Machine Learning in Antibody Diagnostics for Inflammatory Bowel Disease Subtype Classification
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Population and Design
2.2. Analysis of Antibodies
2.3. Statistical Analysis
3. Results
3.1. Antibody Status and Panel Diagnostic in Adult CD and UC Patients
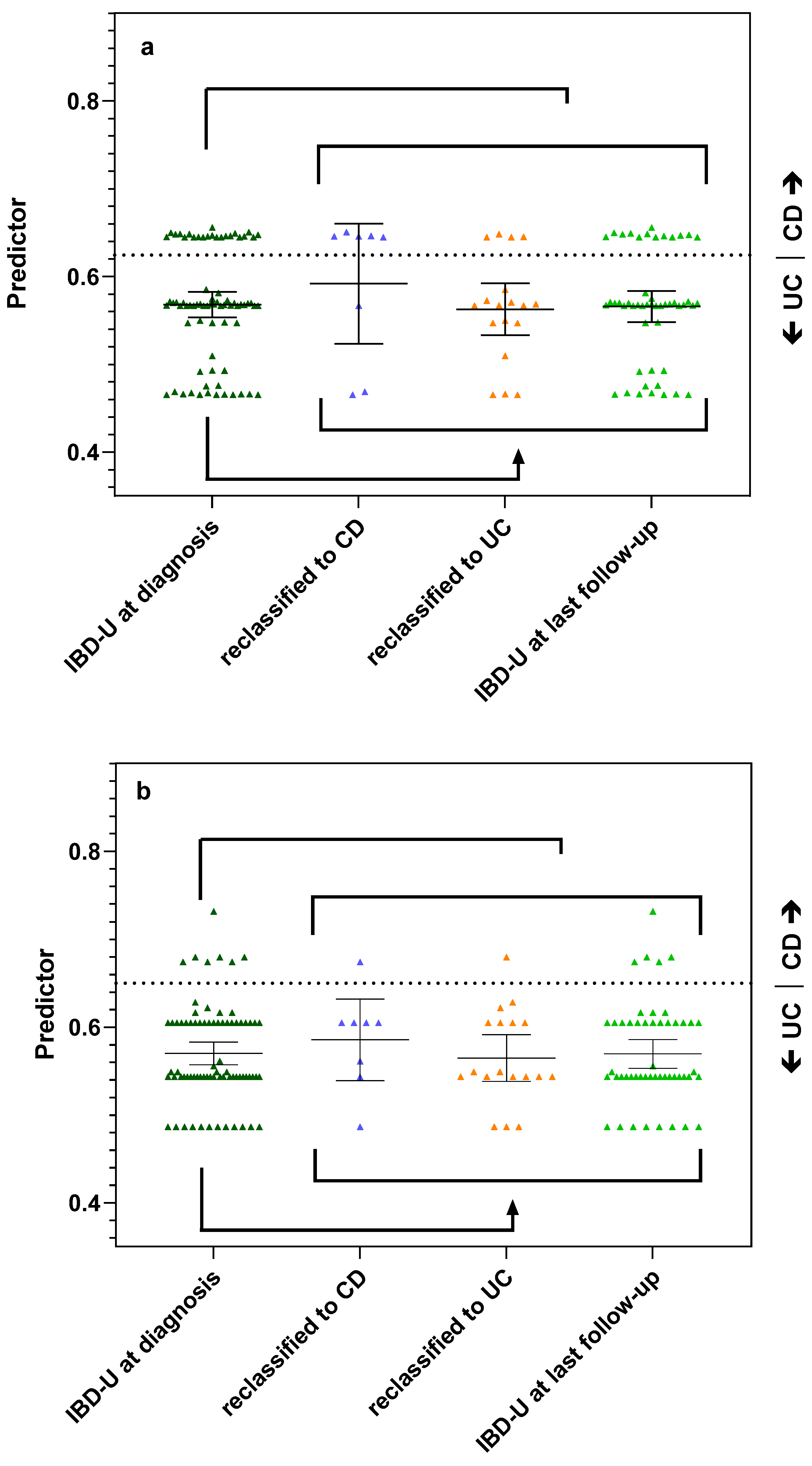
3.2. Antibody Status and Panel Diagnostics in IBD-U Patients
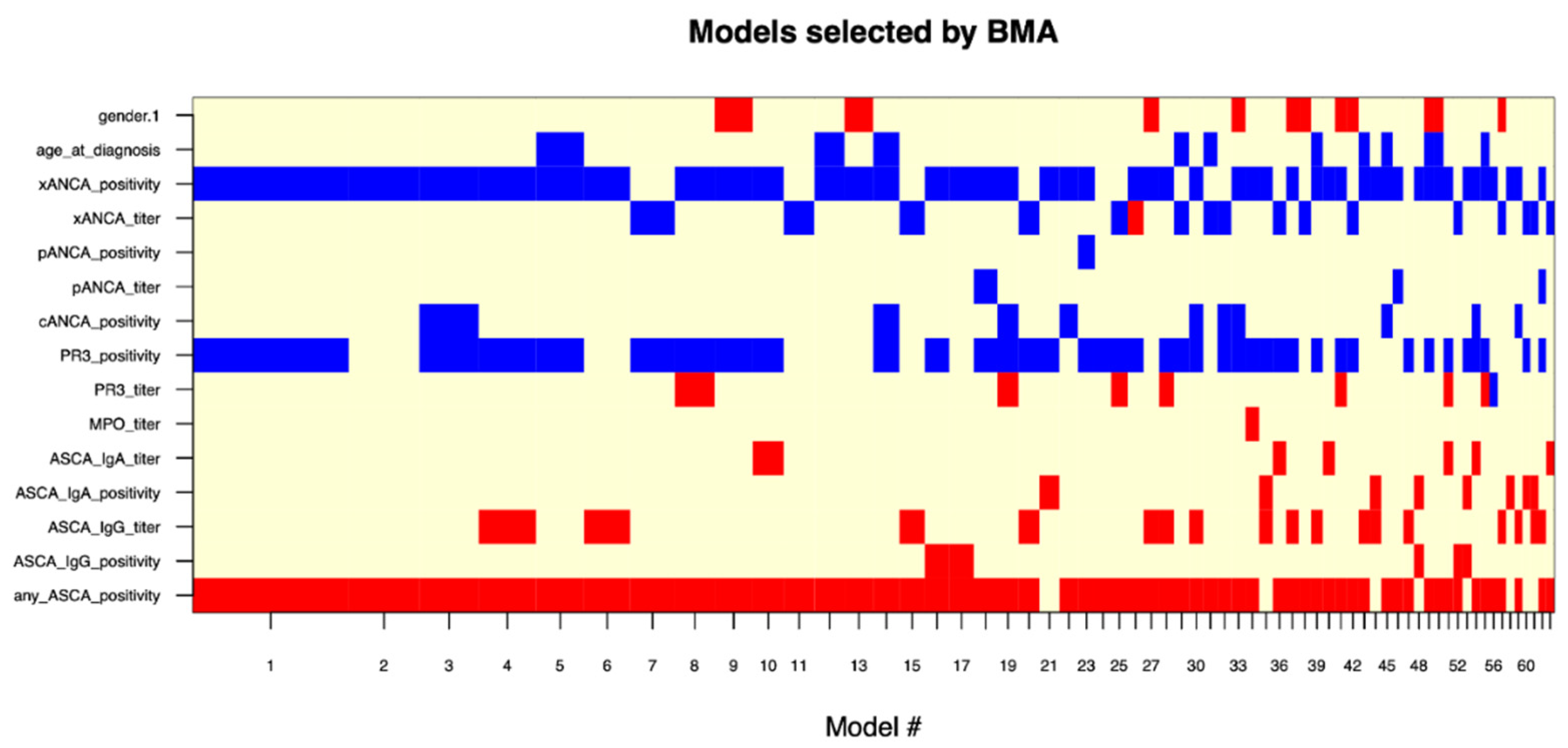
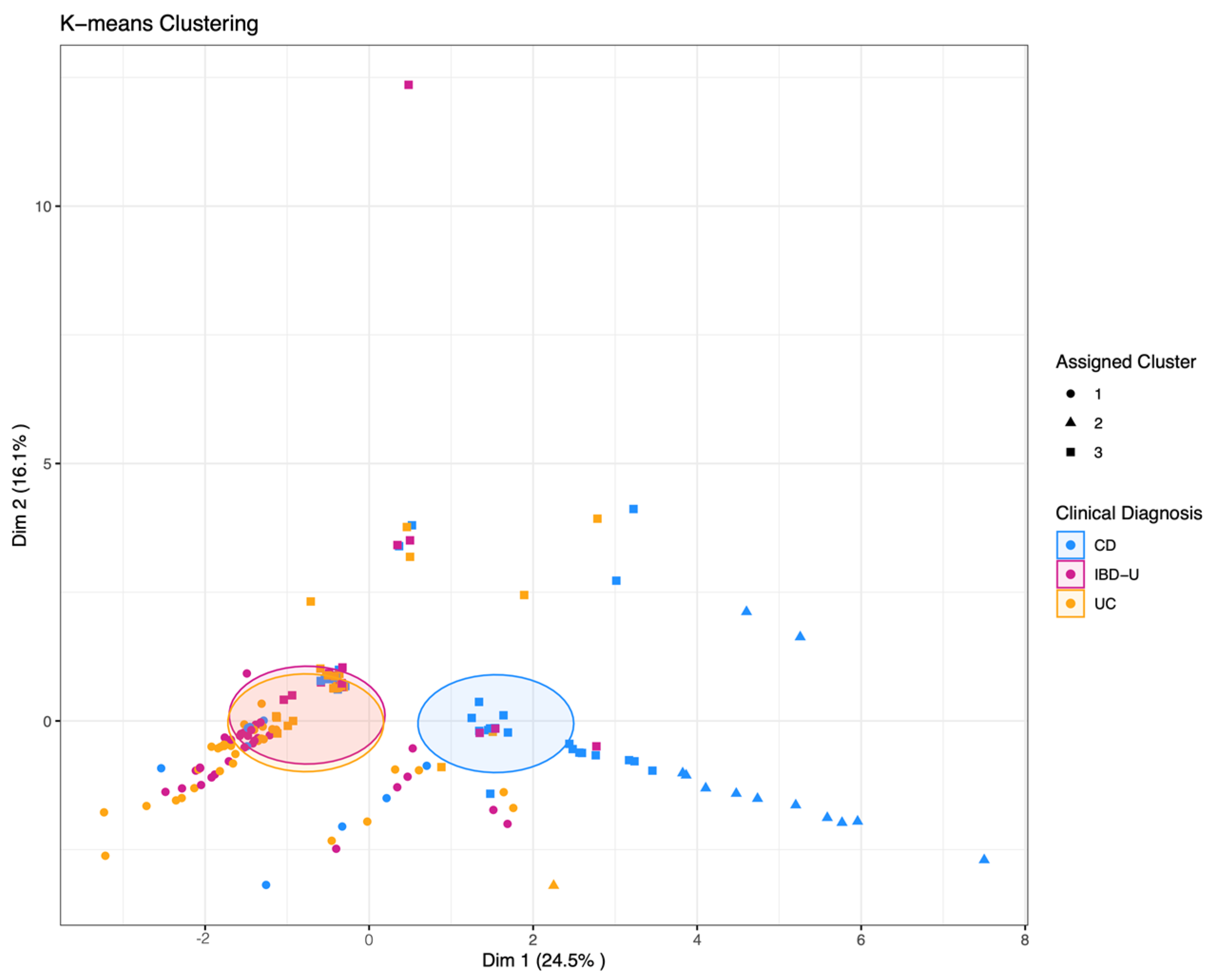
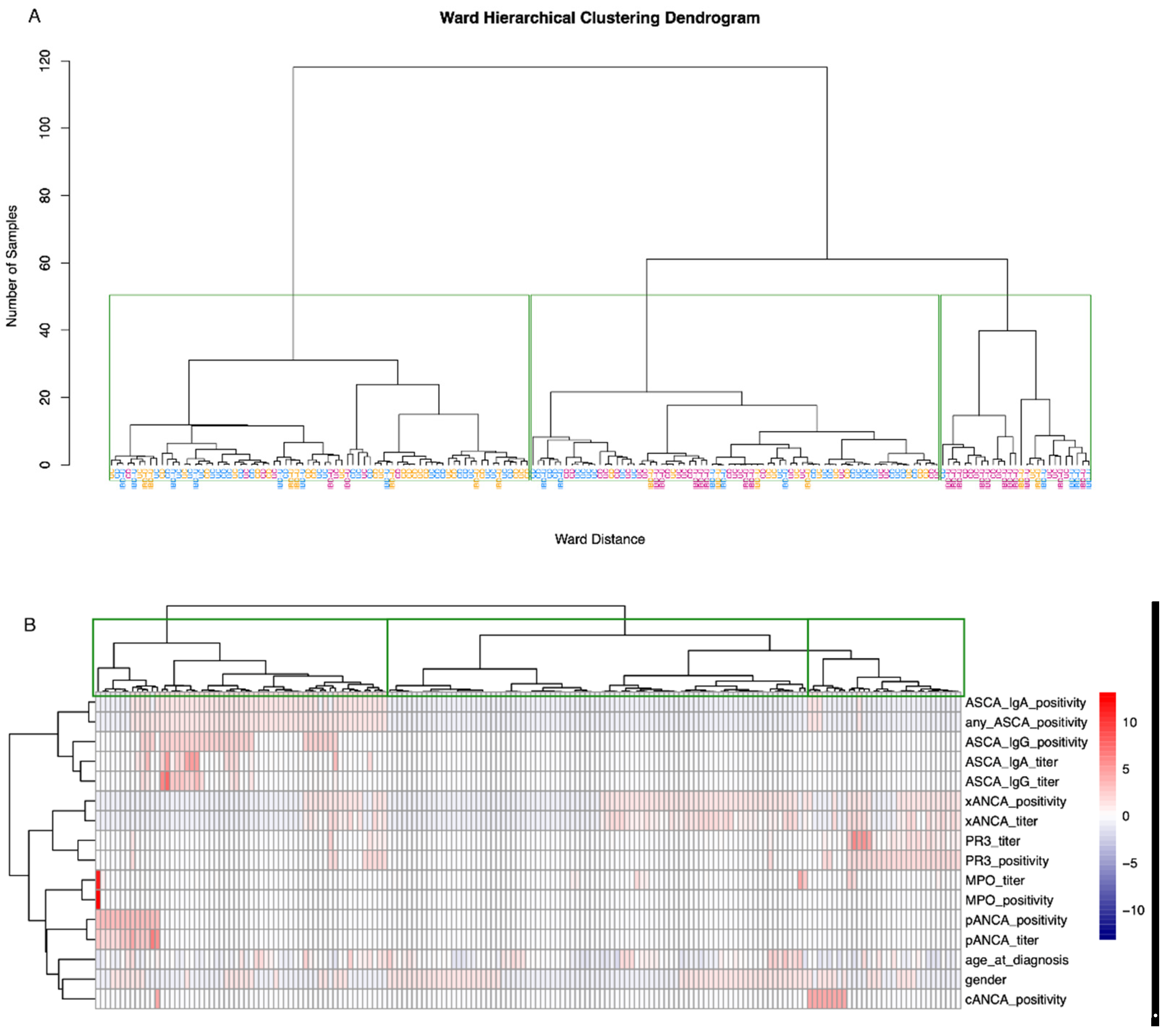
3.3. Supervised and Unsupervised Machine Learning for Bi- and Multiclass Prediction Models (CD vs. UC and CD vs. UC vs. IBD-U)
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Cleynen, I.; Boucher, G.; Jostins, L.; Schumm, L.P.; Zeissig, S.; Ahmad, T.; Andersen, V.; Andrews, J.M.; Annese, V.; Brand, S.; et al. Inherited determinants of Crohn’s disease and ulcerative colitis phenotypes: A genetic association study. Lancet 2016, 387, 156–167. [Google Scholar] [CrossRef]
- Birimberg-Schwartz, L.; Zucker, D.M.; Akriv, A.; Cucchiara, S.; Cameron, F.L.; Wilson, D.C.; Lazowska, I.; Yianni, L.; Paul, S.P.; Romano, C.; et al. Development and Validation of Diagnostic Criteria for IBD Subtypes Including IBD-unclassified in Children: A Multicentre Study From the Pediatric IBD Porto Group of ESPGHAN. J. Crohn’s Colitis 2017, 11, 1078–1084. [Google Scholar] [CrossRef] [PubMed]
- Hedrick, T.L.; Friel, C.M. Colonic crohn disease. Clin. Colon Rectal Surg. 2013, 26, 84–89. [Google Scholar] [CrossRef] [PubMed]
- Subramanian, S.; Ekbom, A.; Rhodes, J.M. Recent advances in clinical practice: A systematic review of isolated colonic Crohn’s disease: The third IBD? Gut 2017, 66, 362–381. [Google Scholar] [CrossRef] [PubMed]
- Berger, T.D.; Lee, H.M.; Padmanaban, L.R.; Wine, E.; Yerushalmy-Feler, A.; Hojsak, I.; Kazeka, D.; Serban, D.E.; Yogev, D.; Ledder, O.; et al. Clinical Features and Outcomes of Paediatric Patients With Isolated Colonic Crohn Disease. J. Pediatr. Gastroenterol. Nutr. 2022, 74, 258–266. [Google Scholar] [CrossRef] [PubMed]
- Everhov, A.H.; Sachs, M.C.; Malmborg, P.; Nordenvall, C.; Myrelid, P.; Khalili, H.; Elmberg, M.; Ekbom, A.; Askling, J.; Jakobsson, G.; et al. Changes in inflammatory bowel disease subtype during follow-up and over time in 44,302 patients. Scand. J. Gastroenterol. 2019, 54, 55–63. [Google Scholar] [CrossRef]
- Chandradevan, R.; Hofmekler, T.; Mondal, K.; Harun, N.; Venkateswaran, S.; Somineni, H.K.; Ballengee, C.R.; Kim, M.O.; Griffiths, A.; Noe, J.D.; et al. Evolution of Pediatric Inflammatory Bowel Disease Unclassified (IBD-U): Incorporated With Serological and Gene Expression Profiles. Inflamm. Bowel Dis. 2018, 24, 2285–2290. [Google Scholar] [CrossRef]
- Prenzel, F.; Uhlig, H.H. Frequency of indeterminate colitis in children and adults with IBD—A metaanalysis. J. Crohn’s Colitis 2009, 3, 277–281. [Google Scholar] [CrossRef]
- Burisch, J.; Zammit, S.C.; Ellul, P.; Turcan, S.; Duricova, D.; Bortlik, M.; Andersen, K.W.; Andersen, V.; Kaimakliotis, I.P.; Fumery, M.; et al. Disease course of inflammatory bowel disease unclassified in a European population-based inception cohort: An Epi-IBD study. J. Gastroenterol. Hepatol. 2019, 34, 996–1003. [Google Scholar] [CrossRef]
- Horn, M.P.; Peter, A.M.; Righini Grunder, F.; Leichtle, A.B.; Spalinger, J.; Schibli, S.; Sokollik, C. PR3-ANCA and panel diagnostics in pediatric inflammatory bowel disease to distinguish ulcerative colitis from Crohn’s disease. PLoS ONE 2018, 13, e0208974. [Google Scholar] [CrossRef]
- Pittet, V.; Juillerat, P.; Mottet, C.; Felley, C.; Ballabeni, P.; Burnand, B.; Michetti, P.; Vader, J.P.; Swiss IBD Cohort Study Group. Cohort profile: The Swiss Inflammatory Bowel Disease Cohort Study (SIBDCS). Int. J. Epidemiol. 2009, 38, 922–931. [Google Scholar] [CrossRef] [PubMed]
- Pittet, V.; Michetti, P.; Mueller, C.; Braegger, C.P.; von Kanel, R.; Schoepfer, A.; Macpherson, A.J.; Rogler, G. Cohort Profile Update: The Swiss Inflammatory Bowel Disease Cohort Study (SIBDCS). Int. J. Epidemiol. 2019, 48, 385–386f. [Google Scholar] [CrossRef] [PubMed]
- Gomollon, F.; Dignass, A.; Annese, V.; Tilg, H.; Van Assche, G.; Lindsay, J.O.; Peyrin-Biroulet, L.; Cullen, G.J.; Daperno, M.; Kucharzik, T.; et al. 3rd European Evidence-based Consensus on the Diagnosis and Management of Crohn’s Disease 2016: Part 1: Diagnosis and Medical Management. J. Crohn’s Colitis 2017, 11, 3–25. [Google Scholar] [CrossRef]
- Silverberg, M.S.; Satsangi, J.; Ahmad, T.; Arnott, I.D.; Bernstein, C.N.; Brant, S.R.; Caprilli, R.; Colombel, J.F.; Gasche, C.; Geboes, K.; et al. Toward an integrated clinical, molecular and serological classification of inflammatory bowel disease: Report of a Working Party of the 2005 Montreal World Congress of Gastroenterology. Can. J. Gastroenterol. 2005, 19 (Suppl. A), 5A–36A. [Google Scholar] [CrossRef]
- Satsangi, J.; Silverberg, M.S.; Vermeire, S.; Colombel, J.F. The Montreal classification of inflammatory bowel disease: Controversies, consensus, and implications. Gut 2006, 55, 749–753. [Google Scholar] [CrossRef]
- Miller, A.J. Selection of Subsets of Regression Variables. J. R. Stat. Soc. 1984, 147, 36. [Google Scholar] [CrossRef]
- Raftery, A.E.; Madigan, D.; Hoeting, J.A. Bayesian Model Averaging for Linear Regression Models. J. Am. Stat. Assoc. 1997, 92, 179–191. [Google Scholar] [CrossRef]
- Rinawi, F.; Assa, A.; Eliakim, R.; Mozer-Glassberg, Y.; Nachmias Friedler, V.; Niv, Y.; Rosenbach, Y.; Silbermintz, A.; Zevit, N.; Shamir, R. The natural history of pediatric-onset IBD-unclassified and prediction of Crohn’s disease reclassification: A 27-year study. Scand. J. Gastroenterol. 2017, 52, 558–563. [Google Scholar] [CrossRef]
- Joossens, S.; Reinisch, W.; Vermeire, S.; Sendid, B.; Poulain, D.; Peeters, M.; Geboes, K.; Bossuyt, X.; Vandewalle, P.; Oberhuber, G.; et al. The value of serologic markers in indeterminate colitis: A prospective follow-up study. Gastroenterology 2002, 122, 1242–1247. [Google Scholar] [CrossRef]
- Birimberg-Schwartz, L.; Wilson, D.C.; Kolho, K.L.; Karolewska-Bochenek, K.; Afzal, N.A.; Spray, C.; Romano, C.; Lionetti, P.; Hauer, A.C.; Martinez-Vinson, C.; et al. pANCA and ASCA in Children with IBD-Unclassified, Crohn’s Colitis, and Ulcerative Colitis-A Longitudinal Report from the IBD Porto Group of ESPGHAN. Inflamm. Bowel Dis. 2016, 22, 1908–1914. [Google Scholar] [CrossRef]
- Hamdeh, S.; Aziz, M.; Altayar, O.; Olyaee, M.; Murad, M.H.; Hanauer, S.B. Early vs Late Use of Anti-TNFa Therapy in Adult Patients With Crohn Disease: A Systematic Review and Meta-Analysis. Inflamm. Bowel Dis. 2020, 26, 1808–1818. [Google Scholar] [CrossRef] [PubMed]
- Verstockt, B.; Salas, A.; Sands, B.E.; Abraham, C.; Leibovitzh, H.; Neurath, M.F.; Vande Casteele, N.; Alimentiv Translational Research Consortium (ATRC). IL-12 and IL-23 pathway inhibition in inflammatory bowel disease. Nat. Rev. Gastroenterol. Hepatol. 2023, 20, 433–446. [Google Scholar] [CrossRef] [PubMed]
- Mahler, M.; Bogdanos, D.P.; Pavlidis, P.; Fritzler, M.J.; Csernok, E.; Damoiseaux, J.; Bentow, C.; Shums, Z.; Forbes, A.; Norman, G.L. PR3-ANCA: A promising biomarker for ulcerative colitis with extensive disease. Clin. Chim. Acta 2013, 424, 267–273. [Google Scholar] [CrossRef] [PubMed]

| Analyte | BeLu * | SIBDCS |
|---|---|---|
| Intercept | 0.5937 | 0.425704 |
| PR-3 ANCA positivity | −0.4085 | −0.22856 |
| xANCA positivity | −0.328 | −0.25171 |
| pANCA positivity | −0.6299 | n.a. |
| ASCA IgG, Titer | 0.0052 | n.a. |
| ASCA IgG positivity | n.a. | 0.277831 |
| ASCA IgA positivity | n.a. | 0.301052 |
| All IBD-U | IBD-U w/o Reclassification | IBD-U w/Reclassification | ||||
|---|---|---|---|---|---|---|
| number of patients | 76 | 50 | 26 | |||
| males, n (%) | 38 (50) | 24 (48) | 14 (53.8) | |||
| age at diagnostic, median (IQR), y | 20 (12–31) | 23 (13–37) | 17.5 (11–28) | |||
| age at serum sampling, median (IQR), y | 23.5 (14–40) | 28 (14–46) | 17.5 (12–30) | |||
| disease duration at serum sampling, median (IQR), y | 2 (1–5) | 3 (1–6.5) | 1 (0–5) | |||
| disease duration at last follow-up, median (IQR), y | 6.5 (4–12) | 7 (4–11) | 6 (4–16) | |||
| disease duration at reclassification, median (IQR), y | n.a. | n.a. | 3.5 (3–9) | |||
| need of surgery, n (%) | 6 (7.9) | 3 (6.0) | 3 (11.5) | |||
| ever treated with biologicals, n (%) | 43 (56.6) | 27 (54) | 16 (61.5) | |||
| Disease location at diagnosis and last follow-up, n (%) | diagnosis | follow-up | diagnosis | follow-up | diagnosis | follow-up |
| E1: proctitis | 3 (3.9) | 4 (5.3) | 2 (2.6) | 1 (1.3) | 1 (1.3) | 3 (3.9) |
| E2: left-sided colitis | 15 (19.7) | 18 (23.7) | 11 (14.5) | 13 (17.1) | 4 (5.3) | 5 (6.6) |
| E3: extensive (pancolitis) | 41 (53.9) | 41 (53.9) | 24 (31.6) | 30 (39.5) | 17 (22.4) | 11 (14.5) |
| unknown | 17 (22.4) | 6 (7.9) | 13 (17.1) | 6 (7.9) | 4 (5.3) | 0 (0) |
| L1: ileal | n.a. | 1 (1.3) | n.a. | n.a. | n.a. | 1 (1.3) |
| L2: colonic | n.a. | 4 (5.3) | n.a. | n.a. | n.a. | 4 (5.3) |
| L3: ileo-colonic | n.a. | 0 (0) | n.a. | n.a. | n.a. | 0 (0) |
| L4: upper GI disease | n.a. | 0 (0) | n.a. | n.a. | n.a. | 0 (0) |
| no endoscopy | n.a. | 2 (2.6) | 0 (0) | 0 (0) | 0 (0) | 2 (2.6) |
| All IBD-U | IBD-U w/o Reclassification | IBD-U w/Re- Classification * | IBD-U → CD * | IBD-U → UC * | |
|---|---|---|---|---|---|
| number of patients | 76 | 50 | 26 | 8 | 18 |
| ANCA positive | |||||
| cANCA, n (%) | 5 (6.6) | 4 (8.0) | 1 (3.8) | 0 (0) | 1 (5.6) |
| (atypical) pANCA, n (%) | 3 (3.9) | 3 (6.0) | 0 (0) | 0 (0) | 0 (0) |
| xANCA, n (%) | 46 (60.5) | 32 (64.0) | 14 (53.8) | 3 (37.5) | 11 (60.5) |
| PR3-ANCA, n (%) | 20 (26.3) | 11 (22.0) | 9 (34.6) | 2 (25.0) | 7 (38.9) |
| PR3-ANCA U/mL, Median, IQR | 1.3 (0–5.2) | 0.9 (0.4–3.1) | 1.9 (0.8–6.9) | 1.2 (0.5–6.2) | 2.8 (1.0–7.0) |
| MPO-ANCA, n (%) | 1 (1.3) | 1 (2.0) | 0 (0) | 0 (0) | 0 (0) |
| MPO-ANCA U/mL, Median | 0 | 0 | 0 | 0 | 0 |
| ASCA positive | |||||
| IgA, n (%) | 13 (17.1) | 8 (16.0) | 5 (19.2) | 2 (25.0) | 3 (16.7) |
| IgA U/mL, Median, IQR | 2.0 (1–3.9) | 2.3 (1–4) | 1.7 (0–3.9) | 2.5 (1.4–6.7) | 1.6 (0–2.5) |
| IgG, n (%) | 6 (7.9) | 4 (7.9) | 2 (7.7) | 0 (0) | 2 (11.1) |
| IgG U/mL, Median, IQR | 1.4 (0.6–3.1) | 1.5 (0.6–3.0) | 1.1 (0.6–3.4) | 1.3 (0.6–2.9) | 0.8 (0.6–3.5) |
| Antibody combinations (n (%)) | |||||
| xANCA neg, PR3-ANCA neg, all ASCA neg | 21 (27.6) | 13 (26.0) | 8 (30.8) | 4 (50.0) | 4 (22.2) |
| xANCA pos, PR3-ANCA pos | 15 (19.7) | 9 (18.0) | 6 (23.1) | 2 (25.0) | 4 (22.2) |
| xANCA neg, all ASCA neg | 25 (32.9) | 15 (30.0) | 10 (38.5) | 4 (50.0) | 6 (33.3) |
| xANCA neg, any ASCA pos | 5 (6.6) | 3 (6.0) | 2 (7.7) | 1 (12.5) | 1 (5.6) |
| xANCA pos, all ASCA neg | 37 (48.7) | 26 (52.0) | 11 (42.3) | 2 (25.0) | 9 (50.0) |
| xANCA pos, any ASCA pos | 9 (11.8) | 6 (12.0) | 3 (11.5) | 1 (12.5) | 2 (11.1) |
| PR3-ANCA neg, all ASCA neg | 46 (60.5) | 31 (62.0) | 15 (57.7) | 5 (62.5) | 10 (55.6) |
| PR3-ANCA neg, any ASCA pos | 10 (13.2) | 8 (16.0) | 2 (7.7) | 1 (12.5) | 1 (5.6) |
| PR3-ANCA pos, all ASCA neg | 16 (21.1) | 10 (20.0) | 6 (23.1) | 1 (12.5) | 5 (27.8) |
| PR3-ANCA pos, any ASCA pos | 4 (5.3) | 1 (2.0) | 3 (11.5) | 1 (12.5) | 2 (11.1) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sokollik, C.; Pahud de Mortanges, A.; Leichtle, A.B.; Juillerat, P.; Horn, M.P., on behalf of the Swiss IBD Cohort Study Group. Machine Learning in Antibody Diagnostics for Inflammatory Bowel Disease Subtype Classification. Diagnostics 2023, 13, 2491. https://doi.org/10.3390/diagnostics13152491
Sokollik C, Pahud de Mortanges A, Leichtle AB, Juillerat P, Horn MP on behalf of the Swiss IBD Cohort Study Group. Machine Learning in Antibody Diagnostics for Inflammatory Bowel Disease Subtype Classification. Diagnostics. 2023; 13(15):2491. https://doi.org/10.3390/diagnostics13152491
Chicago/Turabian StyleSokollik, Christiane, Aurélie Pahud de Mortanges, Alexander B. Leichtle, Pascal Juillerat, and Michael P. Horn on behalf of the Swiss IBD Cohort Study Group. 2023. "Machine Learning in Antibody Diagnostics for Inflammatory Bowel Disease Subtype Classification" Diagnostics 13, no. 15: 2491. https://doi.org/10.3390/diagnostics13152491
APA StyleSokollik, C., Pahud de Mortanges, A., Leichtle, A. B., Juillerat, P., & Horn, M. P., on behalf of the Swiss IBD Cohort Study Group. (2023). Machine Learning in Antibody Diagnostics for Inflammatory Bowel Disease Subtype Classification. Diagnostics, 13(15), 2491. https://doi.org/10.3390/diagnostics13152491






