Landmark-Assisted Anatomy-Sensitive Retinal Vessel Segmentation Network
Abstract
1. Introduction
- TransUNet is more in line with anatomical retinal vessel segmentation due to its special structure. We use transformers as the segmentation backbone to benefit from the advantages of convolutional layers in extracting local features and multi-head self-attention in modeling global relations. Meanwhile, we reform the skip connections in TransUNet to decode deep semantics more easily and accurately.
- A self-supervised landmark-assisted segmentation framework is proposed to further improve the accuracy of retinal vessel segmentation. In particular, we propose a strategy for contrastive learning to improve the plausibility and accuracy of landmark representations of anatomical topology. We utilize landmarks that sparsely represent retinal vessel morphology to guide the model towards learning the content, rather than the style that is not conducive to segmentation. Furthermore, landmarks enhance the richness of explicit descriptions of retinal vascular anatomy, which is friendly for the model to learn based on fewer samples.
- We implement the proposed network on the DRIVE, CHASE-DB1, and STARE datasets, and extensive experimental results show that our method achieves state-of-the-art performance in most cases.
2. Related Work
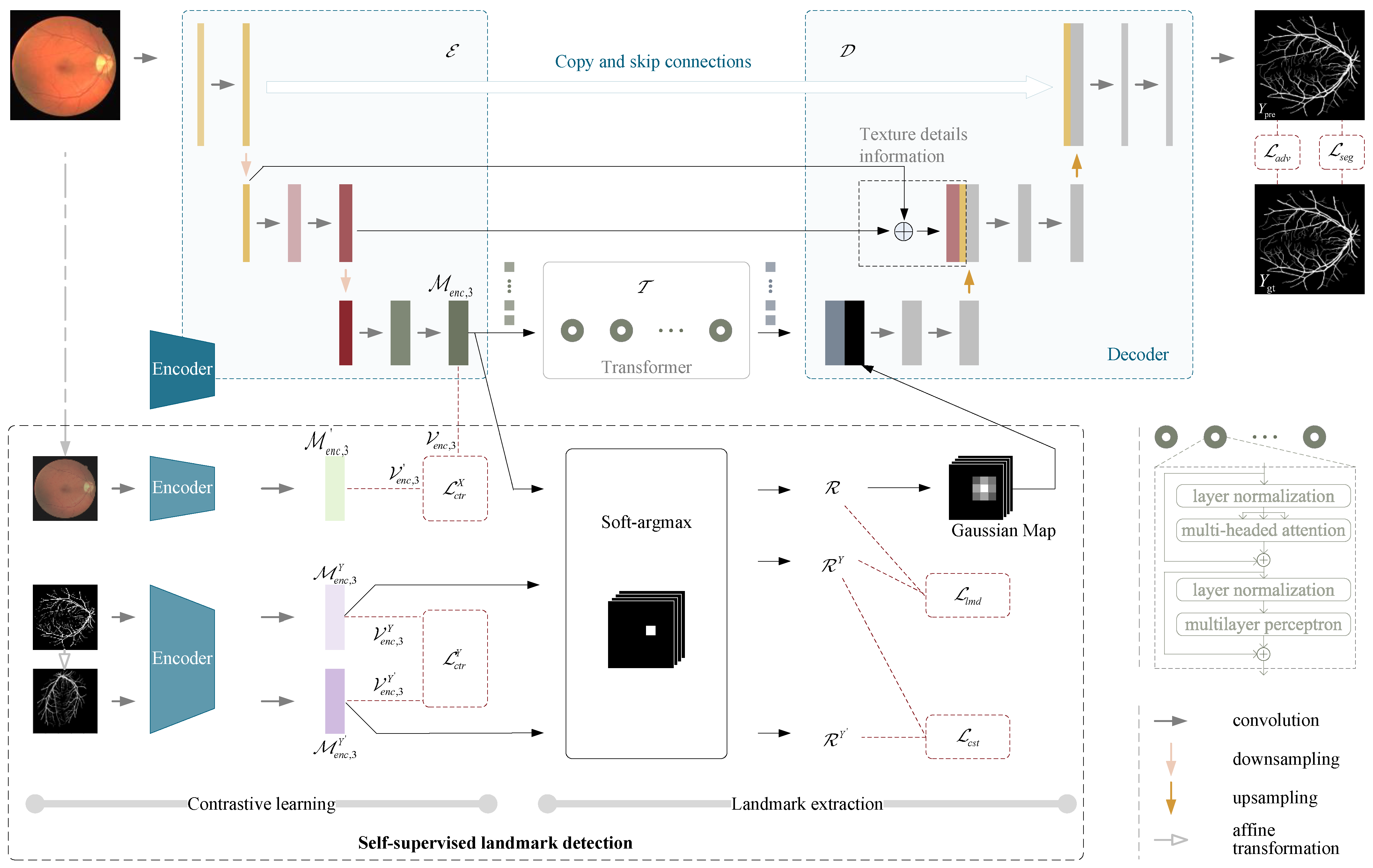
3. Methods and Materials
3.1. Datasets
3.2. TransUNet
3.3. Convolutional Encoder
3.4. Transformer Module
3.5. Convolutional Decoder
3.6. Self-Supervised Landmark Detection
Coordinate Extraction for Landmarks
3.7. Landmark Auxiliary Guided Segmentation
3.8. Implementation Details
4. Results and Discussions
4.1. Evaluation Metrics
4.2. Comparison with the State-of-the-Art Methods
4.2.1. Quantitative Analysis
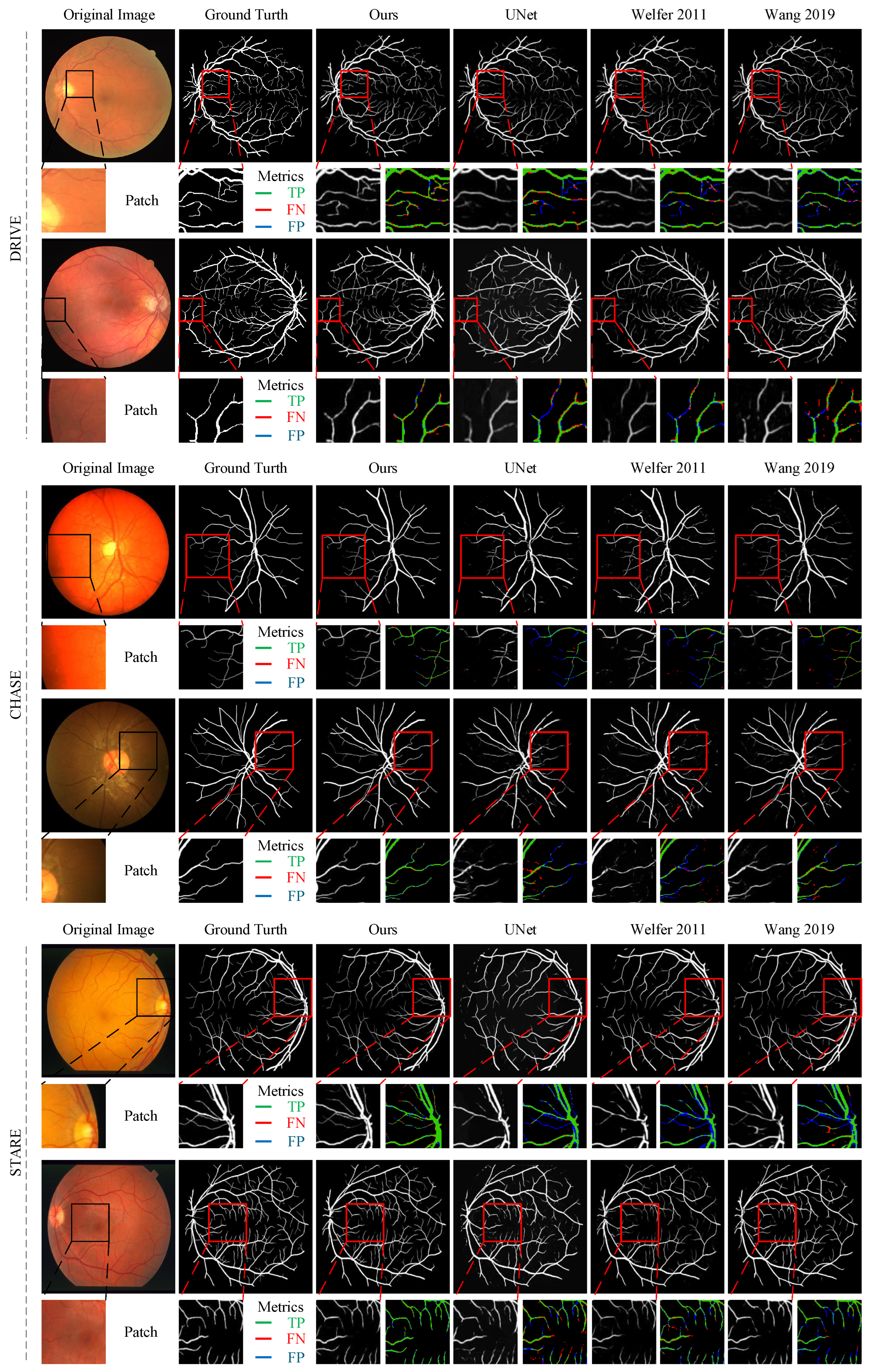
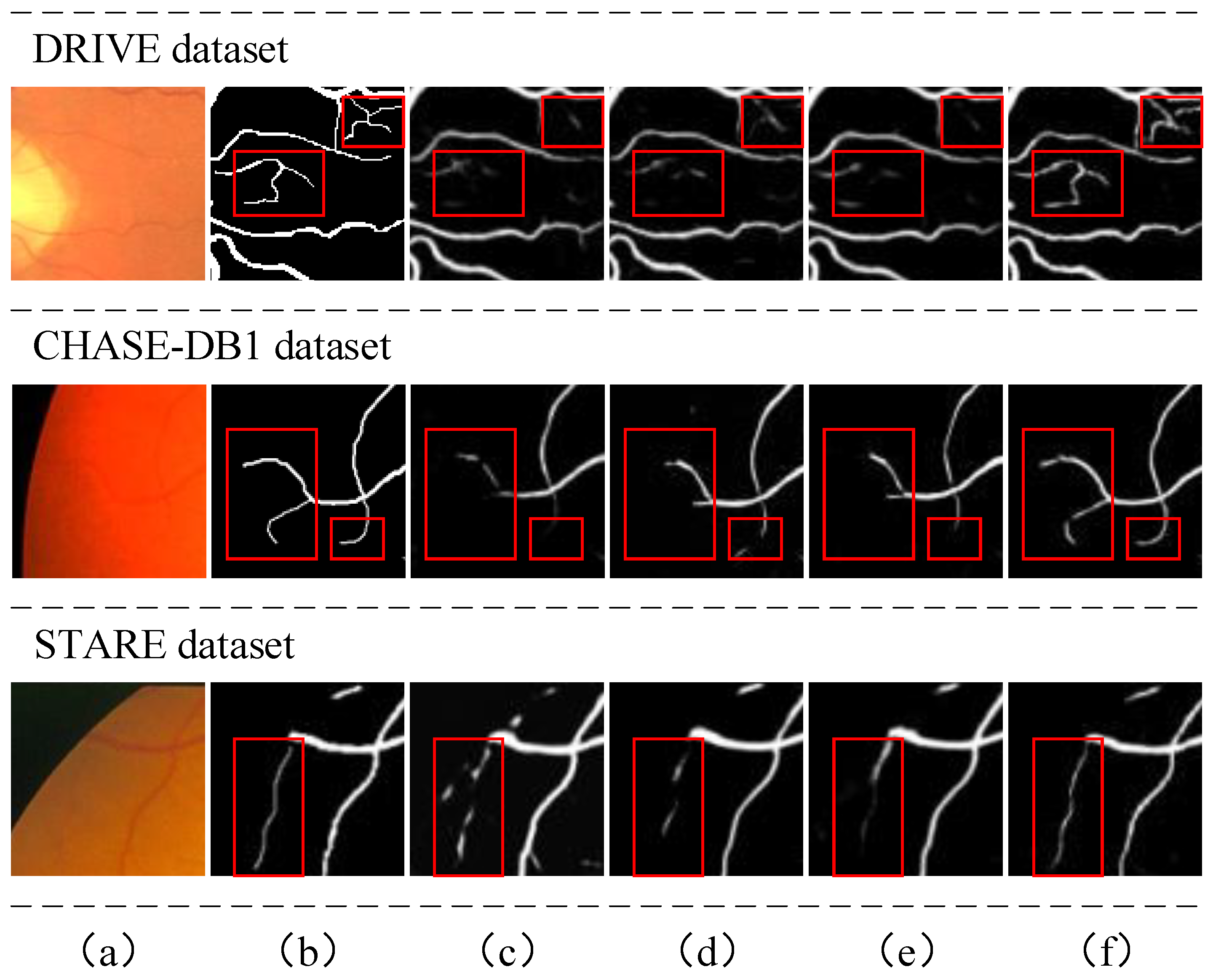
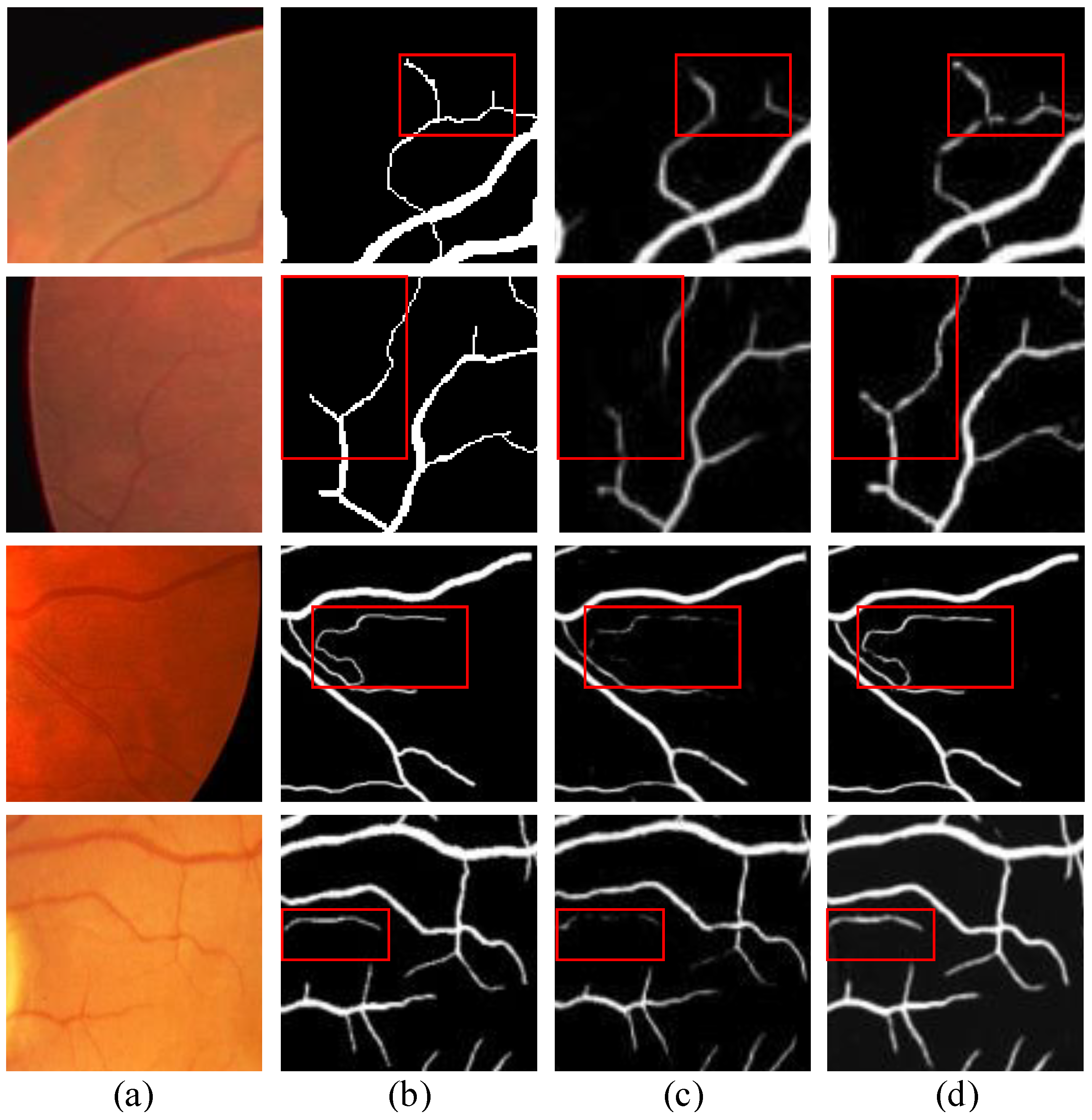
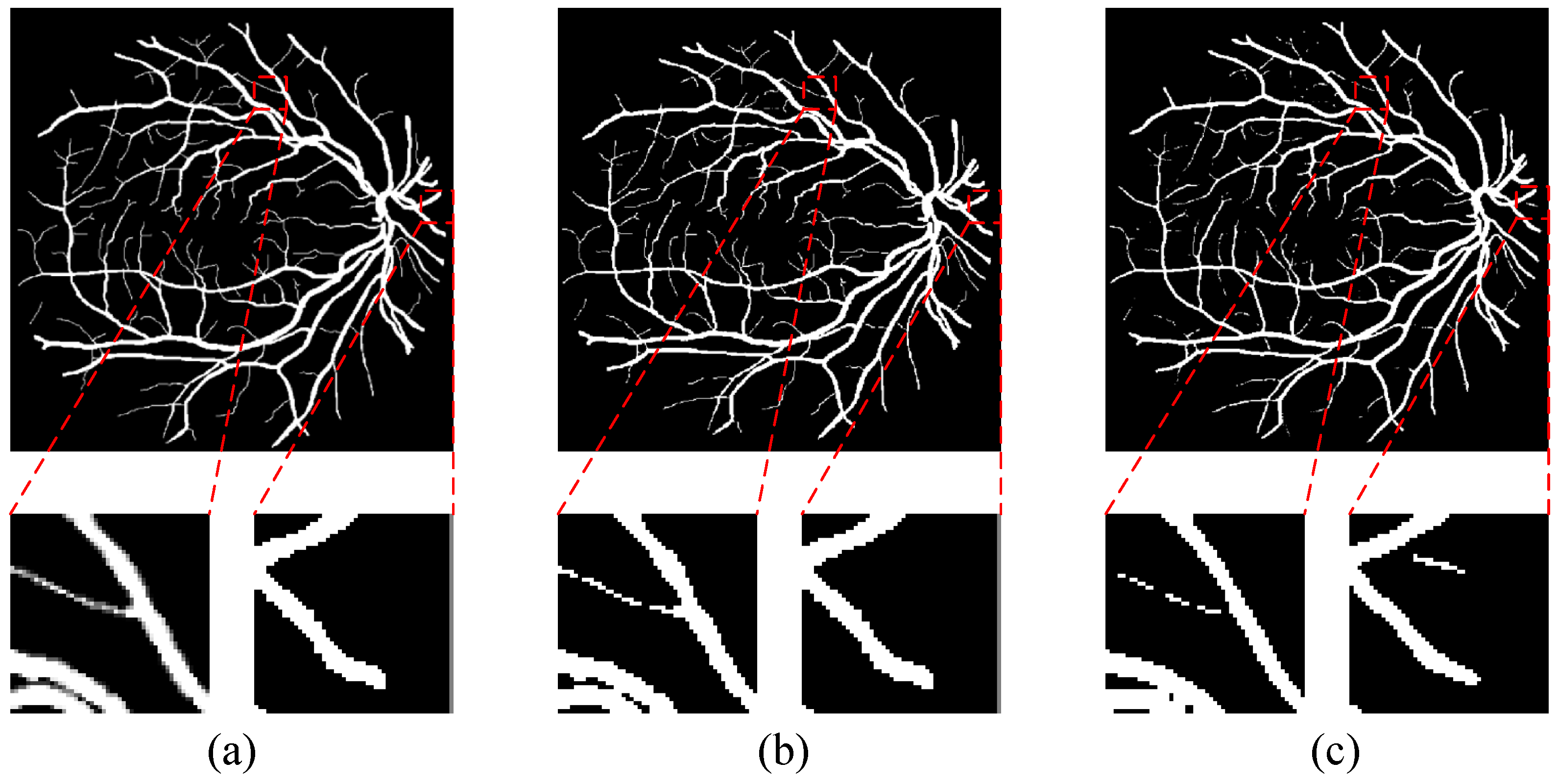
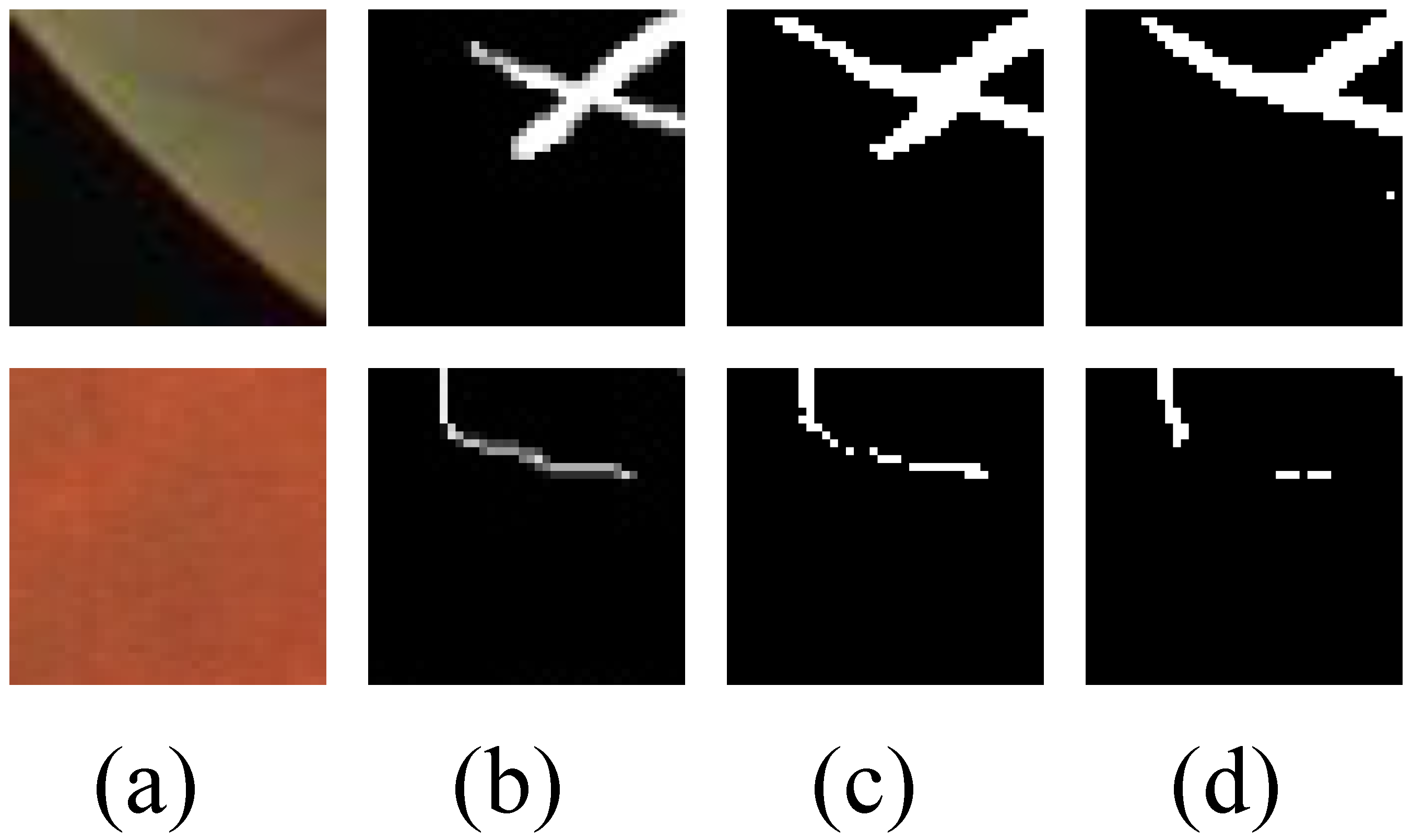
4.2.2. Qualitative Analysis
4.3. Ablation Experiments
4.3.1. Effect of TransUNet
4.3.2. Effect of Self-Supervised Landmark Detection
4.4. Effect of Image Size
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Grélard, F.; Baldacci, F.; Vialard, A.; Domenger, J.P. New methods for the geometrical analysis of tubular organs. Med. Image Anal. 2017, 42, 89–101. [Google Scholar] [CrossRef] [PubMed]
- Saroj, S.K.; Kumar, R.; Singh, N.P. Fréchet PDF based Matched Filter Approach for Retinal Blood Vessels Segmentation. Comput. Methods Programs Biomed. 2020, 194, 105490. [Google Scholar] [CrossRef] [PubMed]
- Mapayi, T.; Owolawi, P.A. Automatic Retinal Vascular Network Detection using Multi-Thresholding Approach based on Otsu. In Proceedings of the 2019 International Multidisciplinary Information Technology and Engineering Conference (IMITEC), Vanderbijlpark, South Africa, 21–22 November 2019; pp. 1–5. [Google Scholar]
- Welfer, D.; Scharcanski, J.; Marinho, D.R. Fovea center detection based on the retina anatomy and mathematical morphology. Comput. Methods Programs Biomed. 2011, 104, 397–409. [Google Scholar] [CrossRef] [PubMed]
- Jin, Q.; Meng, Z.; Pham, T.D.; Chen, Q.; Wei, L.; Su, R. DUNet: A deformable network for retinal vessel segmentation. Knowl. Based Syst. 2019, 178, 149–162. [Google Scholar] [CrossRef]
- Wu, C.; Zou, Y.; Yang, Z. U-GAN: Generative Adversarial Networks with U-Net for Retinal Vessel Segmentation. In Proceedings of the 2019 14th International Conference on Computer Science & Education (ICCSE), Toronto, ON, USA, 19–21 August 2019; pp. 642–646. [Google Scholar]
- Ronneberger, O.; Fischer, P.; Brox, T. U-Net: Convolutional Networks for Biomedical Image Segmentation; Springer: Cham, Switzerland, 2015. [Google Scholar]
- Wang, B.; Wang, S.; Qiu, S.; Wei, W.; Wang, H.; He, H. CSU-Net: A Context Spatial U-Net for Accurate Blood Vessel Segmentation in Fundus Images. IEEE J. Biomed. Health Inform. 2021, 25, 1128–1138. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Verma, M.; Nakashima, Y.; Nagahara, H.; Kawasaki, R. IterNet: Retinal Image Segmentation Utilizing Structural Redundancy in Vessel Networks. In Proceedings of the 2020 IEEE Winter Conference on Applications of Computer Vision (WACV), Snowmass Village, CO, USA, 1–5 March 2020; pp. 3645–3654. [Google Scholar]
- Cao, H.; Wang, Y.; Chen, J.; Jiang, D.; Zhang, X.; Tian, Q.; Wang, M. Swin-Unet: Unet-like Pure Transformer for Medical Image Segmentation. In Computer Vision—ECCV 2022 Workshops; Springer: Cham, Switzerland, 2023. [Google Scholar]
- Xia, X.; Huang, Z.; Huang, Z.; Shu, L.; Li, L. A CNN-Transformer Hybrid Network for Joint Optic Cup and Optic Disc Segmentation in Fundus Images. In Proceedings of the 2022 International Conference on Computer Engineering and Artificial Intelligence (ICCEAI), Shijiazhuang, China, 22–24 July 2022; pp. 482–486. [Google Scholar]
- Chen, J.; Lu, Y.; Yu, Q.; Luo, X.; Adeli, E.; Wang, Y.; Lu, L.; Yuille, A.L.; Zhou, Y. TransUNet: Transformers Make Strong Encoders for Medical Image Segmentation. arXiv 2021, arXiv:2102.04306. [Google Scholar]
- Sun, Y.; Wang, X.; Tang, X. Deep Convolutional Network Cascade for Facial Point Detection. In Proceedings of the 2013 IEEE Conference on Computer Vision and Pattern Recognition, Portland, OR, USA, 23–28 June 2013; pp. 3476–3483. [Google Scholar]
- Zhang, K.; Zhang, Z.; Li, Z.; Qiao, Y. Joint Face Detection and Alignment Using Multitask Cascaded Convolutional Networks. IEEE Signal Process. Lett. 2016, 23, 1499–1503. [Google Scholar] [CrossRef]
- Kowalski, M.; Naruniec, J.; Trzcinski, T. Deep Alignment Network: A Convolutional Neural Network for Robust Face Alignment. In Proceedings of the 2017 IEEE Conference on Computer Vision and Pattern Recognition Workshops (CVPRW), Honolulu, HI, USA, 21–26 July 2017; pp. 2034–2043. [Google Scholar]
- Shi, H.; Wang, Z. Improved Stacked Hourglass Network with Offset Learning for Robust Facial Landmark Detection. In Proceedings of the 2019 9th International Conference on Information Science and Technology (ICIST), Kopaonik, Serbia, 10–13 March 2019; pp. 58–64. [Google Scholar]
- Siarohin, A.; Lathuiliere, S.; Tulyakov, S.; Ricci, E.; Sebe, N. Animating arbitrary objects via deep motion transfer. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), Long Beach, CA, USA, 15–20 June 2019; pp. 2377–2386. [Google Scholar]
- Jiang, Y.; Tan, N.; Peng, T.; Zhang, H. Retinal Vessels Segmentation Based on Dilated Multi-Scale Convolutional Neural Network. IEEE Access 2019, 7, 76342–76352. [Google Scholar] [CrossRef]
- Orlando, J.I.; Prokofyeva, E.; Blaschko, M.B. A Discriminatively Trained Fully Connected Conditional Random Field Model for Blood Vessel Segmentation in Fundus Images. IEEE Trans. Biomed. Eng. 2017, 64, 16–27. [Google Scholar] [CrossRef]
- Zhang, J.; Chen, Y.; Bekkers, E.; Wang, M.; Dashtbozorg, B.; ter Haar Romeny, B.M. Retinal vessel delineation using a brain-inspired wavelet transform and random forest. Pattern Recognit. 2017, 69, 107–123. [Google Scholar] [CrossRef]
- Srinidhi, C.L.; Aparna, P.; Rajan, J. A visual attention guided unsupervised feature learning for robust vessel delineation in retinal images. Biomed. Signal Process. Control. 2018, 44, 110–126. [Google Scholar] [CrossRef]
- Yan, Z.; Yang, X.; Cheng, K.-T. Joint Segment-Level and Pixel-Wise Losses for Deep Learning Based Retinal Vessel Segmentation. IEEE Trans. Biomed. Eng. 2018, 65, 1912–1923. [Google Scholar] [CrossRef] [PubMed]
- Xu, R.; Jiang, G.; Ye, X.; Chen, Y. Retinal vessel segmentation via multiscaled deep-guidance. In Pacific Rim Conference on Multimedia; Springer: Berlin, Germany, 2018; pp. 158–168. [Google Scholar]
- Zhuang, J. LadderNet: Multi-Path Networks Based on U-Net for Medical Image Segmentation. arXiv 2018, arXiv:1810.07810. [Google Scholar]
- Alom, M.Z.; Yakopcic, C.; Hasan, M.; Taha, T.M.; Asari, V.K. Recurrent residual U-Net for medical image segmentation. J. Med. Imaging 2019, 6, 14006. [Google Scholar] [CrossRef] [PubMed]
- Guo, S.; Wang, K.; Kang, H.; Zhang, Y.; Gao, Y.; Li, T. BTS-DSN: Deeply Supervised Neural Network with Short Connections for Retinal Vessel Segmentation. Int. J. Med. Inform. 2018, 126, 105–113. [Google Scholar] [CrossRef]
- Wang, B.; Qiu, S.; He, H. Dual encoding U-Net for retinal vessel segmentation, Medical Image Computing and Computer Assisted Intervention. Med. Image Comput. Comput. Assist. Interv. 2019, 22, 84–92. [Google Scholar]
- Zhou, Z.; Siddiquee, M.M.R.; Tajbakhsh, N.; Liang, J. UNet++: Redesigning Skip Connections to Exploit Multiscale Features in Image Segmentation. IEEE Trans. Med. Imaging 2020, 39, 1856–1867. [Google Scholar] [CrossRef]
- Xu, R.; Ye, X.; Jiang, G.; Liu, T.; Tanaka, S. Retinal Vessel Segmentation via a Semantics and Multi-Scale Aggregation Network. In Proceedings of the ICASSP 2020-2020 IEEE International Conference on Acoustics, Speech and Signal Processing (ICASSP), Barcelona, Spain, 4–8 May 2020. [Google Scholar]
- Wang, D.; Haytham, A.; Pottenburgh, J.; Saeedi, O.; Tao, Y. Hard attention net for automatic retinal vessel segmentation. IEEE J. Biomed. Health Inform. 2020, 24, 3384–3396. [Google Scholar] [CrossRef]
- Mou, L.; Zhao, Y.; Fu, H.; Liu, Y.; Cheng, J.; Zheng, Y.; Su, P.; Yang, J.; Chen, L.; Frangi, A.F.; et al. CS2-Net: Deep learning segmentation of curvilinear structures in medical imaging. Med. Image Anal. 2021, 67, 101874. [Google Scholar] [CrossRef]
- Zhang, Y.; He, M.; Chen, Z.; Hu, K.; Li, X.; Gao, X. Bridge-Net: Context-involved U-Net with patch-based loss weight mapping for retinal blood vessel segmentation. Exp. Syst. Appl. 2022, 195, 116526. [Google Scholar] [CrossRef]
- Liu, Y.; Shen, J.; Yang, L.; Bian, G.; Yu, H. ResDO-UNet: A deep residual network for accurate retinal vessel segmentation from fundus images. Biomed. Signal Process. Control. 2023, 79, 104087. [Google Scholar] [CrossRef]
- Rukundo, O. Effects of image size on deep learning. Electronics 2023, 12, 985. [Google Scholar] [CrossRef]

| DRIVE | CHASE-DB1 | STARE | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Method | Year | Acc | Se | Sp | F1 | Acc | Se | Sp | F1 | Acc | Se | Sp | F1 |
| U-Net [7] | 2015 | 0.9536 | 0.7653 | 0.9811 | 0.8078 | 0.9604 | 0.7870 | 0.9777 | 0.7828 | 0.9588 | 0.7639 | 0.9796 | 0.7817 |
| Orlando et al. [19] | 2017 | 0.9454 | 0.7897 | 0.9684 | 0.7857 | 0.9467 | 0.7565 | 0.9655 | 0.7332 | 0.9519 | 0.7680 | 0.9738 | 0.7644 |
| Zhang et al. [20] | 2017 | 0.9466 | 0.7861 | 0.9712 | 0.7953 | 0.9502 | 0.7644 | 0.9716 | 0.7581 | 0.9547 | 0.7882 | 0.9729 | 0.7815 |
| Srinidhi et al. [21] | 2018 | 0.9589 | 0.8644 | 0.9667 | 0.7607 | 0.9474 | 0.8297 | 0.9663 | 0.7189 | 0.9502 | 0.8325 | 0.9746 | 0.7698 |
| Yan et al. [22] | 2018 | 0.9542 | 0.7653 | 0.9818 | - | 0.9610 | 0.7633 | 0.9809 | - | 0.9612 | 0.7581 | 0.9846 | - |
| Xu et al. [23] | 2018 | 0.9557 | 0.8026 | 0.9780 | 0.8189 | 0.9613 | 0.7899 | 0.9785 | 0.7856 | 0.9499 | 0.8196 | 0.9661 | 0.7982 |
| Zhuang et al. [24] | 2018 | 0.9561 | 0.7856 | 0.9810 | 0.8202 | 0.9536 | 0.7978 | 0.9818 | 0.8031 | - | - | - | - |
| Alom et al. [25] | 2019 | 0.9556 | 0.7792 | 0.9813 | 0.8171 | 0.9634 | 0.7756 | 0.9820 | 0.7928 | 0.9712 | 0.8292 | 0.9862 | 0.8475 |
| Jin et al. [5] | 2019 | 0.9566 | 0.7963 | 0.9800 | 0.8237 | 0.9610 | 0.8155 | 0.9752 | 0.7883 | 0.9641 | 0.7595 | 0.9878 | 0.8143 |
| Jiang et al. [18] | 2019 | 0.9709 | 0.7839 | 0.9890 | 0.8246 | 0.9721 | 0.7839 | 0.9894 | 0.8062 | 0.9781 | 0.8249 | 0.9904 | 0.8482 |
| Guo et al. [26] | 2019 | 0.9561 | 0.7891 | 0.9804 | 0.8249 | 0.9627 | 0.7888 | 0.9801 | 0.7983 | - | - | - | - |
| Wang et al. [27] | 2019 | 0.9567 | 0.7940 | 0.9816 | 0.8270 | 0.9661 | 0.8074 | 0.9821 | 0.8037 | - | - | - | - |
| Zhou et al. [28] | 2020 | 0.9535 | 0.7473 | 0.9835 | 0.8035 | 0.9506 | 0.6361 | 0.9894 | 0.7390 | 0.9605 | 0.7776 | 0.9832 | 0.8132 |
| Xu et al. [29] | 2020 | 0.9557 | 0.7953 | 0.9807 | 0.8252 | 0.9650 | 0.8455 | 0.9769 | 0.8138 | 0.9590 | 0.8378 | 0.9741 | 0.8308 |
| Wang et al. [30] | 2020 | 0.9581 | 0.7991 | 0.9813 | 0.8293 | 0.9670 | 0.8329 | 0.9813 | 0.8191 | 0.9673 | 0.8186 | 0.9844 | - |
| Li et al. [9] | 2020 | 0.9573 | 0.7735 | 0.9838 | 0.8205 | 0.9760 | 0.7969 | 0.9881 | 0.8072 | 0.9701 | 0.7715 | 0.9886 | 0.8146 |
| Mou et al. [31] | 2021 | 0.9553 | 0.8154 | 0.9757 | 0.8228 | 0.9651 | 0.8329 | 0.9784 | 0.8141 | 0.9670 | 0.8396 | 0.9813 | 0.8420 |
| Zhang et al. [32] | 2022 | 0.9565 | 0.785 | 0.9618 | 0.82 | - | - | - | - | 0.9668 | 0.8002 | 0.9864 | 0.8289 |
| Liu et al. [33] | 2023 | 0.9561 | 0.7985 | 0.9791 | 0.8229 | 0.9672 | 0.8020 | 0.9794 | 0.8236 | 0.9635 | 0.8039 | 0.9836 | 0.8315 |
| Proposed | 2023 | 0.9577 | 0.8147 | 0.9862 | 0.8329 | 0.9754 | 0.8110 | 0.9881 | 0.8222 | 0.9635 | 0.8518 | 0.9829 | 0.8450 |
| DRIVE | CHASE-DB1 | STARE | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Method | Acc | Se | Sp | F1 | Acc | Se | Sp | F1 | Acc | Se | Sp | F1 |
| U-Net | 0.9536 | 0.7653 | 0.9811 | 0.8078 | 0.9604 | 0.7870 | 0.9777 | 0.7828 | 0.9588 | 0.7639 | 0.9796 | 0.7817 |
| TransUNet | 0.9543 | 0.7874 | 0.9860 | 0.8148 | 0.9681 | 0.7994 | 0.9878 | 0.8079 | 0.9610 | 0.7670 | 0.9879 | 0.8057 |
| TransUNet + SLD | 0.9577 | 0.8147 | 0.9862 | 0.8329 | 0.9754 | 0.8110 | 0.9881 | 0.8222 | 0.9635 | 0.8518 | 0.9829 | 0.8450 |
| DRIVE | CHASE-DB1 | STARE | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Size | Acc | Se | Sp | F1 | Acc | Se | Sp | F1 | Acc | Se | Sp | F1 |
| 512 × 512 | 0.9577 | 0.8147 | 0.9862 | 0.8329 | 0.9754 | 0.8110 | 0.9881 | 0.8222 | 0.9635 | 0.8518 | 0.9829 | 0.8450 |
| 256 × 256 | 0.9688 | 0.8188 | 0.9869 | 0.8455 | 0.9685 | 0.8155 | 0.9889 | 0.8243 | 0.9641 | 0.8188 | 0.9888 | 0.8466 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, H.; Qiu, Y.; Song, C.; Li, J. Landmark-Assisted Anatomy-Sensitive Retinal Vessel Segmentation Network. Diagnostics 2023, 13, 2260. https://doi.org/10.3390/diagnostics13132260
Zhang H, Qiu Y, Song C, Li J. Landmark-Assisted Anatomy-Sensitive Retinal Vessel Segmentation Network. Diagnostics. 2023; 13(13):2260. https://doi.org/10.3390/diagnostics13132260
Chicago/Turabian StyleZhang, Haifeng, Yunlong Qiu, Chonghui Song, and Jiale Li. 2023. "Landmark-Assisted Anatomy-Sensitive Retinal Vessel Segmentation Network" Diagnostics 13, no. 13: 2260. https://doi.org/10.3390/diagnostics13132260
APA StyleZhang, H., Qiu, Y., Song, C., & Li, J. (2023). Landmark-Assisted Anatomy-Sensitive Retinal Vessel Segmentation Network. Diagnostics, 13(13), 2260. https://doi.org/10.3390/diagnostics13132260





