The Translational Impact of Plant-Derived Xeno-miRNA miR-168 in Gastrointestinal Cancers and Preneoplastic Conditions
Abstract
:1. Introduction
2. Materials and Methods
2.1. Ethics
2.2. Surgical Tissue Specimens
2.3. Biopsies from Gastric and Colon Mucosa
2.4. RNA and DNA Isolation
2.5. miR-168 Expression Analysis
2.6. Correlation with Molecular Characteristics
2.7. Statistical Analysis
3. Results
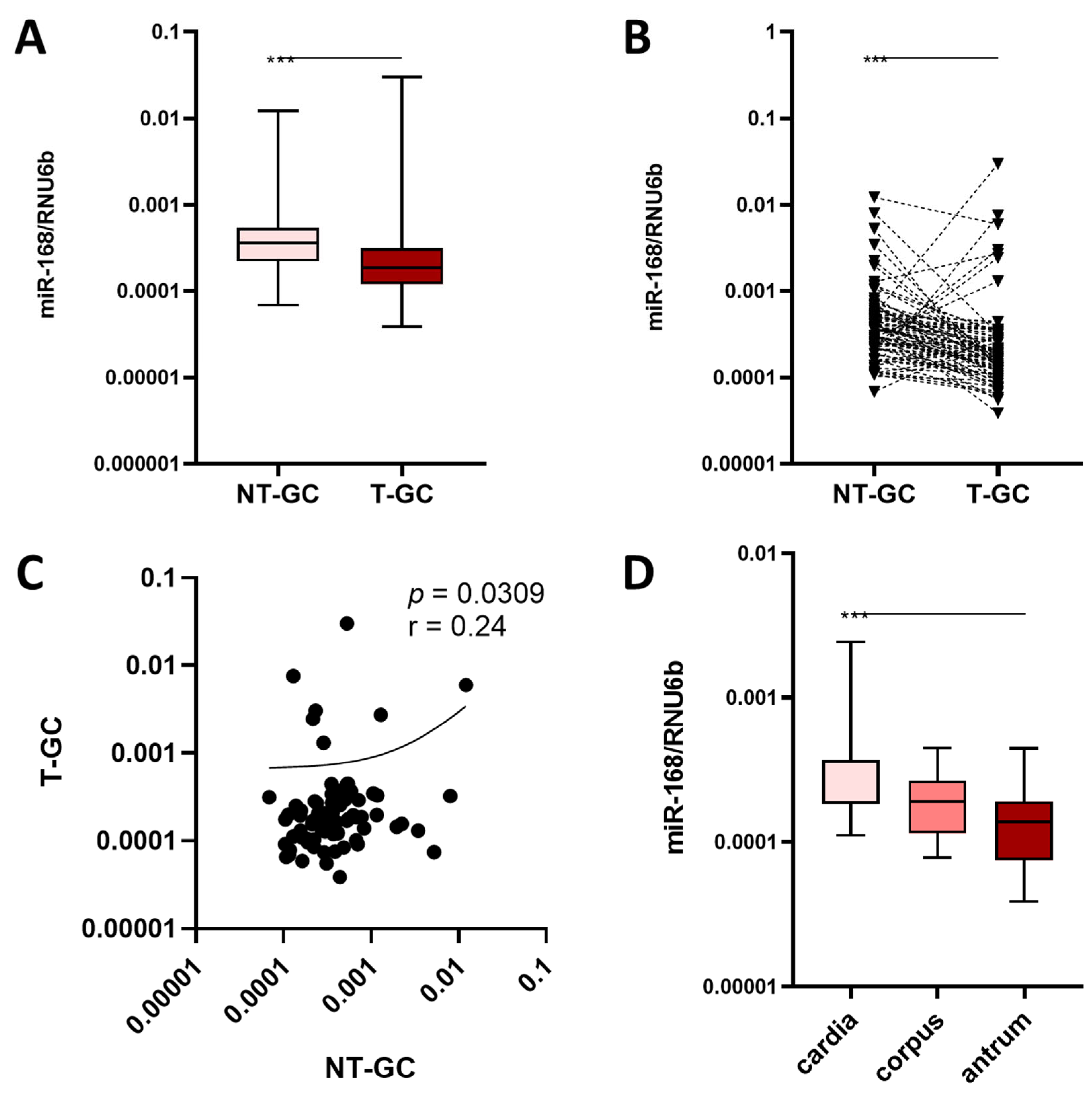
3.1. miR-168 in Nonneoplastic Mucosa
3.2. miR-168 in Neoplastic Mucosa
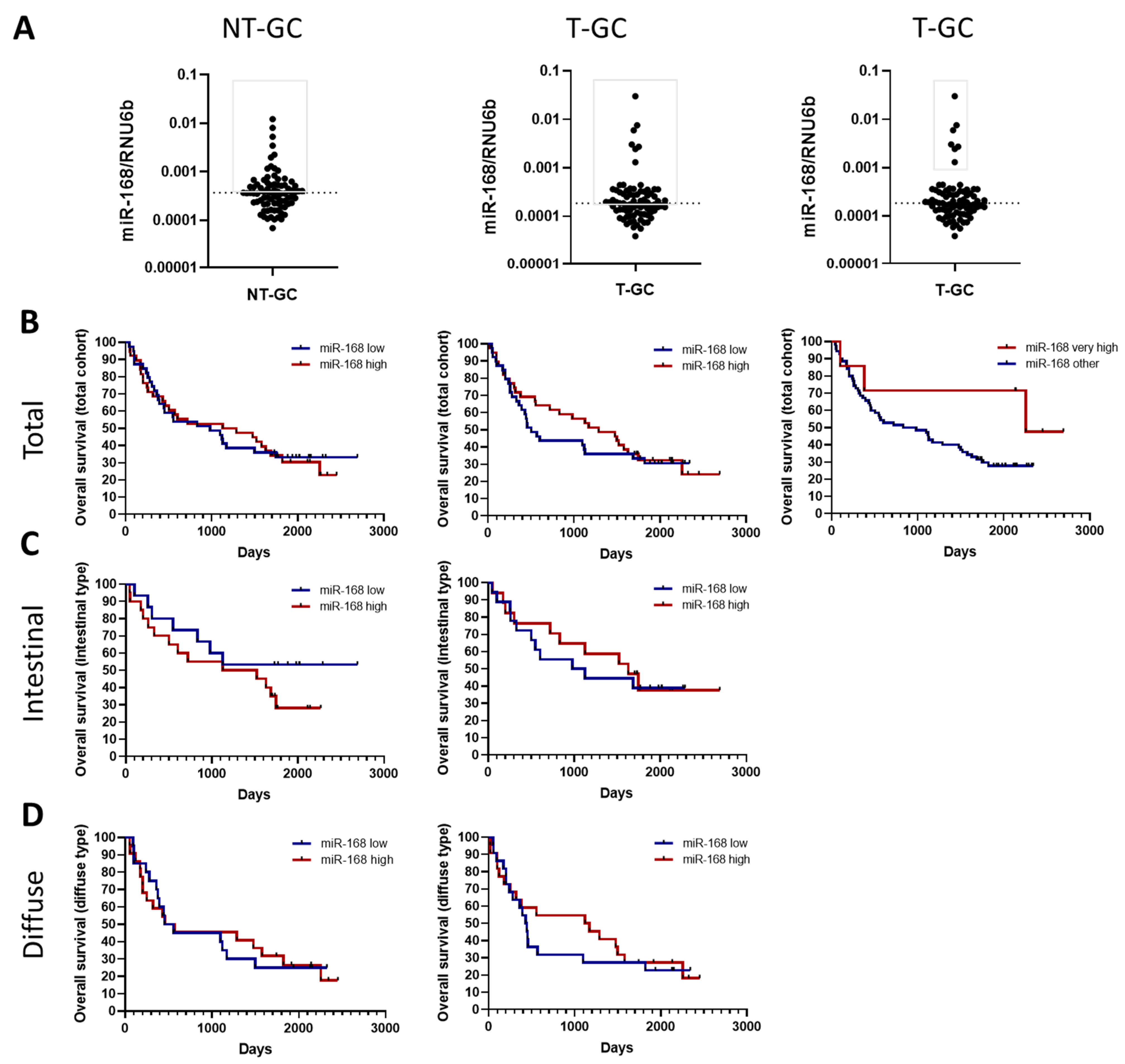
3.3. miR-168 and Overall Survival
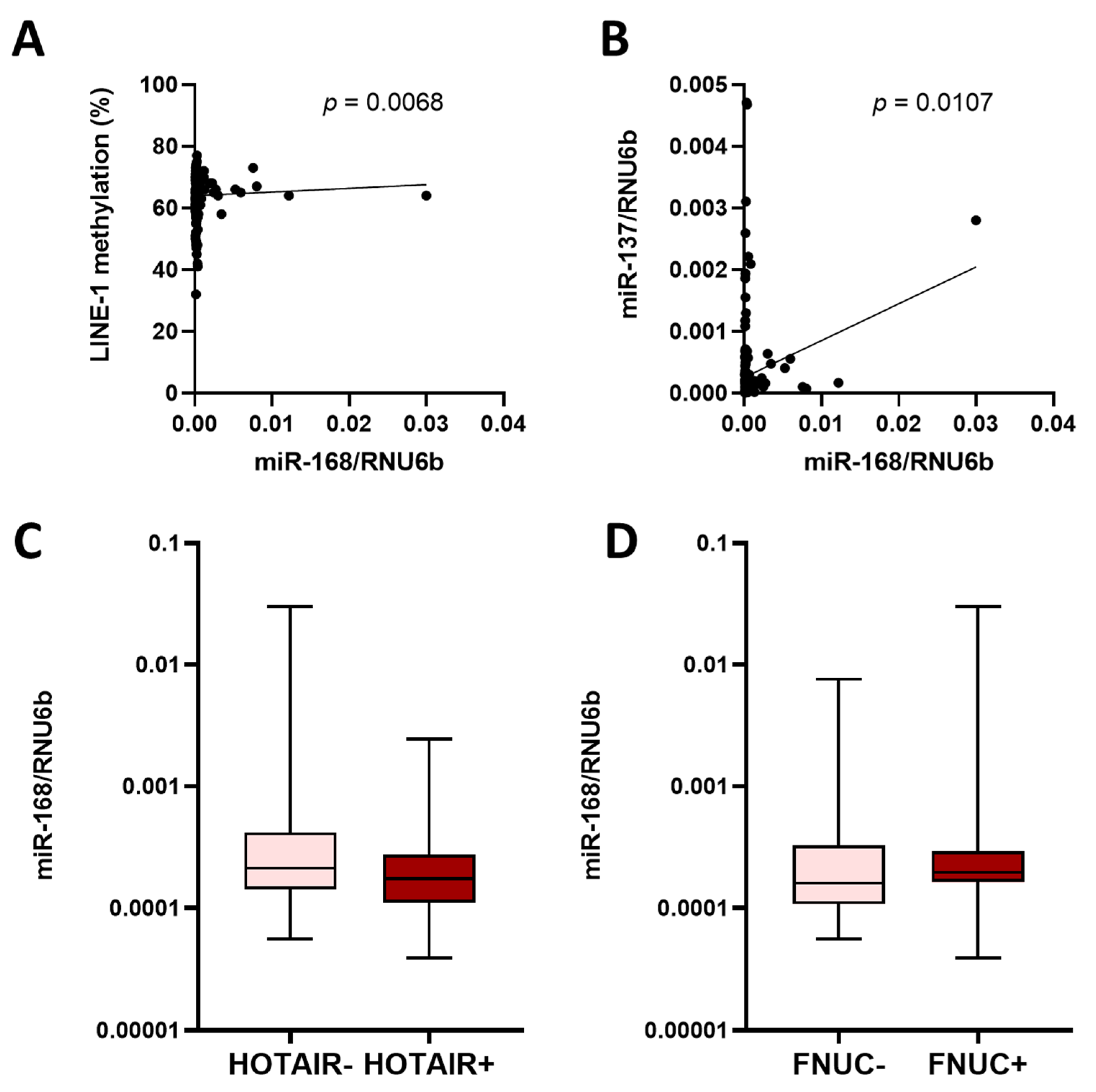
3.4. miR-168 and Other Molecular Features
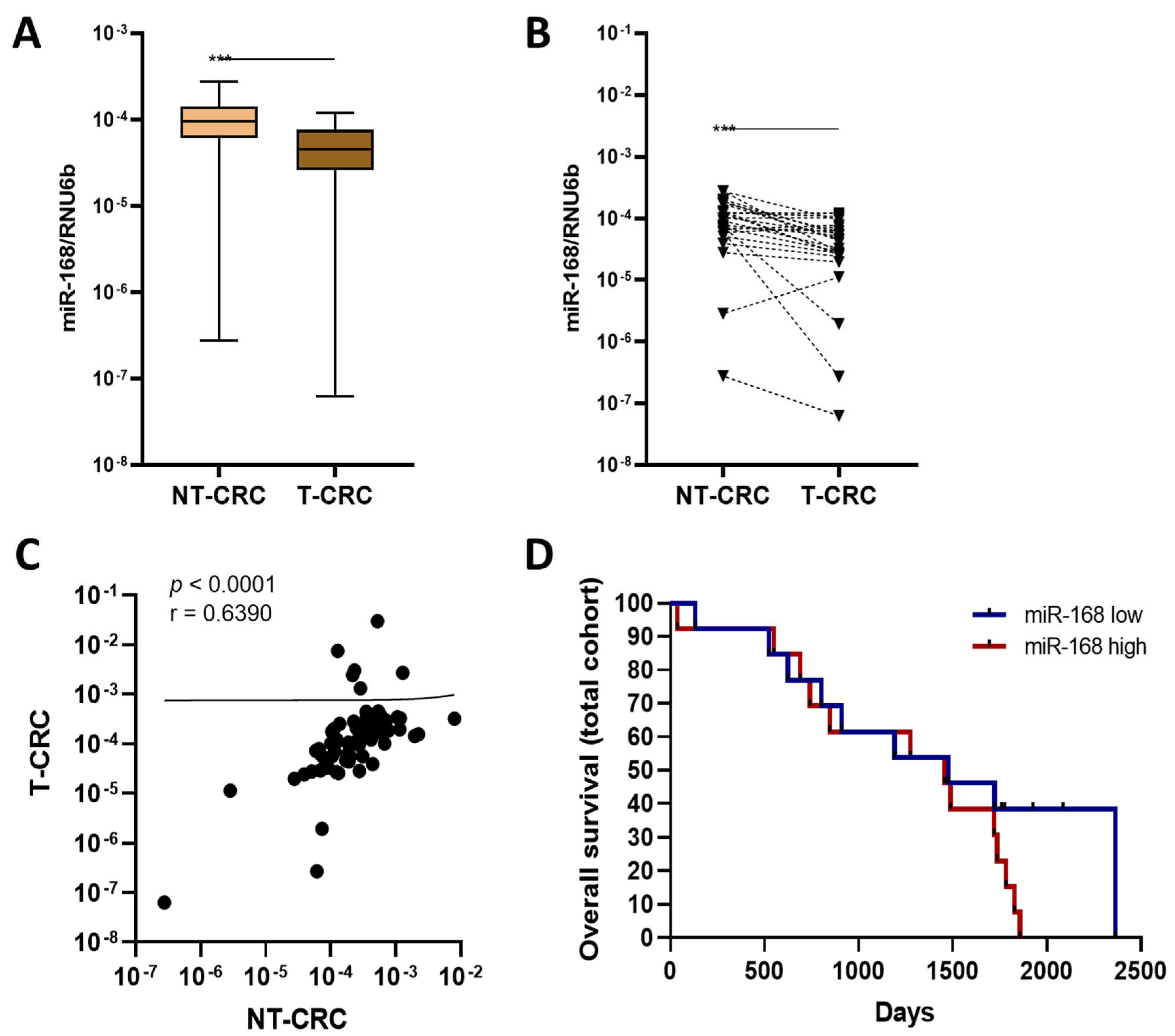
3.5. miR-168 in CRC
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Ha, M.; Kim, V.N. Regulation of microRNA biogenesis. Nat. Rev. Mol. Cell Biol. 2014, 15, 509–524. [Google Scholar] [CrossRef] [PubMed]
- Peng, Y.; Croce, C.M. The role of MicroRNAs in human cancer. Signal Transduct. Target Ther. 2016, 1, 15004. [Google Scholar] [CrossRef] [PubMed]
- Gracias, D.T.; Stelekati, E.; Hope, J.L.; Boesteanu, A.C.; Doering, T.A.; Norton, J.; Mueller, Y.M.; Fraietta, J.A.; Wherry, E.J.; Turner, M.; et al. The microRNA miR-155 controls CD8(+) T cell responses by regulating interferon signaling. Nat. Immunol. 2013, 14, 593–602. [Google Scholar] [CrossRef] [PubMed]
- Mashima, R. Physiological roles of miR-155. Immunology 2015, 145, 323–333. [Google Scholar] [CrossRef]
- Oertli, M.; Engler, D.B.; Kohler, E.; Koch, M.; Meyer, T.F.; Müller, A. MicroRNA-155 is essential for the T cell-mediated control of Helicobacter pylori infection and for the induction of chronic Gastritis and Colitis. J. Immunol. 2011, 187, 3578–3586. [Google Scholar] [CrossRef]
- Calin, G.A.; Croce, C.M. MicroRNA signatures in human cancers. Nat. Rev. Cancer 2006, 6, 857–866. [Google Scholar] [CrossRef]
- Lu, J.; Getz, G.; Miska, E.A.; Alvarez-Saavedra, E.; Lamb, J.; Peck, D.; Sweet-Cordero, A.; Ebert, B.L.; Mak, R.H.; Ferrando, A.A.; et al. MicroRNA expression profiles classify human cancers. Nature 2005, 435, 834–838. [Google Scholar] [CrossRef]
- Schindler, P.; Kupcinskas, J.; Juzenas, S.; Skieceviciene, J.; Salteniene, V.; Schulz, C.; Weigt, J.; Malfertheiner, P.; Link, A. Expression of microRNAs in the ascites of patients with peritoneal carcinomatosis and peritonitis. Cancer Cytopathol. 2018, 126, 353–363. [Google Scholar] [CrossRef]
- Chen, X.; Ba, Y.; Ma, L.; Cai, X.; Yin, Y.; Wang, K.; Guo, J.; Zhang, Y.; Chen, J.; Guo, X.; et al. Characterization of microRNAs in serum: A novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res. 2008, 18, 997–1006. [Google Scholar] [CrossRef]
- Carter, J.V.; Galbraith, N.J.; Yang, D.; Burton, J.F.; Walker, S.P.; Galandiuk, S. Blood-based microRNAs as biomarkers for the diagnosis of colorectal cancer: A systematic review and meta-analysis. Br. J. Cancer 2017, 116, 762–774. [Google Scholar] [CrossRef]
- Shigeyasu, K.; Toden, S.; Zumwalt, T.J.; Okugawa, Y.; Goel, A. Emerging Role of MicroRNAs as Liquid Biopsy Biomarkers in Gastrointestinal Cancers. Clin. Cancer Res. 2017, 23, 2391–2399. [Google Scholar] [CrossRef] [PubMed]
- Rhoades, M.W.; Reinhart, B.J.; Lim, L.P.; Burge, C.B.; Bartel, B.; Bartel, D.P. Prediction of plant microRNA targets. Cell 2002, 110, 513–520. [Google Scholar] [CrossRef] [PubMed]
- Reinhart, B.J.; Weinstein, E.G.; Rhoades, M.W.; Bartel, B.; Bartel, D.P. MicroRNAs in plants. Genes Dev. 2002, 16, 1616–1626. [Google Scholar] [CrossRef] [PubMed]
- Fabris, L.; Calin, G.A. Circulating free xeno-microRNAs—The new kids on the block. Mol. Oncol. 2016, 10, 503–508. [Google Scholar] [CrossRef]
- Zhang, L.; Hou, D.; Chen, X.; Li, D.; Zhu, L.; Zhang, Y.; Li, J.; Bian, Z.; Liang, X.; Cai, X.; et al. Exogenous plant MIR168a specifically targets mammalian LDLRAP1: Evidence of cross-kingdom regulation by microRNA. Cell Res. 2012, 22, 107–126. [Google Scholar] [CrossRef]
- Link, J.; Thon, C.; Schanze, D.; Steponaitiene, R.; Kupcinskas, J.; Zenker, M.; Canbay, A.; Malfertheiner, P.; Link, A. Food-Derived Xeno-microRNAs: Influence of Diet and Detectability in Gastrointestinal Tract-Proof-of-Principle Study. Mol. Nutr. Food Res. 2019, 63, e1800076. [Google Scholar] [CrossRef]
- Dickinson, B.; Zhang, Y.; Petrick, J.S.; Heck, G.; Ivashuta, S.; Marshall, W.S. Lack of detectable oral bioavailability of plant microRNAs after feeding in mice. Nat. Biotechnol. 2013, 31, 965–967. [Google Scholar] [CrossRef]
- Witwer, K.W.; McAlexander, M.A.; Queen, S.E.; Adams, R.J. Real-time quantitative PCR and droplet digital PCR for plant miRNAs in mammalian blood provide little evidence for general uptake of dietary miRNAs: Limited evidence for general uptake of dietary plant xenomiRs. RNA Biol. 2013, 10, 1080–1086. [Google Scholar] [CrossRef]
- Javed, M.; Solanki, M.; Sinha, A.; Shukla, L.I. Position Based Nucleotide Analysis of miR168 Family in Higher Plants and its Targets in Mammalian Transcripts. Microrna 2017, 6, 136–142. [Google Scholar] [CrossRef]
- Amieva, M.; Peek, R.M. Pathobiology of Helicobacter pylori–Induced Gastric Cancer. Gastroenterology 2016, 150, 64–78. [Google Scholar] [CrossRef]
- Wong, M.C.S.; Huang, J.; Chan, P.S.F.; Choi, P.; Lao, X.Q.; Chan, S.M.; Teoh, A.; Liang, P. Global Incidence and Mortality of Gastric Cancer, 1980–2018. JAMA Netw. Open 2021, 4, e2118457. [Google Scholar] [CrossRef] [PubMed]
- Link, A.; Bornschein, J.; Thon, C. Helicobacter Pylori Induced Gastric Carcinogenesis—The Best Molecular Model We Have? Best Pract. Res. Clin. Gastroenterol. 2021, 50–51, 101743. [Google Scholar] [CrossRef] [PubMed]
- Malfertheiner, P.; Camargo, M.C.; El-Omar, E.; Liou, J.M.; Peek, R.; Schulz, C.; Smith, S.I.; Suerbaum, S. Helicobacter pylori infection. Nat. Rev. Dis. Primers 2023, 9, 19. [Google Scholar] [CrossRef] [PubMed]
- Kupcinskas, J.; Steponaitiene, R.; Langner, C.; Smailyte, G.; Skieceviciene, J.; Kupcinskas, L.; Malfertheiner, P.; Link, A. LINE-1 hypomethylation is not a common event in preneoplastic stages of gastric carcinogenesis. Sci. Rep. 2017, 7, 4828. [Google Scholar] [CrossRef] [PubMed]
- Steponaitiene, R.; Kupcinskas, J.; Langner, C.; Balaguer, F.; Venclauskas, L.; Pauzas, H.; Tamelis, A.; Skieceviciene, J.; Kupcinskas, L.; Malfertheiner, P.; et al. Epigenetic silencing of miR-137 is a frequent event in gastric carcinogenesis. Mol. Carcinog. 2016, 55, 376–386. [Google Scholar] [CrossRef]
- Petkevicius, V.; Thon, C.; Steponaitiene, R.; Skieceviciene, J.; Janciauskas, D.; Jechorek, D.; Malfertheiner, P.; Kupcinskas, J.; Link, A. Differential Expression of Long Noncoding RNA HOTAIR in Intestinal Metaplasia and Gastric Cancer. Clin. Transl. Gastroenterol. 2022, 13, e00483. [Google Scholar] [CrossRef]
- Boehm, E.T.; Thon, C.; Kupcinskas, J.; Steponaitiene, R.; Skieceviciene, J.; Canbay, A.; Malfertheiner, P.; Link, A. Fusobacterium nucleatum is associated with worse prognosis in Lauren’s diffuse type gastric cancer patients. Sci. Rep. 2020, 10, 16240. [Google Scholar] [CrossRef]
- Dixon, M.F.; Genta, R.M.; Yardley, J.H.; Correa, P. Classification and grading of gastritis. The updated Sydney System. International Workshop on the Histopathology of Gastritis, Houston 1994. Am. J. Surg. Pathol. 1996, 20, 1161–1181. [Google Scholar] [CrossRef]
- Link, A.; Langner, C.; Schirrmeister, W.; Habendorf, W.; Weigt, J.; Venerito, M.; Tammer, I.; Schlüter, D.; Schlaermann, P.; Meyer, T.F.; et al. Helicobacter pylori vacA genotype is a predominant determinant of immune response to Helicobacter pylori CagA. World J. Gastroenterol. 2017, 23, 4712–4723. [Google Scholar] [CrossRef]
- Schönauen, K.; Le, N.; von Arnim, U.; Schulz, C.; Malfertheiner, P.; Link, A. Circulating and Fecal microRNAs as Biomarkers for Inflammatory Bowel Diseases. Inflamm. Bowel Dis. 2018, 24, 1547–1557. [Google Scholar] [CrossRef]
- Lehr, K.; Nikitina, D.; Vilchez-Vargas, R.; Steponaitiene, R.; Thon, C.; Skieceviciene, J.; Schanze, D.; Zenker, M.; Malfertheiner, P.; Kupcinskas, J.; et al. Microbial composition of tumorous and adjacent gastric tissue is associated with prognosis of gastric cancer. Sci. Rep. 2023, 13, 4640. [Google Scholar] [CrossRef] [PubMed]
- Chen, Q.; Zhang, F.; Dong, L.; Wu, H.; Xu, J.; Li, H.; Wang, J.; Zhou, Z.; Liu, C.; Wang, Y.; et al. SIDT1-dependent absorption in the stomach mediates host uptake of dietary and orally administered microRNAs. Cell Res. 2021, 31, 247–258. [Google Scholar] [CrossRef] [PubMed]
- Fu, D.-G. Epigenetic alterations in gastric cancer (Review). Mol. Med. Rep. 2015, 10, 3223–3230. [Google Scholar] [CrossRef] [PubMed]
- Okugawa, Y.; Grady, W.M.; Goel, A. Epigenetic Alterations in Colorectal Cancer: Emerging Biomarkers. Gastroenterology 2015, 149, 1204–1225.e12. [Google Scholar] [CrossRef] [PubMed]
- Cancer Genome Atlas Research Network. Comprehensive molecular characterization of gastric adenocarcinoma. Nature 2014, 513, 202–209. [Google Scholar] [CrossRef]
- Ungerleider, N.; Bullard, W.; Kara, M.; Wang, X.; Roberts, C.; Renne, R.; Tibbetts, S.; Flemington, E.K. EBV miRNAs are potent effectors of tumor cell transcriptome remodeling in promoting immune escape. PLoS Pathog. 2021, 17, e1009217. [Google Scholar] [CrossRef]
- Kashyap, D.; Rele, S.; Bagde, P.H.; Saini, V.; Chatterjee, D.; Jain, A.K.; Pandey, R.K.; Jha, H.C. Comprehensive insight into altered host cell-signaling cascades upon Helicobacter pylori and Epstein-Barr virus infections in cancer. Arch. Microbiol. 2023, 205, 262. [Google Scholar] [CrossRef]
- Hirabayashi, M.; Georges, D.; Clifford, G.M.; de Martel, C. Estimating the Global Burden of Epstein-Barr Virus-Associated Gastric Cancer: A Systematic Review and Meta-Analysis. Clin. Gastroenterol. Hepatol. 2023, 21, 922–930.e21. [Google Scholar] [CrossRef]
- Teng, Y.; Ren, Y.; Sayed, M.; Hu, X.; Lei, C.; Kumar, A.; Hutchins, E.; Mu, J.; Deng, Z.; Luo, C.; et al. Plant-Derived Exosomal MicroRNAs Shape the Gut Microbiota. Cell Host Microbe 2018, 24, 637–652.e8. [Google Scholar] [CrossRef]
- He, X.; Wang, Y.; Fan, X.; Lei, N.; Tian, Y.; Zhang, D.; Pan, W. A schistosome miRNA promotes host hepatic fibrosis by targeting transforming growth factor beta receptor III. J. Hepatol. 2020, 72, 519–527. [Google Scholar] [CrossRef]
- Zhang, Q.; Xie, Z.; Li, Y.; Zhu, Q.; Shi, H.; Zhao, R.; Yang, X.; Tian, J.; Ma, L. The potential of Lycium barbarum miR166a in kidney cancer treatment. Exp. Cell Res. 2023, 423, 113455. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Wang, F.; Weng, Z.; Sui, X.; Fang, Y.; Tang, X.; Shen, X. Soybean-derived miRNAs specifically inhibit proliferation and stimulate apoptosis of human colonic Caco-2 cancer cells but not normal mucosal cells in culture. Genomics 2020, 112, 2949–2958. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Wang, F.; Song, H.; Weng, Z.; Bao, Y.; Fang, Y.; Tang, X.; Shen, X. Soybean-derived gma-miR159a alleviates colon tumorigenesis by suppressing TCF7/MYC in mice. J. Nutr. Biochem. 2021, 92, 108627. [Google Scholar] [CrossRef] [PubMed]


| All | miR-168-Low | miR-168-High | p | ||||
|---|---|---|---|---|---|---|---|
| n = 81 | % | n = 40 | % | n = 41 | % | ||
| Age | 65.85 | ±11.58 | 64.15 | ±10.76 | 67.50 | ±12.37 | 0.2 * |
| Gender | 0.37 | ||||||
| male | 47 | 58% | 21 | 52.5% | 26 | 63% | |
| female | 34 | 42% | 19 | 47.5% | 15 | 37% | |
| Tumour localization | 0.04 | ||||||
| cardia | 8 | 10% | 2 | 5% | 6 | 15% | |
| corpus | 45 | 56% | 19 | 47.5% | 26 | 63% | |
| antrum | 28 | 34% | 19 | 47.5% | 9 | 22% | |
| UICC | 0.33 | ||||||
| I | 16 | 20% | 5 | 12.5% | 11 | 26.8% | |
| II | 21 | 26% | 11 | 27.5% | 10 | 24.4% | |
| III | 36 | 44% | 23 | 57.5% | 13 | 31.7% | |
| IV | 8 | 10% | 1 | 2.5% | 7 | 17.1% | |
| T | 0.3 | ||||||
| 1 + 2 | 18 | 22% | 6 | 15% | 12 | 29% | |
| 3 | 36 | 45% | 19 | 47.5% | 17 | 42% | |
| 4 | 27 | 33% | 15 | 37.5% | 12 | 29% | |
| N | 0.22 | ||||||
| 0 | 29 | 36% | 11 | 27.59% | 19 | 46.5% | |
| 1 | 15 | 19% | 8 | 20% | 7 | 17% | |
| 2 | 13 | 16% | 6 | 15% | 7 | 17% | |
| 3 | 23 | 28% | 15 | 37.5% | 8 | 19.5% | |
| Unknown | 1 | 1% | 1 | 2.4% | |||
| M | 0.06 | ||||||
| 0 | 73 | 90% | 39 | 97.5% | 34 | 83% | |
| 1 | 8 | 10% | 1 | 2.5% | 7 | 17% | |
| Grading | 0.15 | ||||||
| 1 | 3 | 4% | 0 | 0% | 3 | 7% | |
| 2 | 29 | 36% | 13 | 32.5% | 16 | 39% | |
| 3 | 49 | 60% | 27 | 67.5% | 22 | 54% | |
| Lauren Classification | 0.14 | ||||||
| Diffuse Type | 44 | 54% | 23 | 57.5% | 21 | 51% | |
| Intestinal Type | 26 | 32% | 12 | 30% | 14 | 34% | |
| Mixed Type | 7 | 9% | 5 | 12.5% | 2 | 5% | |
| Unknown | 4 | 5% | 4 | 10% | |||
| H. pylori | 0.02 | ||||||
| Negative | 8 | 10% | 1 | 2.5% | 7 | 17% | |
| Positive | 17 | 21% | 6 | 15% | 11 | 27% | |
| Unknown | 56 | 69% | 33 | 82.5% | 23 | 56% | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Link, J.; Thon, C.; Petkevicius, V.; Steponaitiene, R.; Malfertheiner, P.; Kupcinskas, J.; Link, A. The Translational Impact of Plant-Derived Xeno-miRNA miR-168 in Gastrointestinal Cancers and Preneoplastic Conditions. Diagnostics 2023, 13, 2701. https://doi.org/10.3390/diagnostics13162701
Link J, Thon C, Petkevicius V, Steponaitiene R, Malfertheiner P, Kupcinskas J, Link A. The Translational Impact of Plant-Derived Xeno-miRNA miR-168 in Gastrointestinal Cancers and Preneoplastic Conditions. Diagnostics. 2023; 13(16):2701. https://doi.org/10.3390/diagnostics13162701
Chicago/Turabian StyleLink, Jastin, Cosima Thon, Vytenis Petkevicius, Ruta Steponaitiene, Peter Malfertheiner, Juozas Kupcinskas, and Alexander Link. 2023. "The Translational Impact of Plant-Derived Xeno-miRNA miR-168 in Gastrointestinal Cancers and Preneoplastic Conditions" Diagnostics 13, no. 16: 2701. https://doi.org/10.3390/diagnostics13162701
APA StyleLink, J., Thon, C., Petkevicius, V., Steponaitiene, R., Malfertheiner, P., Kupcinskas, J., & Link, A. (2023). The Translational Impact of Plant-Derived Xeno-miRNA miR-168 in Gastrointestinal Cancers and Preneoplastic Conditions. Diagnostics, 13(16), 2701. https://doi.org/10.3390/diagnostics13162701







