, 107, 951–959. [Google Scholar] [CrossRef] [PubMed]
- Lin, W.Y.; Brock, I.W.; Connley, D.; Cramp, H.; Tucker, R.; Slate, J.; Reed, M.W.; Balasubramanian, S.P.; Cannon-Albright, L.A.; Camp, N.J.; et al. Associations of ATR and CHEK1 single nucleotide polymorphisms with breast cancer. PLoS ONE 2013, 8, e68578. [Google Scholar] [CrossRef]
- Angele, S.; Romestaing, P.; Moullan, N.; Vuillaume, M.; Chapot, B.; Friesen, M.; Jongmans, W.; Cox, D.G.; Pisani, P.; Gerard, J.P.; et al. ATM haplotypes and cellular response to DNA damage: Association with breast cancer risk and clinical radiosensitivity. Cancer Res. 2003, 63, 8717–8725. [Google Scholar] [PubMed]
- Su, D.; Ma, S.; Liu, P.; Jiang, Z.; Lv, W.; Zhang, Y.; Deng, Q.; Smith, S.; Yu, H. Genetic polymorphisms and treatment response in advanced non-small cell lung cancer. Lung Cancer 2007, 56, 281–288. [Google Scholar] [CrossRef]
- Okazaki, T.; Jiao, L.; Chang, P.; Evans, D.B.; Abbruzzese, J.L.; Li, D. Single-nucleotide polymorphisms of DNA damage response genes are associated with overall survival in patients with pancreatic cancer. Clin. Cancer Res. 2008, 14, 2042–2048. [Google Scholar] [CrossRef]
- Merdad, A.; Gari, M.A.; Hussein, S.; Al-Khayat, S.; Tashkandi, H.; Al-Maghrabi, J.; Al-Thubaiti, F.; Hussein, I.R.; Koumosani, T.; Shaer, N.; et al. Characterization of familial breast cancer in Saudi Arabia. BMC Genom. 2015, 16, S3. [Google Scholar] [CrossRef]
- Ho, V.; Chung, L.; Lim, S.H.; Ma, Y.; Wang, B.; Lea, V.; Abubakar, A.; Ng, W.; Lee, M.; Roberts, T.L.; et al. Prognostic Impact of TP53 Mutations and Tumor Mutational Load in Colorectal Cancer. Gastrointest. Dis. 2022, 4, 165–179. [Google Scholar] [CrossRef]
- Rahman, N. Realizing the promise of cancer predisposition genes. Nature 2014, 505, 302–308. [Google Scholar] [CrossRef]
- Heinemann, V.; Stintzing, S.; Kirchner, T.; Boeck, S.; Jung, A. Clinical relevance of EGFR- and KRAS-status in colorectal cancer patients treated with monoclonal antibodies directed against the EGFR. Cancer Treat. Rev. 2009, 35, 262–271. [Google Scholar] [CrossRef] [PubMed]
- Saoudi Gonzalez, N.; López, D.; Gómez, D.; Ros, J.; Baraibar, I.; Salva, F.; Tabernero, J.; Élez, E. Pharmacokinetics and pharmacodynamics of approved monoclonal antibody therapy for colorectal cancer. Expert Opin. Drug Metab. Toxicol. 2022, 18, 755–767. [Google Scholar] [CrossRef] [PubMed]
- Zhao, B.; Wang, L.; Qiu, H.; Zhang, M.; Sun, L.; Peng, P.; Yu, Q.; Yuan, X. Mechanisms of resistance to anti-EGFR therapy in colorectal cancer. Oncotarget 2017, 8, 3980–4000. [Google Scholar] [CrossRef] [PubMed]
- Huang, D.; Sun, W.; Zhou, Y.; Li, P.; Chen, F.; Chen, H.; Xia, D.; Xu, E.; Lai, M.; Wu, Y.; et al. Mutations of key driver genes in colorectal cancer progression and metastasis. Cancer Metastasis Rev. 2018, 37, 173–178. [Google Scholar] [CrossRef] [PubMed]





| Gender | Female | F = 55 |
| Male | M = 52 | |
| Age (yrs) | Mean (sd) | 58 (14.5) |
| Range | 95–20 | |
| Pathological Diagnosis | Adenocarcinoma | 106 |
| Unknown CRC | 1 | |
| Location | Right colon | 56 |
| Left colon | 45 | |
| Transverse colon | 6 | |
| Histological grade | Well-differentiated | 16 |
| Moderately differentiated | 84 | |
| Poorly differentiated | 6 | |
| Unknown | 1 | |
| Microvascular invasion | Present | 30 |
| Absent | 76 | |
| Unknown | 1 | |
| AJCC Stage | 0 | 1 |
| 1 | 10 | |
| 2 | 36 | |
| 3 | 36 | |
| 4 | 22 | |
| Unknown | 2 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Alsolme, E.; Alqahtani, S.; Fageeh, M.; Barakeh, D.; Sharma, N.K.; Mangul, S.; Robinson, H.A.; Fathaddin, A.; Hauser, C.A.E.; Abedalthagafi, M. The Genomic Landscape of Colorectal Cancer in the Saudi Arabian Population Using a Comprehensive Genomic Panel. Diagnostics 2023, 13, 2993. https://doi.org/10.3390/diagnostics13182993
Alsolme E, Alqahtani S, Fageeh M, Barakeh D, Sharma NK, Mangul S, Robinson HA, Fathaddin A, Hauser CAE, Abedalthagafi M. The Genomic Landscape of Colorectal Cancer in the Saudi Arabian Population Using a Comprehensive Genomic Panel. Diagnostics. 2023; 13(18):2993. https://doi.org/10.3390/diagnostics13182993
Chicago/Turabian StyleAlsolme, Ebtehal, Saleh Alqahtani, Musa Fageeh, Duna Barakeh, Nitesh K. Sharma, Serghei Mangul, Heather A. Robinson, Amany Fathaddin, Charlotte A. E. Hauser, and Malak Abedalthagafi. 2023. "The Genomic Landscape of Colorectal Cancer in the Saudi Arabian Population Using a Comprehensive Genomic Panel" Diagnostics 13, no. 18: 2993. https://doi.org/10.3390/diagnostics13182993
APA StyleAlsolme, E., Alqahtani, S., Fageeh, M., Barakeh, D., Sharma, N. K., Mangul, S., Robinson, H. A., Fathaddin, A., Hauser, C. A. E., & Abedalthagafi, M. (2023). The Genomic Landscape of Colorectal Cancer in the Saudi Arabian Population Using a Comprehensive Genomic Panel. Diagnostics, 13(18), 2993. https://doi.org/10.3390/diagnostics13182993






