An Efficient Combination of Convolutional Neural Network and LightGBM Algorithm for Lung Cancer Histopathology Classification
Abstract
1. Introduction
2. Related Work
3. The Datasets
3.1. The Selected Dataset
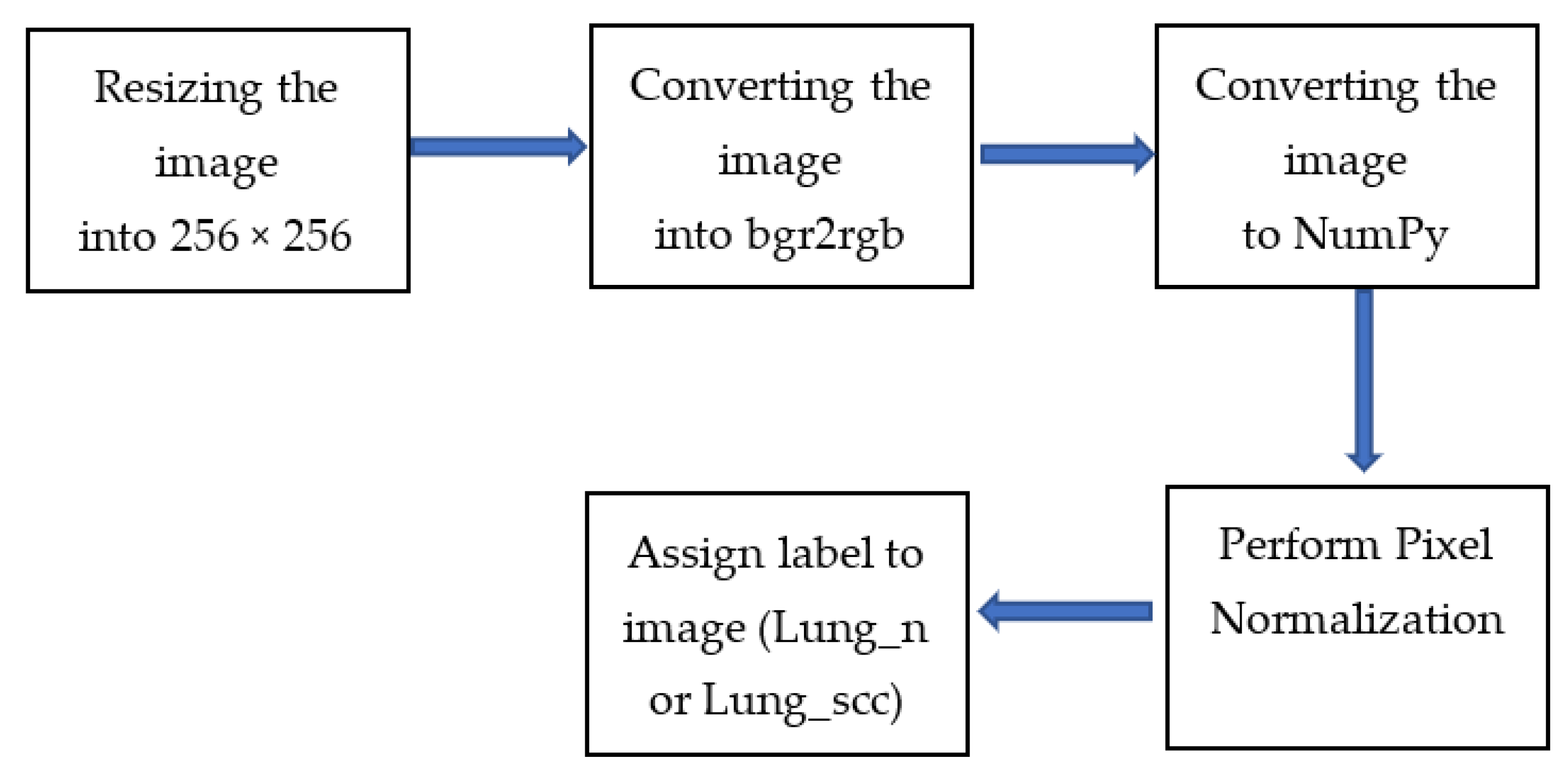
3.2. Data Pre-Processing
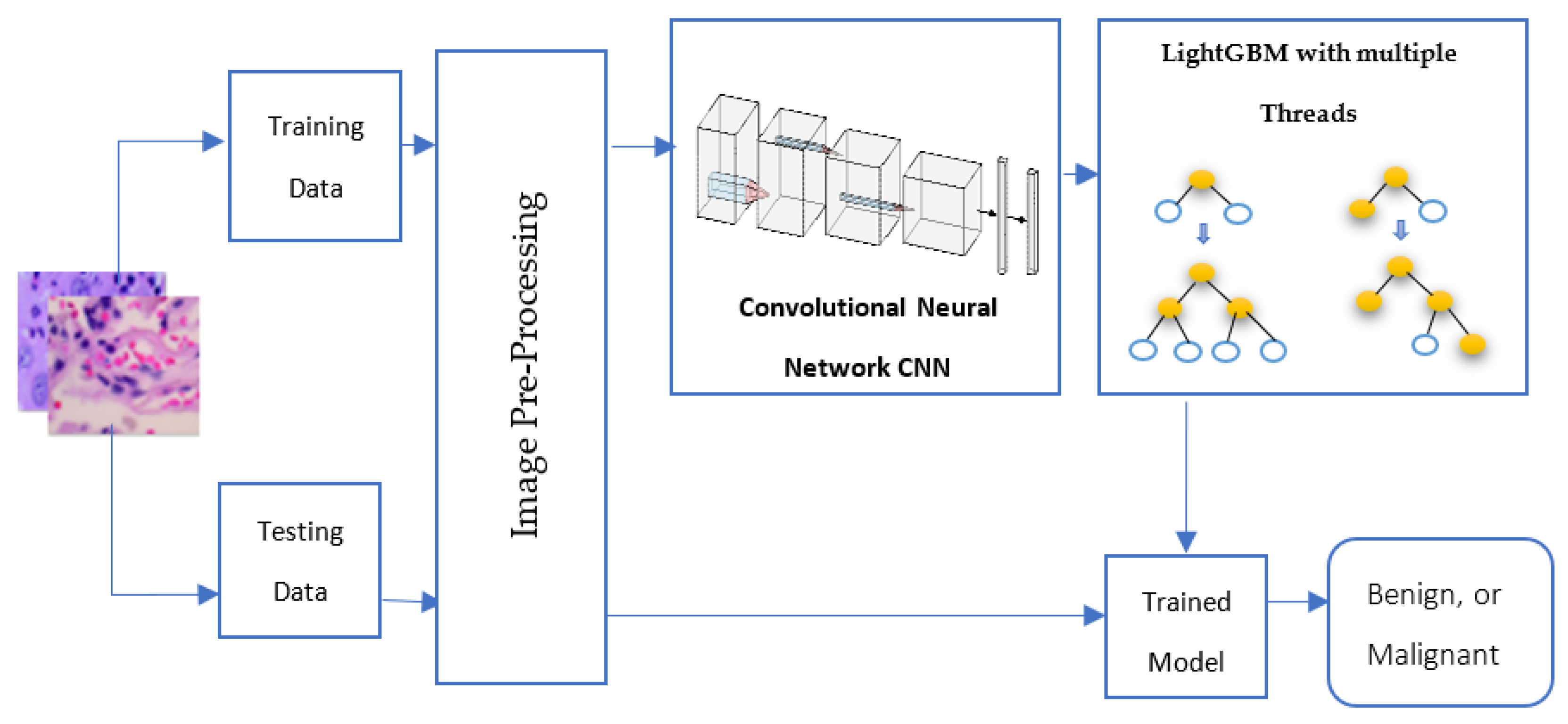
4. The Proposed Approach
4.1. The Proposed Deep Learning Feature Extractor
- Input Layer: With this layer, data were put into and passed into the first convolution layer. In this case, the input is a 256 × 256 pixel image with RGB color channels.
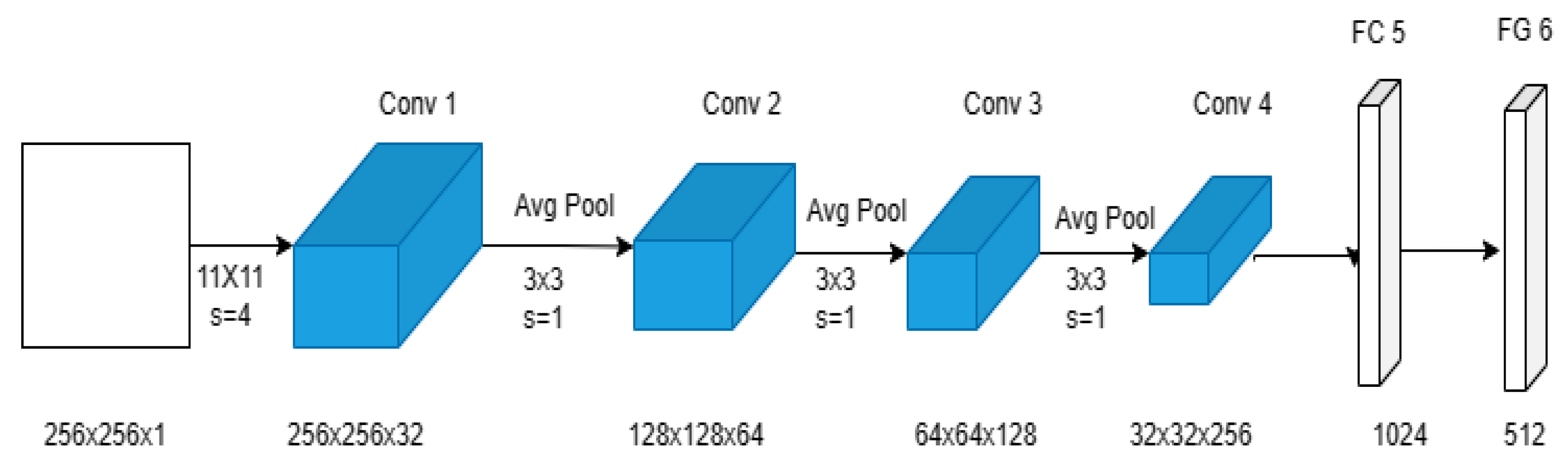
- Convolution Layer: To understand the geographic structure of images, this layer was employed to convolute the input image with trainable filters. This model has four convolution layers (CL), each with kernels of 32, 64, 128, and 256. The first CL layer to be utilized has a kernel size of (11 × 11) and a stride of 4. The padding has also been set to valid. The kernel size for the other CL levels is (3 × 3), with a stride of 1, and the padding has been kept the same. ReLU activation has also been used to boost performance in nonlinear procedures.
- Pooling Layer: The output convolution layer images have been down-sampled using a pooling process. There is a max pooling layer with a (2 × 2) kernel size utilized after each CL layer. The most popular max pooling operation has been employed by all pooling layers.
- Flatten Layer: The output of the convolution layer has been converted using this layer into a 1D tensor.
- Fully connected layer or dense layer: With a simple vector as their input, these layers produce a vector as their output. There are two dense layers in this model. In the first dense layer, there are 1024 neurons. In the second dense, there are 512 neurons.
4.2. LightGBM Model
5. The Experimental Work and Results
5.1. Performance Measures
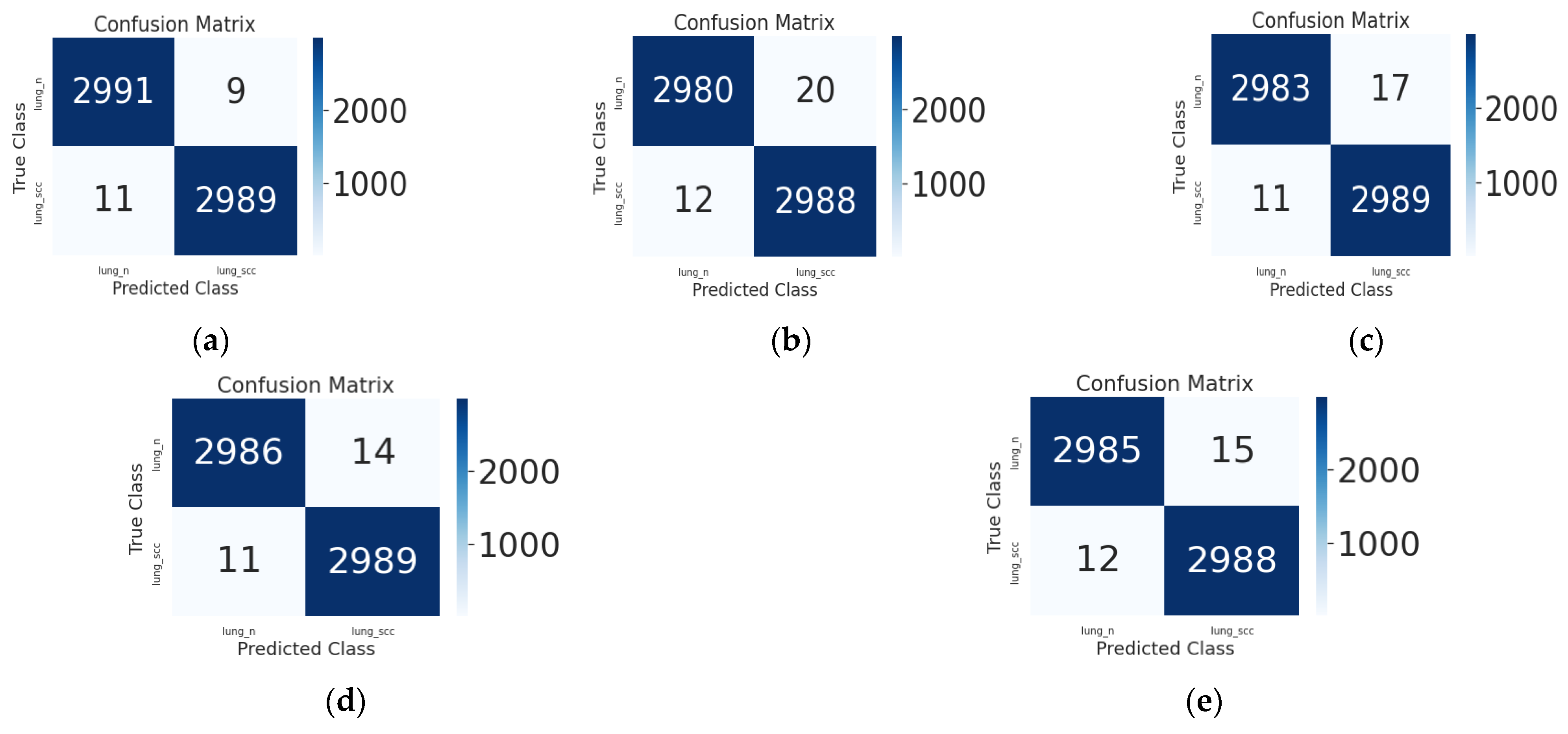
5.2. Quantitative Evaluation and Discussion
6. Conclusions and Future Work
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Araghi, M.; Soerjomataram, I.; Jenkins, M.; Brierley, J.; Morris, E.; Bray, F.; Arnold, M. Global Trends in Colorectal Cancer Mortality: Projections to the Year 2035. Int. J. Cancer 2019, 144, 2992–3000. [Google Scholar] [CrossRef] [PubMed]
- Walser, T.; Cui, X.; Yanagawa, J.; Lee, J.M.; Heinrich, E.; Lee, G.; Sharma, S.; Dubinett, S.M. Smoking and Lung Cancer: The Role of Inflammation. Proc. Am. Thorac. Soc. 2008, 5, 811–815. [Google Scholar] [CrossRef]
- Callaghan, R.C.; Allebeck, P.; Sidorchuk, A. Marijuana Use and Risk of Lung Cancer: A 40-Year Cohort Study. Cancer Causes Control 2013, 24, 1811–1820. [Google Scholar] [CrossRef] [PubMed]
- Squamous-Cell-Carcinoma-of-the-Lung. Available online: https://www.health.harvard.edu/cancer/squamous-cell-carcinoma-of-the-lung (accessed on 28 May 2023).
- Masud, M.; Sikder, N.; Nahid, A.-A.; Bairagi, A.K.; AlZain, M.A. A Machine Learning Approach to Diagnosing Lung and Colon Cancer Using a Deep Learning-Based Classification Framework. Sensors 2021, 21, 748. [Google Scholar] [CrossRef] [PubMed]
- Hamed, E.A.-R.; Salem, M.A.-M.; Badr, N.L.; Tolba, M.F. Lung Cancer Classification Model Using Convolution Neural Network. In Proceedings of the 3rd International Conference on Artificial Intelligence and Computer Vision (AICV2023), Marrakesh, Morocco, 5–7 March 2023; Springer: Berlin/Heidelberg, Germany, 2023; pp. 16–26. [Google Scholar]
- Albawi, S.; Mohammed, T.A.; Al-Zawi, S. Understanding of a Convolutional Neural Network. In Proceedings of the 2017 International Conference on Engineering and Technology (ICET), Antalya, Turkey, 21–23 August 2017; pp. 1–6. [Google Scholar]
- LeCun, Y.; Bengio, Y.; Hinton, G. Deep Learning. Nature 2015, 521, 436–444. [Google Scholar] [CrossRef]
- Jiang, X. Feature Extraction for Image Recognition and Computer Vision. In Proceedings of the 2009 2nd IEEE International Conference on Computer Science and Information Technology, Beijing, China, 8–11 August 2009; pp. 1–15. [Google Scholar]
- Chen, C.; Zhou, K.; Wang, Z.; Xiao, R. Generative Consistency for Semi-Supervised Cerebrovascular Segmentation from TOF-MRA. IEEE Trans. Med. Imaging 2022, 42, 346–353. [Google Scholar] [CrossRef]
- Wu, Q.; Chen, Y.; Huang, N.; Yue, X. Weakly-Supervised Cerebrovascular Segmentation Network with Shape Prior and Model Indicator. In Proceedings of the 2022 International Conference on Multimedia Retrieval, Newark, NJ, USA, 27–30 June 2022; pp. 668–676. [Google Scholar]
- Isensee, F.; Jaeger, P.F.; Kohl, S.A.A.; Petersen, J.; Maier-Hein, K.H. NnU-Net: A Self-Configuring Method for Deep Learning-Based Biomedical Image Segmentation. Nat. Methods 2021, 18, 203–211. [Google Scholar] [CrossRef]
- Talo, M.; Yildirim, O.; Baloglu, U.B.; Aydin, G.; Acharya, U.R. Convolutional Neural Networks for Multi-Class Brain Disease Detection Using MRI Images. Comput. Med. Imaging Graph. 2019, 78, 101673. [Google Scholar] [CrossRef]
- Celik, Y.; Talo, M.; Yildirim, O.; Karabatak, M.; Acharya, U.R. Automated Invasive Ductal Carcinoma Detection Based Using Deep Transfer Learning with Whole-Slide Images. Pattern Recognit. Lett. 2020, 133, 232–239. [Google Scholar] [CrossRef]
- Esteva, A.; Kuprel, B.; Novoa, R.A.; Ko, J.; Swetter, S.M.; Blau, H.M.; Thrun, S. Dermatologist-Level Classification of Skin Cancer with Deep Neural Networks. Nature 2017, 542, 115–118. [Google Scholar] [CrossRef] [PubMed]
- Yoon, S.H.; Lee, K.H.; Kim, J.Y.; Lee, Y.K.; Ko, H.; Kim, K.H.; Park, C.M.; Kim, Y.-H. Chest Radiographic and CT Findings of the 2019 Novel Coronavirus Disease (COVID-19): Analysis of Nine Patients Treated in Korea. Korean J. Radiol. 2020, 21, 494–500. [Google Scholar] [CrossRef] [PubMed]
- Rajpurkar, P.; Irvin, J.; Zhu, K.; Yang, B.; Mehta, H.; Duan, T.; Ding, D.; Bagul, A.; Langlotz, C.; Shpanskaya, K. Chexnet: Radiologist-Level Pneumonia Detection on Chest X-rays with Deep Learning. arXiv 2017, arXiv:1711.05225. [Google Scholar]
- Tan, J.H.; Fujita, H.; Sivaprasad, S.; Bhandary, S.V.; Rao, A.K.; Chua, K.C.; Acharya, U.R. Automated Segmentation of Exudates, Haemorrhages, Microaneurysms Using Single Convolutional Neural Network. Inf. Sci. 2017, 420, 66–76. [Google Scholar] [CrossRef]
- Gaál, G.; Maga, B.; Lukács, A. Attention U-Net Based Adversarial Architectures for Chest X-ray Lung Segmentation. arXiv 2020, arXiv:2003.10304. [Google Scholar]
- Yu, Y.; Li, J.; Li, J.; Xia, Y.; Ding, Z.; Samali, B. Automated Damage Diagnosis of Concrete Jack Arch Beam Using Optimized Deep Stacked Autoencoders and Multi-Sensor Fusion. Dev. Built Environ. 2023, 14, 100128. [Google Scholar] [CrossRef]
- Yu, Y.; Liang, S.; Samali, B.; Nguyen, T.N.; Zhai, C.; Li, J.; Xie, X. Torsional Capacity Evaluation of RC Beams Using an Improved Bird Swarm Algorithm Optimised 2D Convolutional Neural Network. Eng. Struct. 2022, 273, 115066. [Google Scholar] [CrossRef]
- Baranwal, N.; Doravari, P.; Kachhoria, R. Classification of Histopathology Images of Lung Cancer Using Convolutional Neural Network (CNN). In Disruptive Developments in Biomedical Applications; CRC Press: Boca Raton, FL, USA, 2022; pp. 75–89. ISBN 100327269X. [Google Scholar]
- Nishio, M.; Nishio, M.; Jimbo, N.; Nakane, K. Homology-Based Image Processing for Automatic Classification of Histopathological Images of Lung Tissue. Cancers 2021, 13, 1192. [Google Scholar] [CrossRef]
- Mangal, S.; Chaurasia, A.; Khajanchi, A. Convolution Neural Networks for Diagnosing Colon and Lung Cancer Histopathological Images. arXiv 2020, arXiv:2009.03878. [Google Scholar]
- Hossain, M. Early Stage Detection and Classification of Colon Cancer Using Deep Learning and Explainable AI on Histopathological Images. Ph.D. Thesis, Brac University, Dhaka, Bangladesh, 2022. [Google Scholar]
- Tasnim, Z.; Chakraborty, S.; Shamrat, F.; Chowdhury, A.N.; Nuha, H.A.; Karim, A.; Zahir, S.B.; Billah, M.M. Deep Learning Predictive Model for Colon Cancer Patient Using CNN-Based Classification. Int. J. Adv. Comput. Sci. Appl. 2021, 12, 687–696. [Google Scholar] [CrossRef]
- Hlavcheva, D.; Yaloveha, V.; Podorozhniak, A.; Kuchuk, H. Comparison of CNNs for Lung Biopsy Images Classification. In Proceedings of the 2021 IEEE 3rd Ukraine Conference on Electrical and Computer Engineering (UKRCON), Lviv, Ukraine, 26–28 August 2021; pp. 1–5. [Google Scholar]
- Garg, S.; Garg, S. Prediction of Lung and Colon Cancer through Analysis of Histopathological Images by Utilizing Pre-Trained CNN Models with Visualization of Class Activation and Saliency Maps. In Proceedings of the 2020 3rd Artificial Intelligence and Cloud Computing Conference, Kyoto, Japan, 18–20 December 2020; pp. 38–45. [Google Scholar]
- Hatuwal, B.K.; Thapa, H.C. Lung Cancer Detection Using Convolutional Neural Network on Histopathological Images. Int. J. Comput. Trends Technol. 2020, 68, 21–24. [Google Scholar] [CrossRef]
- Setiawan, W.; Suhadi, M.M.; Pramudita, Y.D. Histopathology of Lung Cancer Classification Using Convolutional Neural Network with Gamma Correction. Commun. Math. Biol. Neurosci. 2022, 2022, 81. [Google Scholar]
- Pradhan, M.; Sahu, R.K. Automatic Detection of Lung Cancer Using the Potential of Artificial Intelligence (AI). In Machine Learning and AI Techniques in Interactive Medical Image Analysis; IGI Global: Hershey, PA, USA, 2023; pp. 106–123. [Google Scholar]
- Tummala, S.; Kadry, S.; Nadeem, A.; Rauf, H.T.; Gul, N. An Explainable Classification Method Based on Complex Scaling in Histopathology Images for Lung and Colon Cancer. Diagnostics 2023, 13, 1594. [Google Scholar] [CrossRef] [PubMed]
- Borkowski, A.A.; Bui, M.M.; Thomas, L.B.; Wilson, C.P.; DeLand, L.A.; Mastorides, S.M. Lung and Colon Cancer Histopathological Image Dataset (Lc25000). arXiv 2019, arXiv:1912.12142. [Google Scholar]
- Bhinge, S.D.; Bhutkar, M.A.; Randive, D.S.; Wadkar, G.H.; Kamble, S.Y.; Kalel, P.D.; Kadam, S.S. Formulation and Evaluation of Polyherbal Gel Containing Extracts of Azadirachta Indica, Adhatoda Vasica, Piper Betle, Ocimum Tenuiflorum and Pongamia Pinnata. J. Res. Pharm. 2019, 23, 44–54. [Google Scholar] [CrossRef]
- Ke, G.; Meng, Q.; Finley, T.; Wang, T.; Chen, W.; Ma, W.; Ye, Q.; Liu, T.-Y. Lightgbm: A Highly Efficient Gradient Boosting Decision Tree. Adv. Neural Inf. Process Syst. 2017, 30, 3149–3157. [Google Scholar]
- Friedman, J.H. Greedy Function Approximation: A Gradient Boosting Machine. Ann. Stat. 2001, 29, 1189–1232. [Google Scholar] [CrossRef]
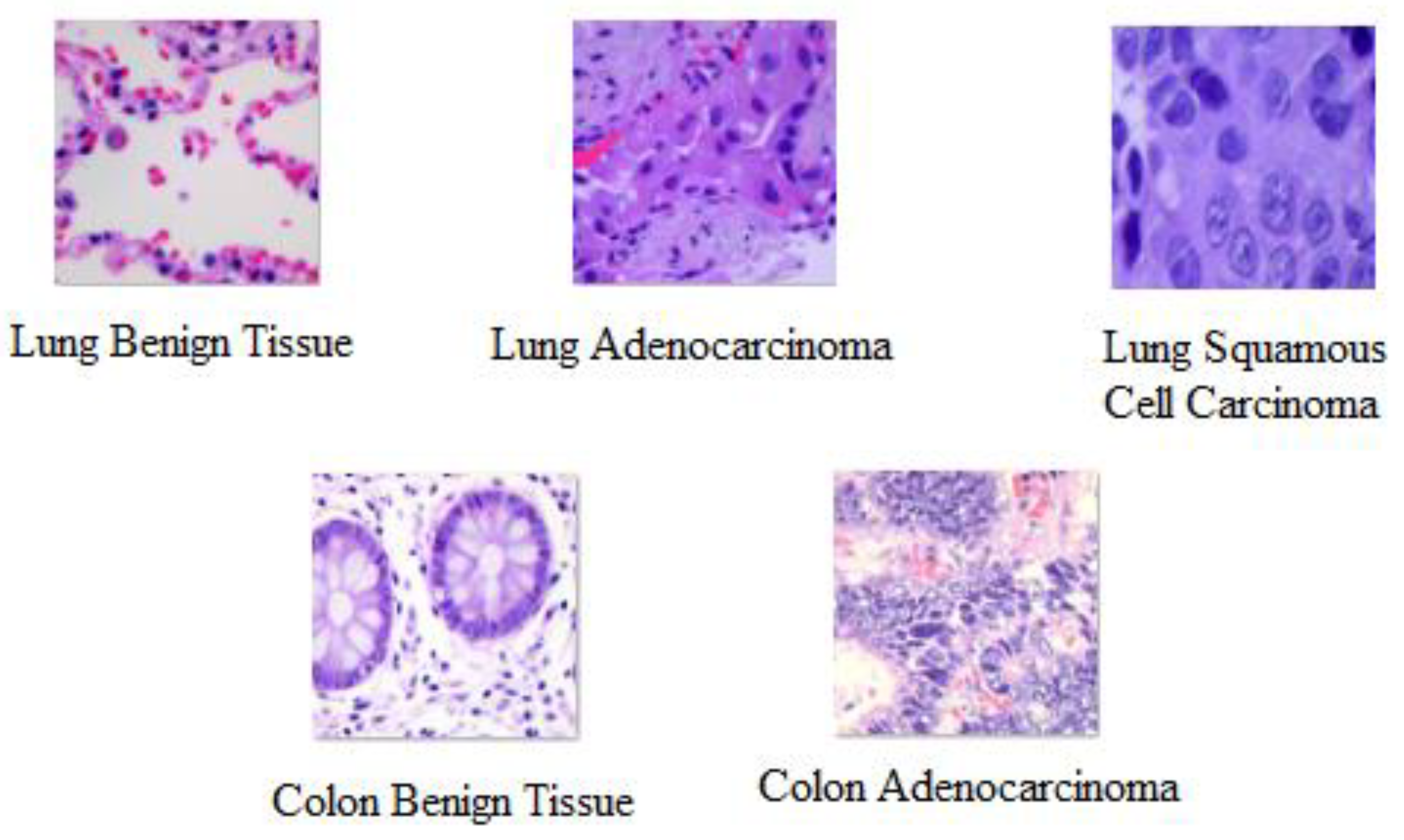


| Cancer Name | Cancer Type | Label Name | Number of Samples |
|---|---|---|---|
| Colon | Adenocarcinoma | Colon_aca | 5000 |
| Colon | Benign Tissue | Colon_n | 5000 |
| Lung | Adenocarcinoma | Lung_aca | 5000 |
| Lung | Benign Tissue | Lung_n | 5000 |
| Lung | Squamous Cell Carcinoma | Lung_scc | 5000 |
| Layer | #Filters/Neurons | Filter Size | Stride | #Nodes | Padding | Activation Function |
|---|---|---|---|---|---|---|
| Conv 1 | 32 | 11 × 11 | 4 × 4 | - | Valid | ReLU |
| Max Pool1 | - | 2 × 2 | - | - | - | - |
| Conv 2 | 64 | 3 × 3 | 1 × 1 | - | - | ReLU |
| Max Pool2 | - | 2 × 2 | - | - | - | - |
| Conv 3 | 128 | 3 × 3 | 1 × 1 | - | - | ReLU |
| Max Pool3 | - | 2 × 2 | - | - | - | - |
| Conv 4 | 256 | 3 × 3 | 1 × 1 | - | - | ReLU |
| Max Pool4 | - | 2 × 2 | - | - | - | - |
| FC 1 | - | - | - | 1024 | - | ReLU |
| FC 2 | - | - | - | 512 | - | ReLU |
| Dropout | Rate = 0.4 | - | - | - | - | - |
| The Type of Cancer | Training Dataset | Testing Dataset |
|---|---|---|
| Lung Benign Tissue | 2000 | 3000 |
| Lung Squamous Cell Carcinoma | 2000 | 3000 |
| Algorithm | Total Parameters | Feature Mapsize | Time of Feature Extraction | Time Consuming during Classification with ML Model | ||||
|---|---|---|---|---|---|---|---|---|
| SVM | RF | AdaBoost | XGBoost | LightGBM Multiple Threads | ||||
| VGG19 | 171 | 55 | 32 | 4 | 5 | 54 | 35 | 30 |
| VGG16 | 165 | 55 | 33 | 2 | 4 | 55 | 33 | 30 |
| AlexNet | 58 | 34 | 5 | 2 | 3 | 54 | 26 | 25 |
| InceptionResNetV2 | 54 | 115 | 29 | 635 | 2 | 6 | 3 | 2 |
| ResNet50 | 24 | 110 | 16 | 1 | 1 | 7 | 3 | 2 |
| Inception_v3 | 22 | 115 | 35 | 1 | 2 | 6 | 4 | 2 |
| GoogleNet | 5 | 55 | 9 | 1 | 4 | 76 | 23 | 16 |
| MobileNet | 3 | 75 | 7 | 7 | 9 | 15 | 13 | 10 |
| Proposed Model | 1 | 64 | 2 | 2 | 2 | 5 | 2 | 1 |
| Proposed CNN + ML | Accuracy | F1-Score | Sensitivity | Specificity | MCC |
|---|---|---|---|---|---|
| SVM | 99.6 | 99.6 | 99.6 | 99.7 | 99.3 |
| RF | 99.5 | 99.5 | 99.6 | 99.3 | 98.9 |
| AbaBoost | 99.5 | 99.5 | 99.6 | 99.4 | 99.1 |
| XGBoost | 99.6 | 99.6 | 99.6 | 99.5 | 99.1 |
| LightGBM multiple Threads | 99.6 | 99.6 | 99.6 | 99.5 | 99.1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hamed, E.A.-R.; Salem, M.A.-M.; Badr, N.L.; Tolba, M.F. An Efficient Combination of Convolutional Neural Network and LightGBM Algorithm for Lung Cancer Histopathology Classification. Diagnostics 2023, 13, 2469. https://doi.org/10.3390/diagnostics13152469
Hamed EA-R, Salem MA-M, Badr NL, Tolba MF. An Efficient Combination of Convolutional Neural Network and LightGBM Algorithm for Lung Cancer Histopathology Classification. Diagnostics. 2023; 13(15):2469. https://doi.org/10.3390/diagnostics13152469
Chicago/Turabian StyleHamed, Esraa A.-R., Mohammed A.-M. Salem, Nagwa L. Badr, and Mohamed F. Tolba. 2023. "An Efficient Combination of Convolutional Neural Network and LightGBM Algorithm for Lung Cancer Histopathology Classification" Diagnostics 13, no. 15: 2469. https://doi.org/10.3390/diagnostics13152469
APA StyleHamed, E. A.-R., Salem, M. A.-M., Badr, N. L., & Tolba, M. F. (2023). An Efficient Combination of Convolutional Neural Network and LightGBM Algorithm for Lung Cancer Histopathology Classification. Diagnostics, 13(15), 2469. https://doi.org/10.3390/diagnostics13152469






