Analysis of Line and Tube Detection Performance of a Chest X-ray Deep Learning Model to Evaluate Hidden Stratification
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethics Approval
2.2. AI Model
2.3. Study Data
2.4. Ground Truth Labelling
2.5. Analysis
2.5.1. Primary Outcome
2.5.2. Secondary Outcome
3. Results
3.1. Primary Outcome
3.2. Secondary Outcome
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Cao, W.; Wang, Q.; Yu, K. Malposition of a Nasogastric Feeding Tube into the Right Pleural Space of a Poststroke Patient. Radiol. Case Rep. 2020, 15, 1988–1991. [Google Scholar] [CrossRef] [PubMed]
- Gimenes, F.R.E.; Pereira, M.C.A.; do Prado, P.R.; de Carvalho, R.E.F.L.; Koepp, J.; de Freitas, L.M.; Teixeira, T.C.A.; Miasso, A.I. Nasogastric/Nasoenteric Tube-Related Incidents in Hospitalised Patients: A Study Protocol of a Multicentre Prospective Cohort Study. BMJ Open 2019, 9, e027967. [Google Scholar] [CrossRef] [PubMed]
- Motta, A.P.G.; Rigobello, M.C.G.; Silveira, R.C.d.C.P.; Gimenes, F.R.E. Nasogastric/Nasoenteric Tube-Related Adverse Events: An Integrative Review. Rev. Lat. Am. Enferm. 2021, 29, e3400. [Google Scholar] [CrossRef] [PubMed]
- Lamont, T.; Beaumont, C.; Fayaz, A.; Healey, F.; Huehns, T.; Law, R.; Lecko, C.; Panesar, S.; Surkitt-Parr, M.; Stroud, M.; et al. Checking Placement of Nasogastric Feeding Tubes in Adults (Interpretation of × Ray Images): Summary of a Safety Report from the National Patient Safety Agency. BMJ 2011, 342, d2586. [Google Scholar] [CrossRef]
- Australian Sentinel Events List. Available online: https://www.safetyandquality.gov.au/our-work/indicators/australian-sentinel-events-list (accessed on 20 September 2021).
- Al-Qahtani, A.S.; Messahel, F.M.; Ouda, W.O.A. Inadvertent Endobronchial Intubation: A Sentinel Event. Saudi J. Anaesth. 2012, 6, 259–262. [Google Scholar] [CrossRef]
- Valentin, A.; Capuzzo, M.; Guidet, B.; Moreno, R.P.; Dolanski, L.; Bauer, P.; Metnitz, P.G.H.; Research Group on Quality Improvement of European Society of Intensive Care Medicine; Sentinel Events Evaluation Study Investigators. Patient Safety in Intensive Care: Results from the Multinational Sentinel Events Evaluation (SEE) Study. Intensive Care Med. 2006, 32, 1591–1598. [Google Scholar] [CrossRef]
- Amorosa, J.K.; Bramwit, M.P.; Mohammed, T.-L.H.; Reddy, G.P.; Brown, K.; Dyer, D.S.; Ginsburg, M.E.; Heitkamp, D.E.; Jeudy, J.; Kirsch, J.; et al. ACR Appropriateness Criteria Routine Chest Radiographs in Intensive Care Unit Patients. J. Am. Coll. Radiol. 2013, 10, 170–174. [Google Scholar] [CrossRef]
- Expert Panel on Thoracic Imaging; McComb, B.L.; Chung, J.H.; Crabtree, T.D.; Heitkamp, D.E.; Iannettoni, M.D.; Jokerst, C.; Saleh, A.G.; Shah, R.D.; Steiner, R.M.; et al. ACR Appropriateness Criteria® Routine Chest Radiography. J. Thorac. Imaging 2016, 31, W13-5. [Google Scholar] [CrossRef]
- Sharma, A.; Sharma, A.; Aryal, D. Is Chest X-ray in Supine Position for Central Lines Confirmation a Gold Standard or a Fallacy? J. Emerg. Crit. Care Med. 2018, 2, 33. [Google Scholar] [CrossRef]
- Ahmad, H.K.; Milne, M.R.; Buchlak, Q.D.; Ektas, N.; Sanderson, G.; Chamtie, H.; Karunasena, S.; Chiang, J.; Holt, X.; Tang, C.H.M.; et al. Machine Learning Augmented Interpretation of Chest X-rays: A Systematic Review. Diagnostics 2023, 13, 743. [Google Scholar] [CrossRef]
- Buchlak, Q.D.; Esmaili, N.; Leveque, J.-C.; Farrokhi, F.; Bennett, C.; Piccardi, M.; Sethi, R.K. Machine Learning Applications to Clinical Decision Support in Neurosurgery: An Artificial Intelligence Augmented Systematic Review. Neurosurg. Rev. 2020, 43, 1235–1253. [Google Scholar] [CrossRef]
- Rajkomar, A.; Dean, J.; Kohane, I. Machine Learning in Medicine. N. Engl. J. Med. 2019, 380, 1347–1358. [Google Scholar] [CrossRef]
- Deo, R.C. Machine Learning in Medicine. Circulation 2015, 132, 1920–1930. [Google Scholar] [CrossRef]
- Ching, T.; Himmelstein, D.S.; Beaulieu-Jones, B.K.; Kalinin, A.A.; Do, B.T.; Way, G.P.; Ferrero, E.; Agapow, P.-M.; Zietz, M.; Hoffman, M.M.; et al. Opportunities and Obstacles for Deep Learning in Biology and Medicine. J. R. Soc. Interface 2018, 15, 20170387. [Google Scholar] [CrossRef]
- Seah, J.C.Y.; Tang, C.H.M.; Buchlak, Q.D.; Holt, X.G.; Wardman, J.B.; Aimoldin, A.; Esmaili, N.; Ahmad, H.; Pham, H.; Lambert, J.F.; et al. Effect of a Comprehensive Deep-Learning Model on the Accuracy of Chest X-ray Interpretation by Radiologists: A Retrospective, Multireader Multicase Study. Lancet Digit Health 2021, 3, e496–e506. [Google Scholar] [CrossRef]
- Jones, C.M.; Buchlak, Q.D.; Oakden-Rayner, L.; Milne, M.; Seah, J.; Esmaili, N.; Hachey, B. Chest Radiographs and Machine Learning—Past, Present and Future. J. Med. Imaging Radiat. Oncol. 2021, 65, 538–544. [Google Scholar] [CrossRef]
- Çallı, E.; Sogancioglu, E.; van Ginneken, B.; van Leeuwen, K.G.; Murphy, K. Deep Learning for Chest X-ray Analysis: A Survey. Med. Image Anal. 2021, 72, 102125. [Google Scholar] [CrossRef]
- Buchlak, Q.D.; Milne, M.R.; Seah, J.; Johnson, A.; Samarasinghe, G.; Hachey, B.; Esmaili, N.; Tran, A.; Leveque, J.-C.; Farrokhi, F.; et al. Charting the Potential of Brain Computed Tomography Deep Learning Systems. J. Clin. Neurosci. 2022, 99, 217–223. [Google Scholar] [CrossRef] [PubMed]
- Huo, Z.; Mao, H.; Zhang, J.; Sykes, A.-M.; Munn, S.; Wandtke, J. Computer-Aided Detection of Malpositioned Endotracheal Tubes in Portable Chest Radiographs. In Medical Imaging 2014: Computer-Aided Diagnosis; SPIE Medical Imaging: San Diego, CA, USA, 2014. [Google Scholar]
- Ramakrishna, B.; Brown, M.; Goldin, J.; Cagnon, C.; Enzmann, D. Catheter Detection and Classification on Chest Radiographs: An Automated Prototype Computer-Aided Detection (CAD) System for Radiologists. In Proceedings of the Medical Imaging 2011: Computer-Aided Diagnosis; Summers, R.M., van Ginneken, B., Eds.; SPIE Medical Imaging: Lake Buena Vista, FL, USA, 2011. [Google Scholar]
- Lakhani, P.; Flanders, A.; Gorniak, R. Endotracheal Tube Position Assessment on Chest Radiographs Using Deep Learning. Radiol. Artif. Intell. 2021, 3, e200026. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.; Mansouri, M.; Tajmir, S.; Lev, M.H.; Do, S. A Deep-Learning System for Fully-Automated Peripherally Inserted Central Catheter (PICC) Tip Detection. J. Digit. Imaging 2018, 31, 393–402. [Google Scholar] [CrossRef] [PubMed]
- Subramanian, V.; Wang, H.; Wu, J.T.; Wong, K.C.L.; Sharma, A.; Syeda-Mahmood, T. Automated Detection and Type Classification of Central Venous Catheters in Chest X-rays. In Medical Image Computing and Computer Assisted Intervention—MICCAI 2019; Lecture Notes in Computer Science; Springer: Cham, Switzerland, 2019; pp. 522–530. [Google Scholar] [CrossRef]
- Yi, X.; Adams, S.; Babyn, P.; Elnajmi, A. Automatic Catheter and Tube Detection in Pediatric X-ray Images Using a Scale-Recurrent Network and Synthetic Data. J. Digit. Imaging 2020, 33, 181–190. [Google Scholar] [CrossRef]
- Wu, J.T.; Wong, K.C.L.; Gur, Y.; Ansari, N.; Karargyris, A.; Sharma, A.; Morris, M.; Saboury, B.; Ahmad, H.; Boyko, O.; et al. Comparison of Chest Radiograph Interpretations by Artificial Intelligence Algorithm vs Radiology Residents. JAMA Netw. Open 2020, 3, e2022779. [Google Scholar] [CrossRef]
- Henderson, R.D.E.; Yi, X.; Adams, S.J.; Babyn, P. Automatic Classification of Multiple Catheters in Neonatal Radiographs with Deep Learning. arXiv 2020, arXiv:2011.07394. [Google Scholar] [CrossRef]
- Abbas, M.; Abdul Salam, A.; Zeb, J. Automatic Detection and Classification of Correct Placement of Tubes on Chest X-rays Using Deep Learning with EfficientNet. In Proceedings of the 2022 2nd International Conference on Digital Futures and Transformative Technologies (ICoDT2), Rawalpindi, Pakistan, 24–26 May 2022. [Google Scholar]
- Drozdov, I.; Dixon, R.; Szubert, B.; Dunn, J.; Green, D.; Hall, N.; Shirandami, A.; Rosas, S.; Grech, R.; Puttagunta, S.; et al. An Artificial Neural Network for Nasogastric Tube Position Decision Support. Radiol. Artif. Intell. 2023, 5, e220165. [Google Scholar] [CrossRef]
- Yi, X.; Adams, S.J.; Henderson, R.D.E.; Babyn, P. Computer-Aided Assessment of Catheters and Tubes on Radiographs: How Good Is Artificial Intelligence for Assessment? Radiol. Artif. Intell. 2020, 2, e190082. [Google Scholar] [CrossRef]
- Aggarwal, R.; Sounderajah, V.; Martin, G.; Ting, D.S.W.; Karthikesalingam, A.; King, D.; Ashrafian, H.; Darzi, A. Diagnostic Accuracy of Deep Learning in Medical Imaging: A Systematic Review and Meta-Analysis. NPJ Digit. Med. 2021, 4, 65. [Google Scholar] [CrossRef]
- Oakden-Rayner, L.; Dunnmon, J.; Carneiro, G.; Re, C. Hidden Stratification Causes Clinically Meaningful Failures in Machine Learning for Medical Imaging. In Proceedings of the ACM Conference on Health, Inference, and Learning, Toronto, ON, Canada, 2–4 April 2020. [Google Scholar]
- Tan, M.; Le, Q.V. EfficientNet: Rethinking Model Scaling for Convolutional Neural Networks. arXiv 2019, arXiv:1905.11946. [Google Scholar]
- Ronneberger, O.; Fischer, P.; Brox, T. U-Net: Convolutional Networks for Biomedical Image Segmentation. In Medical Image Computing and Computer-Assisted Intervention—MICCAI 2015; Lecture Notes in Computer Science; Springer International Publishing: Cham, Switzerland, 2015; pp. 234–241. ISBN 9783319245737. [Google Scholar]
- Johnson, A.E.W.; Pollard, T.J.; Shen, L.; Lehman, L.-W.H.; Feng, M.; Ghassemi, M.; Moody, B.; Szolovits, P.; Anthony Celi, L.; Mark, R.G. MIMIC-III, a Freely Accessible Critical Care Database. Sci. Data 2016, 3, 160035. [Google Scholar] [CrossRef]
- Virtanen, P.; Gommers, R.; Oliphant, T.E.; Haberland, M.; Reddy, T.; Cournapeau, D.; Burovski, E.; Peterson, P.; Weckesser, W.; Bright, J.; et al. SciPy 1.0: Fundamental Algorithms for Scientific Computing in Python. Nat. Methods 2020, 17, 261–272. [Google Scholar] [CrossRef]
- Pedregosa, F.; Varoquaux, G.; Gramfort, A.; Michel, V.; Thirion, B.; Grisel, O.; Blondel, M.; Prettenhofer, P.; Weiss, R.; Dubourg, V.; et al. Scikit-Learn: Machine Learning in Python. J. Mach. Learn. Res. 2011, 12, 2825–2830. [Google Scholar]
- Harris, C.R.; Millman, K.J.; van der Walt, S.J.; Gommers, R.; Virtanen, P.; Cournapeau, D.; Wieser, E.; Taylor, J.; Berg, S.; Smith, N.J.; et al. Array Programming with NumPy. Nature 2020, 585, 357–362. [Google Scholar] [CrossRef] [PubMed]
- Abadi, M.; Agarwal, A.; Barham, P.; Brevdo, E.; Chen, Z.; Citro, C.; Corrado, G.S.; Davis, A.; Dean, J.; Devin, M. Tensorflow: Large-Scale Machine Learning on Heterogeneous Distributed Systems. arXiv 2016, arXiv:1603.04467. [Google Scholar]
- Singh, V.; Danda, V.; Gorniak, R.; Flanders, A.; Lakhani, P. Assessment of Critical Feeding Tube Malpositions on Radiographs Using Deep Learning. J. Digit. Imaging 2019, 32, 651–655. [Google Scholar] [CrossRef] [PubMed]
- Lakhani, P. Deep Convolutional Neural Networks for Endotracheal Tube Position and X-ray Image Classification: Challenges and Opportunities. J. Digit. Imaging 2017, 30, 460–468. [Google Scholar] [CrossRef]
- Zech, J.R.; Badgeley, M.A.; Liu, M.; Costa, A.B.; Titano, J.J.; Oermann, E.K. Variable Generalization Performance of a Deep Learning Model to Detect Pneumonia in Chest Radiographs: A Cross-Sectional Study. PLoS Med. 2018, 15, e1002683. [Google Scholar] [CrossRef] [PubMed]
- Rajpurkar, P.; Joshi, A.; Pareek, A.; Ng, A.Y.; Lungren, M.P. CheXternal: Generalization of Deep Learning Models for Chest X-ray Interpretation to Photos of Chest X-rays and External Clinical Settings. arXiv 2021, arXiv:2102.08660. [Google Scholar]
- Seah, J.; Tang, C.; Buchlak, Q.D.; Milne, M.R.; Holt, X.; Ahmad, H.; Lambert, J.; Esmaili, N.; Oakden-Rayner, L.; Brotchie, P.; et al. Do Comprehensive Deep Learning Algorithms Suffer from Hidden Stratification? A Retrospective Study on Pneumothorax Detection in Chest Radiography. BMJ Open 2021, 11, e053024. [Google Scholar] [CrossRef]

| Dataset Characteristic | Statistics |
|---|---|
| Patients | 2286 |
| Studies | 2568 |
| Images | 4568 |
| Sex | 29% male 28% female 43% unknown * |
| Age | 74 years (SD 15 years) * |
| View Position | 28% PA 33% AP 31% LAT 8% other |
| Finding | Positives | Negatives | Model AUC Mean over Entire Dataset | AUC Mean 95% CI |
|---|---|---|---|---|
| ETT | 262 | 2295 | 0.9999 | 0.9997–1.0000 |
| CVC | 477 | 2080 | 0.9983 | 0.9970–0.9993 |
| NGT | 206 | 2351 | 0.9994 | 0.9984–1.0000 |
| Finding | Category | Size of Category | Positive Cases in Category | Model AUC Mean (95% CI) | TN | FP | FN | TP |
|---|---|---|---|---|---|---|---|---|
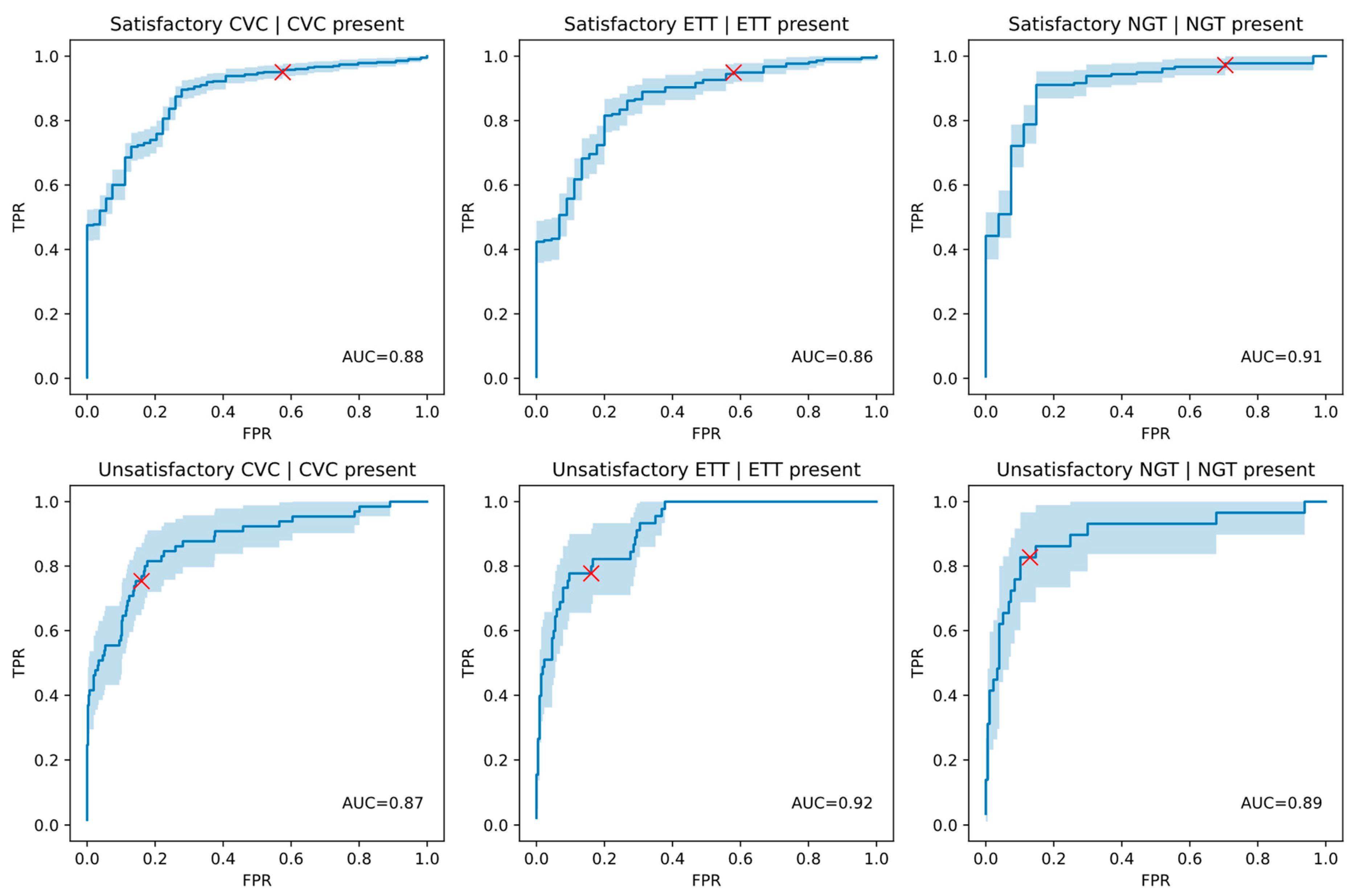
| Satisfactory ETT | ETT | 262 | 209 | 0.8608 (0.8020–0.9128) | 19 | 34 | 11 | 198 |
| Unsatisfactory ETT | ETT | 262 | 53 | 0.9153 (0.8724–0.9519) | 174 | 35 | 18 | 35 |
| Satisfactory CVC | CVC | 477 | 423 | 0.8778 (0.8323–0.9186) | 23 | 31 | 21 | 402 |
| Unsatisfactory CVC | CVC | 477 | 65 | 0.8715 (0.8158–0.9200) | 346 | 66 | 16 | 49 |
| Satisfactory NGT | NGT | 206 | 179 | 0.9051 (0.8409–0.9574) | 8 | 19 | 5 | 174 |
| Unsatisfactory NGT | NGT | 206 | 29 | 0.8943 (0.8062–0.9620) | 154 | 23 | 5 | 24 |
| Finding | Subgroup | Size of Subgroup | Positive Cases in Subgroup | Model AUC Mean (95% CI) | TN | FP | FN | TP |
|---|---|---|---|---|---|---|---|---|
| Satisfactory CVC | Dialysis Catheters | 40 | 36 | 0.9304 (0.8222–1.0000) | 1 | 3 | 2 | 34 |
| Unsatisfactory CVC | Dialysis Catheters | 40 | 6 | 0.9019 (0.6989–1.0000) | 24 | 10 | 1 | 5 |
| Satisfactory CVC | Jugular Lines | 243 | 221 | 0.9139 (0.8509–0.9639) | 10 | 12 | 6 | 215 |
| Unsatisfactory CVC | Jugular Lines | 243 | 32 | 0.8700 (0.7890–0.9379) | 176 | 35 | 7 | 25 |
| Satisfactory CVC | PICCs | 140 | 123 | 0.8227 (0.7220–0.9083) | 7 | 10 | 13 | 110 |
| Unsatisfactory CVC | PICCs | 140 | 23 | 0.7880 (0.6624–0.8947) | 98 | 19 | 9 | 14 |
| Satisfactory CVC | Subclavian Lines | 121 | 107 | 0.8879 (0.7943–0.9639) | 5 | 9 | 2 | 105 |
| Unsatisfactory CVC | Subclavian Lines | 121 | 16 | 0.8892 (0.7840–0.9675) | 87 | 18 | 4 | 12 |
| Satisfactory ETT | Endotracheal Tubes | 211 | 166 | 0.8709 (0.8130–0.9220) | 19 | 26 | 8 | 158 |
| Unsatisfactory ETT | Endotracheal Tubes | 211 | 45 | 0.8923 (0.8387–0.9384) | 131 | 35 | 10 | 35 |
| Satisfactory ETT | Tracheostomies | 51 | 51 | N/A * | 0 | 0 | 3 | 48 |
| Unsatisfactory ETT | Tracheostomies | 51 | 0 | N/A * | 51 | 0 | 0 | 0 |
| Satisfactory NGT | Double Lumen NGTs | 170 | 147 | 0.9091 (0.8388–0.9655) | 7 | 16 | 3 | 144 |
| Unsatisfactory NGT | Double Lumen NGTs | 170 | 25 | 0.8752 (0.7741–0.9538) | 125 | 20 | 5 | 20 |
| Satisfactory NGT | NGTs with Guide Wire | 2 | 1 | N/A † | 1 | 0 | 0 | 1 |
| Unsatisfactory NGT | NGTs with Guide Wire | 2 | 1 | N/A † | 1 | 0 | 0 | 1 |
| Satisfactory NGT | Fine Bore NGTs | 37 | 33 | 0.9091 (0.8000–1.0000) | 1 | 3 | 2 | 31 |
| Unsatisfactory NGT | Fine Bore NGTs | 37 | 4 | 1.0000 (1.0000–1.0000) | 30 | 3 | 0 | 4 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tang, C.H.M.; Seah, J.C.Y.; Ahmad, H.K.; Milne, M.R.; Wardman, J.B.; Buchlak, Q.D.; Esmaili, N.; Lambert, J.F.; Jones, C.M. Analysis of Line and Tube Detection Performance of a Chest X-ray Deep Learning Model to Evaluate Hidden Stratification. Diagnostics 2023, 13, 2317. https://doi.org/10.3390/diagnostics13142317
Tang CHM, Seah JCY, Ahmad HK, Milne MR, Wardman JB, Buchlak QD, Esmaili N, Lambert JF, Jones CM. Analysis of Line and Tube Detection Performance of a Chest X-ray Deep Learning Model to Evaluate Hidden Stratification. Diagnostics. 2023; 13(14):2317. https://doi.org/10.3390/diagnostics13142317
Chicago/Turabian StyleTang, Cyril H. M., Jarrel C. Y. Seah, Hassan K. Ahmad, Michael R. Milne, Jeffrey B. Wardman, Quinlan D. Buchlak, Nazanin Esmaili, John F. Lambert, and Catherine M. Jones. 2023. "Analysis of Line and Tube Detection Performance of a Chest X-ray Deep Learning Model to Evaluate Hidden Stratification" Diagnostics 13, no. 14: 2317. https://doi.org/10.3390/diagnostics13142317
APA StyleTang, C. H. M., Seah, J. C. Y., Ahmad, H. K., Milne, M. R., Wardman, J. B., Buchlak, Q. D., Esmaili, N., Lambert, J. F., & Jones, C. M. (2023). Analysis of Line and Tube Detection Performance of a Chest X-ray Deep Learning Model to Evaluate Hidden Stratification. Diagnostics, 13(14), 2317. https://doi.org/10.3390/diagnostics13142317






