Artificial Intelligence in the Advanced Diagnosis of Bladder Cancer-Comprehensive Literature Review and Future Advancement
Abstract
1. Introduction
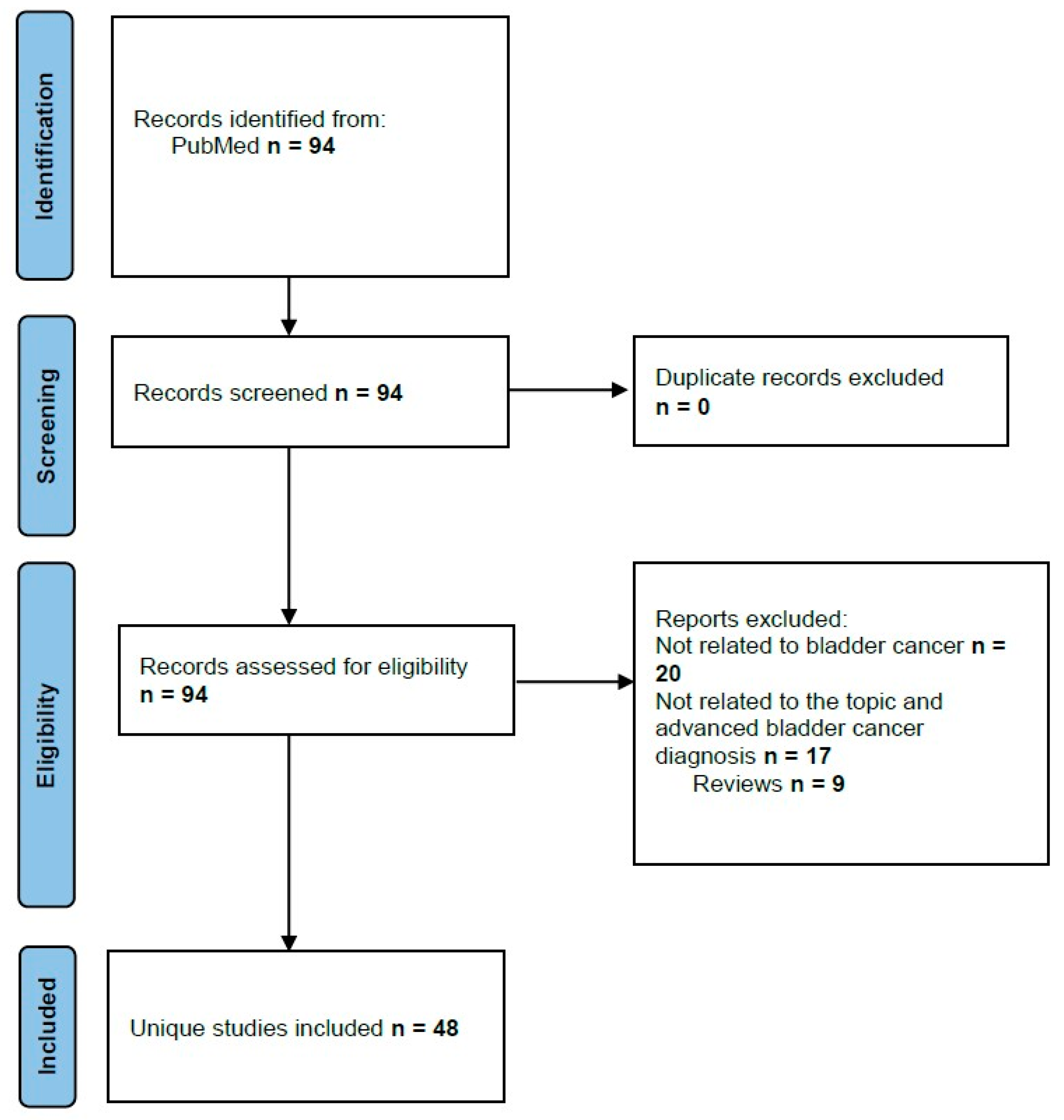
2. Materials and Methods
3. Results
3.1. Generalities of Artificial Intelligence
3.1.1. Machine Learning
3.1.2. Deep Learning
3.2. Bladder Cancer Diagnosis
3.2.1. Bladder Tumor Detection through Cystoscopy
3.2.2. Bladder Tumor Detection through Urine Cytology
3.2.3. Bladder Tumor Detection through Urine Metabolomes
3.2.4. Bladder Cancer Segmentation Research
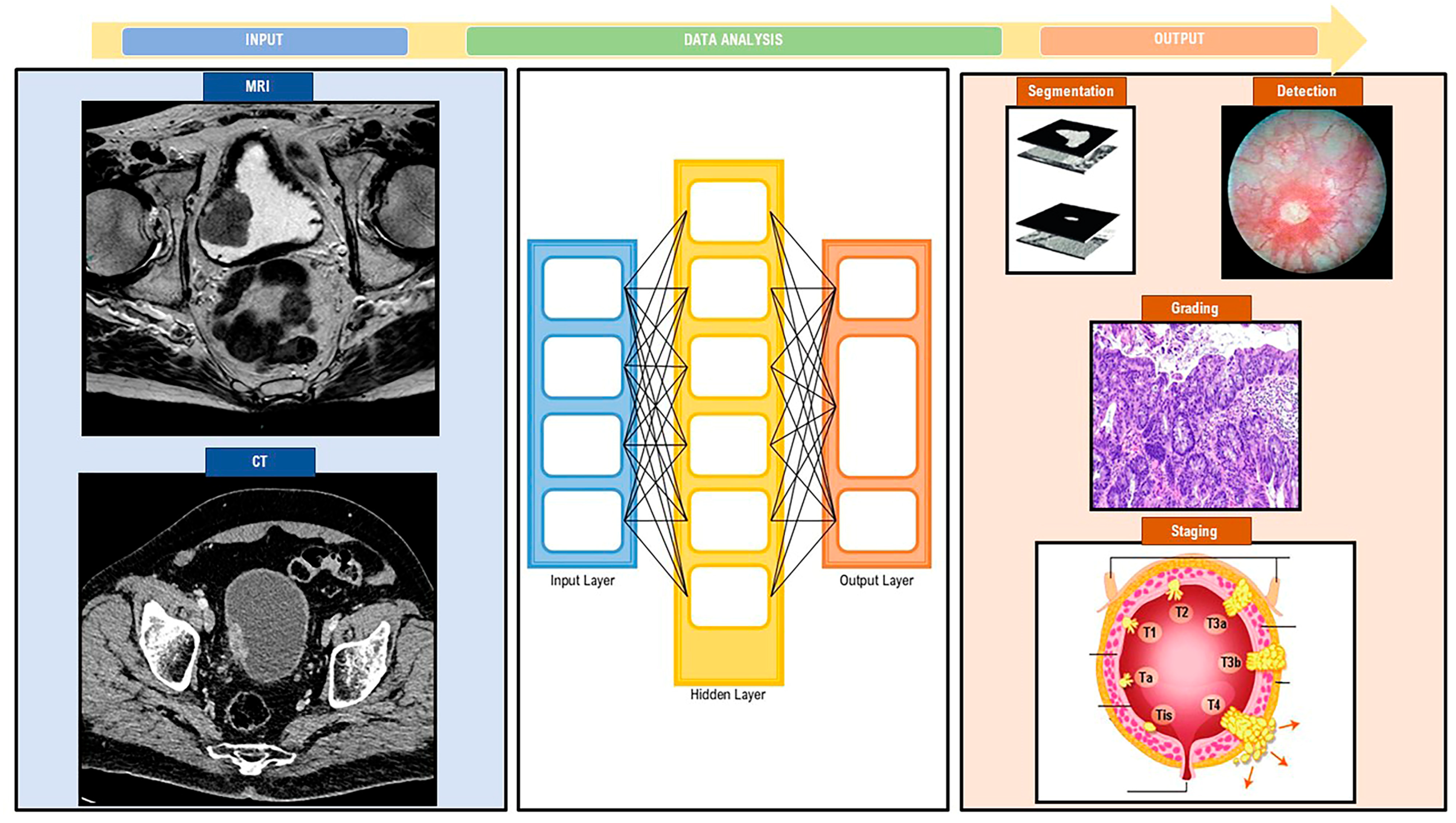
3.2.5. Bladder Cancer Imaging and Artificial Intelligence
3.2.6. Bladder Cancer Grading and Artificial Intelligence
3.2.7. Bladder Cancer and Histopathology
3.2.8. Bladder Cancer Staging and Artificial Intelligence
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Human Intelligence—Cognitive-Contextual Theories|Britannica. Available online: https://www.britannica.com/science/human-intelligence-psychology (accessed on 22 February 2023).
- Stilgoe, J. Machine learning, social learning and the governance of self-driving cars. Soc. Stud. Sci. 2018, 48, 25–56. [Google Scholar] [CrossRef]
- O’Connor, S. Open artificial intelligence platforms in nursing education: Tools for academic progress or abuse? Nurse Educ. Pract. 2023, 66, 103537. [Google Scholar] [CrossRef] [PubMed]
- Borhani, S.; Borhani, R.; Kajdacsy-Balla, A. Artificial intelligence: A promising frontier in bladder cancer diagnosis and outcome prediction. Crit. Rev. Oncol. Hematol. 2022, 171, 103601. [Google Scholar] [CrossRef] [PubMed]
- Tran, K.A.; Kondrashova, O.; Bradley, A.; Williams, E.D.; Pearson, J.V.; Waddell, N. Deep learning in cancer diagnosis, prognosis and treatment selection. Genome Med. 2021, 13, 152. [Google Scholar] [CrossRef] [PubMed]
- Eun, S.-J.; Kim, J.; Kim, K.H. Applications of artificial intelligence in urological setting: A hopeful path to improved care. J. Exerc. Rehabil. 2021, 17, 308–312. [Google Scholar] [CrossRef]
- Goldenberg, S.L.; Nir, G.; Salcudean, S.E. A new era: Artificial intelligence and machine learning in prostate cancer. Nat. Rev. Urol. 2019, 16, 391–403. [Google Scholar] [CrossRef]
- Tătaru, O.S.; Vartolomei, M.D.; Rassweiler, J.J.; Virgil, O.; Lucarelli, G.; Porpiglia, F.; Amparore, D.; Manfredi, M.; Carrieri, G.; Falagario, U.; et al. Artificial Intelligence and Machine Learning in Prostate Cancer Patient Management-Current Trends and Future Perspectives. Diagn. Basel Switz. 2021, 11, 354. [Google Scholar] [CrossRef]
- Thakur, N.; Alam, M.R.; Abdul-Ghafar, J.; Chong, Y. Recent Application of Artificial Intelligence in Non-Gynecological Cancer Cytopathology: A Systematic Review. Cancers 2022, 14, 3529. [Google Scholar] [CrossRef]
- IBM. What is Machine Learning? Available online: https://www.ibm.com/topics/machine-learning (accessed on 1 July 2023).
- Mahoto, N.A.; Shaikh, A.; Sulaiman, A.; Reshan, M.S.A.; Rajab, A.; Rajab, K. A machine learning based data modeling for medical diagnosis. Biomed. Signal Process. Control 2023, 81, 104481. [Google Scholar] [CrossRef]
- Brodie, A.; Dai, N.; Teoh, J.Y.-C.; Decaestecker, K.; Dasgupta, P.; Vasdev, N. Artificial intelligence in urological oncology: An update and future applications. Urol. Oncol. 2021, 39, 379–399. [Google Scholar] [CrossRef]
- Jang, H.-J.; Cho, K.-O. Applications of deep learning for the analysis of medical data. Arch. Pharm. Res. 2019, 42, 492–504. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Sung, J.J.Y. Potentials of AI in medical image analysis in Gastroenterology and Hepatology. J. Gastroenterol. Hepatol. 2021, 36, 31–38. [Google Scholar] [CrossRef]
- LeCun, Y.; Bengio, Y.; Hinton, G. Deep learning. Nature 2015, 521, 436–444. [Google Scholar] [CrossRef]
- Naik, N.; Tokas, T.; Shetty, D.K.; Hameed, B.M.Z.; Shastri, S.; Shah, M.J.; Ibrahim, S.; Rai, B.P.; Chłosta, P.; Somani, B.K. Role of Deep Learning in Prostate Cancer Management: Past, Present and Future Based on a Comprehensive Literature Review. J. Clin. Med. 2022, 11, 3575. [Google Scholar] [CrossRef] [PubMed]
- Jones, W.; Alasoo, K.; Fishman, D.; Parts, L. Computational biology: Deep learning. Emerg. Top. Life Sci. 2017, 1, 257–274. [Google Scholar] [CrossRef] [PubMed]
- Bandyk, M.G.; Gopireddy, D.R.; Lall, C.; Balaji, K.C.; Dolz, J. MRI and CT bladder segmentation from classical to deep learning based approaches: Current limitations and lessons. Comput. Biol. Med. 2021, 134, 104472. [Google Scholar] [CrossRef]
- Suarez-Ibarrola, R.; Hein, S.; Reis, G.; Gratzke, C.; Miernik, A. Current and future applications of machine and deep learning in urology: A review of the literature on urolithiasis, renal cell carcinoma, and bladder and prostate cancer. World J. Urol. 2020, 38, 2329–2347. [Google Scholar] [CrossRef]
- Haenssle, H.A.; Fink, C.; Schneiderbauer, R.; Toberer, F.; Buhl, T.; Blum, A.; Kalloo, A.; Hassen, A.B.H.; Thomas, L.; Enk, A.; et al. Man against machine: Diagnostic performance of a deep learning convolutional neural network for dermoscopic melanoma recognition in comparison to 58 dermatologists. Ann. Oncol. Off. J. Eur. Soc. Med. Oncol. 2018, 29, 1836–1842. [Google Scholar] [CrossRef]
- Poplin, R.; Varadarajan, A.V.; Blumer, K.; Liu, Y.; McConnell, M.V.; Corrado, G.S.; Peng, L.; Webster, D.R. Prediction of cardiovascular risk factors from retinal fundus photographs via deep learning. Nat. Biomed. Eng. 2018, 2, 158–164. [Google Scholar] [CrossRef]
- Vobugari, N.; Raja, V.; Sethi, U.; Gandhi, K.; Raja, K.; Surani, S.R. Advancements in Oncology with Artificial Intelligence-A Review Article. Cancers 2022, 14, 1349. [Google Scholar] [CrossRef]
- Miranda, E.; Suñé, J. Memristors for Neuromorphic Circuits and Artificial Intelligence Applications. Mater. Basel Switz. 2020, 13, 938. [Google Scholar] [CrossRef] [PubMed]
- Stoliar, P.; Schneegans, O.; Rozenberg, M.J. A Functional Spiking Neural Network of Ultra Compact Neurons. Front. Neurosci. 2021, 15, 635098. [Google Scholar] [CrossRef] [PubMed]
- Russakovsky, O.; Deng, J.; Su, H.; Krause, J.; Satheesh, S.; Ma, S.; Huang, Z.; Karpathy, A.; Khosla, A.; Bernstein, M. Imagenet large scale visual recognition challenge. Int. J. Comput. Vis. 2015, 115, 211–252. [Google Scholar] [CrossRef]
- Esteva, A.; Robicquet, A.; Ramsundar, B.; Kuleshov, V.; DePristo, M.; Chou, K.; Cui, C.; Corrado, G.; Thrun, S.; Dean, J. A guide to deep learning in healthcare. Nat. Med. 2019, 25, 24–29. [Google Scholar] [CrossRef] [PubMed]
- Walsh, S.; de Jong, E.E.C.; van Timmeren, J.E.; Ibrahim, A.; Compter, I.; Peerlings, J.; Sanduleanu, S.; Refaee, T.; Keek, S.; Larue, R.T.H.M.; et al. Decision Support Systems in Oncology. JCO Clin. Cancer Inform. 2019, 3, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Rasley, J.; Rajbhandari, S.; Ruwase, O.; He, Y. Deepspeed: System optimizations enable training deep learning models with over 100 billion parameters. In Proceedings of the 26th ACM SIGKDD International Conference on Knowledge Discovery & Data Mining, San Diego, CA, USA, 23–27 August 2020; pp. 3505–3506. [Google Scholar]
- Mintz, Y.; Brodie, R. Introduction to artificial intelligence in medicine. Minim. Invasive Ther. Allied Technol. MITAT Off. J. Soc. Minim. Invasive Ther. 2019, 28, 73–81. [Google Scholar] [CrossRef]
- Saxe, A.; Nelli, S.; Summerfield, C. If deep learning is the answer, what is the question? Nat. Rev. Neurosci. 2021, 22, 55–67. [Google Scholar] [CrossRef]
- Kausch, I.; Sommerauer, M.; Montorsi, F.; Stenzl, A.; Jacqmin, D.; Jichlinski, P.; Jocham, D.; Ziegler, A.; Vonthein, R. Photodynamic diagnosis in non-muscle-invasive bladder cancer: A systematic review and cumulative analysis of prospective studies. Eur. Urol. 2010, 57, 595–606. [Google Scholar] [CrossRef]
- Jichlinski, P.; Leisinger, H.-J. Fluorescence cystoscopy in the management of bladder cancer: A help for the urologist! Urol. Int. 2005, 74, 97–101. [Google Scholar] [CrossRef]
- Eminaga, O.; Eminaga, N.; Semjonow, A.; Breil, B. Diagnostic Classification of Cystoscopic Images Using Deep Convolutional Neural Networks. JCO Clin. Cancer Inform. 2018, 2, 1–8. [Google Scholar] [CrossRef]
- Lorencin, I.; Anđelić, N.; Španjol, J.; Car, Z. Using multi-layer perceptron with Laplacian edge detector for bladder cancer diagnosis. Artif. Intell. Med. 2020, 102, 101746. [Google Scholar] [CrossRef]
- Lorencin, I.; Baressi Šegota, S.; Anđelić, N.; Mrzljak, V.; Ćabov, T.; Španjol, J.; Car, Z. On Urinary Bladder Cancer Diagnosis: Utilization of Deep Convolutional Generative Adversarial Networks for Data Augmentation. Biology 2021, 10, 175. [Google Scholar] [CrossRef]
- Ikeda, A.; Nosato, H.; Kochi, Y.; Kojima, T.; Kawai, K.; Sakanashi, H.; Murakawa, M.; Nishiyama, H. Support System of Cystoscopic Diagnosis for Bladder Cancer Based on Artificial Intelligence. J. Endourol. 2020, 34, 352–358. [Google Scholar] [CrossRef]
- Yang, R.; Du, Y.; Weng, X.; Chen, Z.; Wang, S.; Liu, X. Automatic recognition of bladder tumours using deep learning technology and its clinical application. Int. J. Med. Robot. Comput. Assist. Surg. 2021, 17, e2194. [Google Scholar] [CrossRef]
- Du, Y.; Yang, R.; Chen, Z.; Wang, L.; Weng, X.; Liu, X. A deep learning network-assisted bladder tumour recognition under cystoscopy based on Caffe deep learning framework and EasyDL platform. Int. J. Med. Robot. Comput. Assist. Surg. 2021, 17, 1–8. [Google Scholar] [CrossRef]
- Shkolyar, E.; Jia, X.; Chang, T.C.; Trivedi, D.; Mach, K.E.; Meng, M.Q.-H.; Xing, L.; Liao, J.C. Augmented Bladder Tumor Detection Using Deep Learning. Eur. Urol. 2019, 76, 714–718. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.; Chen, X.; Pan, J.; Dong, W.; Diao, X.; Zhang, R.; Zhang, Y.; Zhang, Y.; Qian, G.; Chen, H.; et al. An Artificial Intelligence System for the Detection of Bladder Cancer via Cystoscopy: A Multicenter Diagnostic Study. J. Natl. Cancer Inst. 2022, 114, 220–227. [Google Scholar] [CrossRef] [PubMed]
- Ali, N.; Bolenz, C.; Todenhöfer, T.; Stenzel, A.; Deetmar, P.; Kriegmair, M.; Knoll, T.; Porubsky, S.; Hartmann, A.; Popp, J.; et al. Deep learning-based classification of blue light cystoscopy imaging during transurethral resection of bladder tumors. Sci. Rep. 2021, 11, 11629. [Google Scholar] [CrossRef]
- Yoo, J.W.; Koo, K.C.; Chung, B.H.; Baek, S.Y.; Lee, S.J.; Park, K.H.; Lee, K.S. Deep learning diagnostics for bladder tumor identification and grade prediction using RGB method. Sci. Rep. 2022, 12, 17699. [Google Scholar] [CrossRef]
- Nojima, S.; Terayama, K.; Shimoura, S.; Hijiki, S.; Nonomura, N.; Morii, E.; Okuno, Y.; Fujita, K. A deep learning system to diagnose the malignant potential of urothelial carcinoma cells in cytology specimens. Cancer Cytopathol. 2021, 129, 984–995. [Google Scholar] [CrossRef] [PubMed]
- Awan, R.; Benes, K.; Azam, A.; Song, T.-H.; Shaban, M.; Verrill, C.; Tsang, Y.W.; Snead, D.; Minhas, F.; Rajpoot, N. Deep learning based digital cell profiles for risk stratification of urine cytology images. Cytom. Part J. Int. Soc. Anal. Cytol. 2021, 99, 732–742. [Google Scholar] [CrossRef]
- Vaickus, L.J.; Suriawinata, A.A.; Wei, J.W.; Liu, X. Automating the Paris System for urine cytopathology-A hybrid deep-learning and morphometric approach. Cancer Cytopathol. 2019, 127, 98–115. [Google Scholar] [CrossRef] [PubMed]
- Sanghvi, A.B.; Allen, E.Z.; Callenberg, K.M.; Pantanowitz, L. Performance of an artificial intelligence algorithm for reporting urine cytopathology. Cancer Cytopathol. 2019, 127, 658–666. [Google Scholar] [CrossRef] [PubMed]
- Khosravi, P.; Kazemi, E.; Imielinski, M.; Elemento, O.; Hajirasouliha, I. Deep Convolutional Neural Networks Enable Discrimination of Heterogeneous Digital Pathology Images. EBioMedicine 2018, 27, 317–328. [Google Scholar] [CrossRef] [PubMed]
- Sokolov, I.; Dokukin, M.E.; Kalaparthi, V.; Miljkovic, M.; Wang, A.; Seigne, J.D.; Grivas, P.; Demidenko, E. Noninvasive diagnostic imaging using machine-learning analysis of nanoresolution images of cell surfaces: Detection of bladder cancer. Proc. Natl. Acad. Sci. USA 2018, 115, 12920–12925. [Google Scholar] [CrossRef] [PubMed]
- Lilli, L.; Giarnieri, E.; Scardapane, S. A Calibrated Multiexit Neural Network for Detecting Urothelial Cancer Cells. Comput. Math. Methods Med. 2021, 2021, 5569458. [Google Scholar] [CrossRef]
- Shao, C.-H.; Chen, C.-L.; Lin, J.-Y.; Chen, C.-J.; Fu, S.-H.; Chen, Y.-T.; Chang, Y.-S.; Yu, J.-S.; Tsui, K.-H.; Juo, C.-G.; et al. Metabolite marker discovery for the detection of bladder cancer by comparative metabolomics. Oncotarget 2017, 8, 38802–38810. [Google Scholar] [CrossRef] [PubMed]
- Kouznetsova, V.L.; Kim, E.; Romm, E.L.; Zhu, A.; Tsigelny, I.F. Recognition of early and late stages of bladder cancer using metabolites and machine learning. Metabolomics Off. J. Metabolomic Soc. 2019, 15, 94. [Google Scholar] [CrossRef]
- Duan, C.; Liang, Z.; Bao, S.; Zhu, H.; Wang, S.; Zhang, G.; Chen, J.J.; Lu, H. A coupled level set framework for bladder wall segmentation with application to MR cystography. IEEE Trans. Med. Imaging 2010, 29, 903–915. [Google Scholar] [CrossRef]
- Cha, K.H.; Hadjiiski, L.; Samala, R.K.; Chan, H.-P.; Caoili, E.M.; Cohan, R.H. Urinary bladder segmentation in CT urography using deep-learning convolutional neural network and level sets. Med. Phys. 2016, 43, 1882. [Google Scholar] [CrossRef]
- Dolz, J.; Xu, X.; Rony, J.; Yuan, J.; Liu, Y.; Granger, E.; Desrosiers, C.; Zhang, X.; Ben Ayed, I.; Lu, H. Multiregion segmentation of bladder cancer structures in MRI with progressive dilated convolutional networks. Med. Phys. 2018, 45, 5482–5493. [Google Scholar] [CrossRef]
- Li, R.; Chen, H.; Gong, G.; Wang, L. Bladder Wall Segmentation in MRI Images via Deep Learning and Anatomical Constraints. Annu. Int. Conf. IEEE Eng. Med. Biol. Soc. IEEE Eng. Med. Biol. Soc. Annu. Int. Conf. 2020, 2020, 1629–1632. [Google Scholar] [CrossRef]
- Niazi, M.K.K.; Yazgan, E.; Tavolara, T.E.; Li, W.; Lee, C.T.; Parwani, A.; Gurcan, M.N. Semantic segmentation to identify bladder layers from H&E Images. Diagn. Pathol. 2020, 15, 87. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.; Hadjiiski, L.M.; Wei, J.; Chan, H.-P.; Cha, K.H.; Cohan, R.H.; Caoili, E.M.; Samala, R.; Zhou, C.; Lu, Y. U-Net based deep learning bladder segmentation in CT urography. Med. Phys. 2019, 46, 1752–1765. [Google Scholar] [CrossRef]
- Zhang, Q.; Liang, Y.; Zhang, Y.; Tao, Z.; Li, R.; Bi, H. A comparative study of attention mechanism based deep learning methods for bladder tumor segmentation. Int. J. Med. Inf. 2023, 171, 104984. [Google Scholar] [CrossRef] [PubMed]
- Schwaibold, H.E.; Sivalingam, S.; May, F.; Hartung, R. The value of a second transurethral resection for T1 bladder cancer. BJU Int. 2006, 97, 1199–1201. [Google Scholar] [CrossRef]
- Ferro, M.; Musi, G.; Marchioni, M.; Maggi, M.; Veccia, A.; Del Giudice, F.; Barone, B.; Crocetto, F.; Lasorsa, F.; Antonelli, A.; et al. Radiogenomics in Renal Cancer Management-Current Evidence and Future Prospects. Int. J. Mol. Sci. 2023, 24, 4615. [Google Scholar] [CrossRef] [PubMed]
- Ferro, M.; de Cobelli, O.; Musi, G.; Del Giudice, F.; Carrieri, G.; Busetto, G.M.; Falagario, U.G.; Sciarra, A.; Maggi, M.; Crocetto, F.; et al. Radiomics in prostate cancer: An up-to-date review. Ther. Adv. Urol. 2022, 14, 17562872221109020. [Google Scholar] [CrossRef]
- Ferro, M.; de Cobelli, O.; Vartolomei, M.D.; Lucarelli, G.; Crocetto, F.; Barone, B.; Sciarra, A.; Del Giudice, F.; Muto, M.; Maggi, M.; et al. Prostate Cancer Radiogenomics-From Imaging to Molecular Characterization. Int. J. Mol. Sci. 2021, 22, 9971. [Google Scholar] [CrossRef]
- Lambin, P.; Rios-Velazquez, E.; Leijenaar, R.; Carvalho, S.; van Stiphout, R.G.P.M.; Granton, P.; Zegers, C.M.L.; Gillies, R.; Boellard, R.; Dekker, A.; et al. Radiomics: Extracting more information from medical images using advanced feature analysis. Eur. J. Cancer Oxf. Engl. 1990 2012, 48, 441–446. [Google Scholar] [CrossRef]
- Xu, X.; Zhang, X.; Tian, Q.; Zhang, G.; Liu, Y.; Cui, G.; Meng, J.; Wu, Y.; Liu, T.; Yang, Z.; et al. Three-dimensional texture features from intensity and high-order derivative maps for the discrimination between bladder tumors and wall tissues via MRI. Int. J. Comput. Assist. Radiol. Surg. 2017, 12, 645–656. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.; Zheng, J.; Li, Y.; Wu, Z.; Shi, S.; Huang, M.; Yu, H.; Dong, W.; Huang, J.; Lin, T. Development and Validation of an MRI-Based Radiomics Signature for the Preoperative Prediction of Lymph Node Metastasis in Bladder Cancer. EBioMedicine 2018, 34, 76–84. [Google Scholar] [CrossRef] [PubMed]
- Zheng, J.; Kong, J.; Wu, S.; Li, Y.; Cai, J.; Yu, H.; Xie, W.; Qin, H.; Wu, Z.; Huang, J.; et al. Development of a noninvasive tool to preoperatively evaluate the muscular invasiveness of bladder cancer using a radiomics approach. Cancer 2019, 125, 4388–4398. [Google Scholar] [CrossRef]
- Kozikowski, M.; Suarez-Ibarrola, R.; Osiecki, R.; Bilski, K.; Gratzke, C.; Shariat, S.F.; Miernik, A.; Dobruch, J. Role of Radiomics in the Prediction of Muscle-invasive Bladder Cancer: A Systematic Review and Meta-analysis. Eur. Urol. Focus 2022, 8, 728–738. [Google Scholar] [CrossRef] [PubMed]
- Taguchi, S.; Tambo, M.; Watanabe, M.; Machida, H.; Kariyasu, T.; Fukushima, K.; Shimizu, Y.; Okegawa, T.; Yokoyama, K.; Fukuhara, H. Prospective Validation of Vesical Imaging-Reporting and Data System Using a Next-Generation Magnetic Resonance Imaging Scanner-Is Denoising Deep Learning Reconstruction Useful? J. Urol. 2021, 205, 686–692. [Google Scholar] [CrossRef]
- Sarkar, S.; Min, K.; Ikram, W.; Tatton, R.W.; Riaz, I.B.; Silva, A.C.; Bryce, A.H.; Moore, C.; Ho, T.H.; Sonpavde, G.; et al. Performing Automatic Identification and Staging of Urothelial Carcinoma in Bladder Cancer Patients Using a Hybrid Deep-Machine Learning Approach. Cancers 2023, 15, 1673. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Xu, X.; Tian, Q.; Li, B.; Wu, Y.; Yang, Z.; Liang, Z.; Liu, Y.; Cui, G.; Lu, H. Radiomics assessment of bladder cancer grade using texture features from diffusion-weighted imaging. J. Magn. Reson. Imaging 2017, 46, 1281–1288. [Google Scholar] [CrossRef]
- Wang, H.; Hu, D.; Yao, H.; Chen, M.; Li, S.; Chen, H.; Luo, J.; Feng, Y.; Guo, Y. Radiomics analysis of multiparametric MRI for the preoperative evaluation of pathological grade in bladder cancer tumors. Eur. Radiol. 2019, 29, 6182–6190. [Google Scholar] [CrossRef]
- Jansen, I.; Lucas, M.; Bosschieter, J.; de Boer, O.J.; Meijer, S.L.; van Leeuwen, T.G.; Marquering, H.A.; Nieuwenhuijzen, J.A.; de Bruin, D.M.; Savci-Heijink, C.D. Automated Detection and Grading of Non-Muscle-Invasive Urothelial Cell Carcinoma of the Bladder. Am. J. Pathol. 2020, 190, 1483–1490. [Google Scholar] [CrossRef]
- Smith, S.C.; Baras, A.S.; Dancik, G.; Ru, Y.; Ding, K.-F.; Moskaluk, C.A.; Fradet, Y.; Lehmann, J.; Stöckle, M.; Hartmann, A.; et al. A 20-gene model for molecular nodal staging of bladder cancer: Development and prospective assessment. Lancet Oncol. 2011, 12, 137–143. [Google Scholar] [CrossRef]
- Seiler, R.; Lam, L.L.; Erho, N.; Takhar, M.; Mitra, A.P.; Buerki, C.; Davicioni, E.; Skinner, E.C.; Daneshmand, S.; Black, P.C. Prediction of Lymph Node Metastasis in Patients with Bladder Cancer Using Whole Transcriptome Gene Expression Signatures. J. Urol. 2016, 196, 1036–1041. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.-X.; Huang, J.; Liu, Z.-W.; Chen, H.-G.; Guo, P.; Cai, Q.-Q.; Zheng, J.-J.; Qin, H.-D.; Zheng, Z.-S.; Chen, X.; et al. A Genomic-clinicopathologic Nomogram for the Preoperative Prediction of Lymph Node Metastasis in Bladder Cancer. EBioMedicine 2018, 31, 54–65. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Chen, P.; McGough, M.; Xing, F.; Wang, C.; Bui, M.; Xie, Y.; Sapkota, M.; Cui, L.; Dhillon, J. Pathologist-level interpretable whole-slide cancer diagnosis with deep learning. Nat. Mach. Intell. 2019, 1, 236–245. [Google Scholar] [CrossRef]
- Velmahos, C.S.; Badgeley, M.; Lo, Y.-C. Using deep learning to identify bladder cancers with FGFR-activating mutations from histology images. Cancer Med. 2021, 10, 4805–4813. [Google Scholar] [CrossRef] [PubMed]
- Garapati, S.S.; Hadjiiski, L.; Cha, K.H.; Chan, H.-P.; Caoili, E.M.; Cohan, R.H.; Weizer, A.; Alva, A.; Paramagul, C.; Wei, J.; et al. Urinary bladder cancer staging in CT urography using machine learning. Med. Phys. 2017, 44, 5814–5823. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Zhang, X.; Tian, Q.; Wang, H.; Cui, L.-B.; Li, S.; Tang, X.; Li, B.; Dolz, J.; Ayed, I.B.; et al. Quantitative Identification of Nonmuscle-Invasive and Muscle-Invasive Bladder Carcinomas: A Multiparametric MRI Radiomics Analysis. J. Magn. Reson. Imaging 2019, 49, 1489–1498. [Google Scholar] [CrossRef]
- Yin, P.-N.; Kc, K.; Wei, S.; Yu, Q.; Li, R.; Haake, A.R.; Miyamoto, H.; Cui, F. Histopathological distinction of non-invasive and invasive bladder cancers using machine learning approaches. BMC Med. Inform. Decis. Mak. 2020, 20, 162. [Google Scholar] [CrossRef]
- Yang, Y.; Zou, X.; Wang, Y.; Ma, X. Application of deep learning as a noninvasive tool to differentiate muscle-invasive bladder cancer and non-muscle-invasive bladder cancer with CT. Eur. J. Radiol. 2021, 139, 109666. [Google Scholar] [CrossRef]
- Li, J.; Qiu, Z.; Cao, K.; Deng, L.; Zhang, W.; Xie, C.; Yang, S.; Yue, P.; Zhong, J.; Lyu, J.; et al. Predicting muscle invasion in bladder cancer based on MRI: A comparison of radiomics, and single-task and multi-task deep learning. Comput. Methods Programs Biomed. 2023, 233, 107466. [Google Scholar] [CrossRef]
- Li, J.; Cao, K.; Lin, H.; Deng, L.; Yang, S.; Gao, Y.; Liang, M.; Lin, C.; Zhang, W.; Xie, C.; et al. Predicting muscle invasion in bladder cancer by deep learning analysis of MRI: Comparison with vesical imaging-reporting and data system. Eur. Radiol. 2023, 33, 2699–2709. [Google Scholar] [CrossRef]
- Xu, Y.; Lou, J.; Gao, Z.; Zhan, M. Computed Tomography Image Features under Deep Learning Algorithm Applied in Staging Diagnosis of Bladder Cancer and Detection on Ceramide Glycosylation. Comput. Math. Methods Med. 2022, 2022, 7979523. [Google Scholar] [CrossRef]
- Zou, Y.; Cai, L.; Chen, C.; Shao, Q.; Fu, X.; Yu, J.; Wang, L.; Chen, Z.; Yang, X.; Yuan, B.; et al. Multi-task deep learning based on T2-Weighted Images for predicting Muscular-Invasive Bladder Cancer. Comput. Biol. Med. 2022, 151, 106219. [Google Scholar] [CrossRef]
- Mossanen, M.; Gore, J.L. The burden of bladder cancer care: Direct and indirect costs. Curr. Opin. Urol. 2014, 24, 487–491. [Google Scholar] [CrossRef]
- Sciarra, A.; Di Lascio, G.; Del Giudice, F.; Leoncini, P.P.; Salciccia, S.; Gentilucci, A.; Porreca, A.; Chung, B.I.; Di Pierro, G.; Busetto, G.M.; et al. Comparison of the clinical usefulness of different urinary tests for the initial detection of bladder cancer: A systematic review. Curr. Urol. 2021, 15, 22–32. [Google Scholar] [CrossRef] [PubMed]
- Anastasi, E.; Maggi, M.; Tartaglione, S.; Angeloni, A.; Gennarini, G.; Leoncini, P.P.; Sperduti, I.; Di Lascio, G.; De Stefano, V.; Di Pierro, G.B.; et al. Predictive value of MCM5 (ADXBLADDER) analysis in urine of men evaluated for the initial diagnosis of bladder cancer: A comparative prospective study. Diagn. Cytopathol. 2020, 48, 1034–1040. [Google Scholar] [CrossRef]
- Ferro, M.; Barone, B.; Crocetto, F.; Lucarelli, G.; Busetto, G.M.; Del Giudice, F.; Maggi, M.; Crocerossa, F.; Cantiello, F.; Damiano, R.; et al. Predictive clinico-pathological factors to identify BCG, unresponsive patients, after re-resection for T1 high grade non-muscle invasive bladder cancer. Urol. Oncol. 2022, 40, 490.e13–490.e20. [Google Scholar] [CrossRef]
- Busetto, G.M.; Porreca, A.; Del Giudice, F.; Maggi, M.; D’Agostino, D.; Romagnoli, D.; Musi, G.; Lucarelli, G.; Palmer, K.; Colonna di Paliano, A.; et al. SARS-CoV-2 Infection and High-Risk Non-Muscle-Invasive Bladder Cancer: Are There Any Common Features? Urol. Int. 2020, 104, 510–522. [Google Scholar] [CrossRef] [PubMed]
- Busetto, G.M.; D’Agostino, D.; Colicchia, M.; Palmer, K.; Artibani, W.; Antonelli, A.; Bianchi, L.; Bocciardi, A.; Brunocilla, E.; Carini, M.; et al. Robot-Assisted, Laparoscopic, and Open Radical Cystectomy: Pre-Operative Data of 1400 Patients From The Italian Radical Cystectomy Registry. Front. Oncol. 2022, 12, 895460. [Google Scholar] [CrossRef] [PubMed]
- Krajewski, W.; Aumatell, J.; Subiela, J.D.; Nowak, Ł.; Tukiendorf, A.; Moschini, M.; Basile, G.; Poletajew, S.; Małkiewicz, B.; Del Giudice, F.; et al. Accuracy of the CUETO, EORTC 2016 and EAU 2021 scoring models and risk stratification tables to predict outcomes in high-grade non-muscle-invasive urothelial bladder cancer. Urol. Oncol. 2022, 40, 491.e11–491.e19. [Google Scholar] [CrossRef]
- Ferro, M.; Del Giudice, F.; Carrieri, G.; Busetto, G.M.; Cormio, L.; Hurle, R.; Contieri, R.; Arcaniolo, D.; Sciarra, A.; Maggi, M.; et al. The Impact of SARS-CoV-2 Pandemic on Time to Primary, Secondary Resection and Adjuvant Intravesical Therapy in Patients with High-Risk Non-Muscle Invasive Bladder Cancer: A Retrospective Multi-Institutional Cohort Analysis. Cancers 2021, 13, 5276. [Google Scholar] [CrossRef]
- Del Giudice, F.; Busetto, G.M.; Gross, M.S.; Maggi, M.; Sciarra, A.; Salciccia, S.; Ferro, M.; Sperduti, I.; Flammia, S.; Canale, V.; et al. Efficacy of three BCG strains (Connaught, TICE and RIVM) with or without secondary resection (re-TUR) for intermediate/high-risk non-muscle-invasive bladder cancers: Results from a retrospective single-institution cohort analysis. J. Cancer Res. Clin. Oncol. 2021, 147, 3073–3080. [Google Scholar] [CrossRef]
- Sarker, I.H. Deep Learning: A Comprehensive Overview on Techniques, Taxonomy, Applications and Research Directions. Sn Comput. Sci. 2021, 2, 420. [Google Scholar] [CrossRef] [PubMed]
- Pawar, S.P.; Talbar, S.N. Two-Stage Hybrid Approach of Deep Learning Networks for Interstitial Lung Disease Classification. BioMed Res. Int. 2022, 2022, 7340902. [Google Scholar] [CrossRef]
- Abbaspour, S.; Fotouhi, F.; Sedaghatbaf, A.; Fotouhi, H.; Vahabi, M.; Linden, M. A Comparative Analysis of Hybrid Deep Learning Models for Human Activity Recognition. Sensors 2020, 20, 5707. [Google Scholar] [CrossRef] [PubMed]
- Haq, E.U.; Jianjun, H.; Huarong, X.; Li, K.; Weng, L. A Hybrid Approach Based on Deep CNN and Machine Learning Classifiers for the Tumor Segmentation and Classification in Brain MRI. Comput. Math. Methods Med. 2022, 2022, 6446680. [Google Scholar] [CrossRef] [PubMed]
- Muehlematter, U.J.; Daniore, P.; Vokinger, K.N. Approval of artificial intelligence and machine learning-based medical devices in the USA and Europe (2015-20): A comparative analysis. Lancet Digit. Health 2021, 3, e195–e203. [Google Scholar] [CrossRef]
- Benjamens, S.; Dhunnoo, P.; Meskó, B. The state of artificial intelligence-based FDA-approved medical devices and algorithms: An online database. NPJ Digit. Med. 2020, 3, 118. [Google Scholar] [CrossRef]
- Linardatos, P.; Papastefanopoulos, V.; Kotsiantis, S. Explainable AI: A Review of Machine Learning Interpretability Methods. Entropy 2020, 23, 18. [Google Scholar] [CrossRef]
- Aggarwal, R.; Farag, S.; Martin, G.; Ashrafian, H.; Darzi, A. Patient Perceptions on Data Sharing and Applying Artificial Intelligence to Health Care Data: Cross-sectional Survey. J. Med. Internet Res. 2021, 23, e26162. [Google Scholar] [CrossRef]
- Esmaeilzadeh, P.; Mirzaei, T.; Dharanikota, S. Patients’ Perceptions Toward Human–Artificial Intelligence Interaction in Health Care: Experimental Study. J. Med. Internet Res. 2021, 23, e25856. [Google Scholar] [CrossRef]

| Term | Brief Explanation |
|---|---|
| Perceptron | a super-simplified version of a biological neuron, which takes different inputs and weighs them up to produce a single output [15] |
| Backpropagation | an algorithm that is used to train neural networks [15] |
| Artificial Neural Networks (ANN) | a computational model (i.e., algorithms or physical hardware) which mimics the human brain to process data and create patterns for decision-making [17] |
| Convolutional Neural Networks (CNN) | a neural network utilizing numerous identical copies of the same neuron, thus allowing a network to learn a neuron once and use it in several places. It is particularly useful for digitized images and pattern recognition [29] |
| Recurrent Neural Network (RNN) | a neural network utilizing sequential information, thus relying on previous computations [29] |
| Supervised Neural Network | a neural network for which, to produce an ideal output, a prior provided output is required. It is ‘trained’ on a given pre-defined dataset and provides outputs depending on the input it has received [30] |
| Unsupervised Neural Network | a neural network for which no labels are required. This involves giving a program with an unlabeled data set (i.e., that it has not been previously trained for). It is used to discover patterns and trends by clustering. [30] |
| Authors/Year | INPUT/ N of Patients | AI Algorithm/ Models | OUTPUT | Summary | Performance |
|---|---|---|---|---|---|
| Smith et al., 2001 [73] | Gene expression Training: 156 pt Validation: 185 pt | WNN | Histopathology: pN stage | Using WNN to develop a gene expression model to predict pathological node status | AUC = 0.67 |
| Seiler et al., 2016 [74] | Gene expression Training: 133 pt Validation: 66 pt | k-NN | Histopathology: pN stage | Using k-NN to develop a gene classifier to predict pathological lymph node metastasis in MIBC | AUC = 0.82 |
| Cha et al., 2016 [53] | CT Urography Training: 81 pt Validation: 92 pt | CNN | Segmentation | Using CNN to segment bladder and ROIs | JSC: 0.76 |
| Xu et al., 2017 [64] | T2w MRI images 62 cancer lesions, 62 controls | SVM | Histopathology: Presence of Cancer | Extracting Radiomics feature to differentiate cancer and non-cancer areas | AUC: 0.94 |
| Shao et al., 2017 [50] | Urine metabolomes 87 BCa pt, 65 control | DT | Histopathology: Presence of Cancer | Evaluate urine metabolite associated with BCa | AUC: 0.77 |
| Zhang et al., 2017 [70] | MRI radiomics features 61 pt | SVM | Histopathology: Grading | Using SVM to discriminate low grade and high grade bladder Ca on MRI | AUC: 0.86 |
| Garapati et al., 2017 [78] | CT images texture analysis 76 pt | LDA CNN SVM RF | Histopathology: Staging | Comparing 4 AI algorithms to discriminate bladder Ca < T2 and ≥T2 | AUC: 0.89–0.97 |
| Vaickus et al., 2018 [45] | Urine cytology 51 negative, 60 atypical, 52 suspicious, and 54 positive cases | CNN (AlexNet/ResNet) | Citology: Detection | A hybrid deep-learning and morphometric algorithm to automate the PARIS system | ACC: >95% |
| Eminaga et al., 2018 [33] | Cystoscopy images | CNN | Cistoscopy: Detection | Detect cancerous features from cystoscopy images using CNN models | ACC: 0.99 |
| Khosravi et al., 2018 [47] | IHC digital slides | CNN | Histopathology: Detection | differentiate 4 biomarkers of BCa on IHC | ACC: 0.99 |
| Sokolov et al., 2018 [48] | High resolutions images using atomic force microscopy. 25 cancer lesions, 43 control | ML | Histopathology: Detection | Non-invasive detection of BCa | ACC: 0.94 |
| Wu et al., 2018 [65] | T2w MRI images Training: 69 pt Validation: 34 pt | LASSO, LR | Histopathology: pN stage | Building a nomogram with mpMRI radiomic features | AUC: 0.84 |
| Wu et al., 2018 [75] | Gene expression Training: 178 pt Validation: 246 pt | LR | Histopathology: pN stage | Utilizing LR to develop a genomic clinicopathologic nomogram for predicting LN metastasis | AUC: 0.89 |
| Dolz et al., 2018 [54] | MRI images Training: 60 pt | CNN | Segmentation | Inner, outer wall, and tumor region segmentation | DSC: 0.69 |
| Shkolyar et al., 2019 [39] | Cystoscopy images Training: 95 pt | CNN | Cistoscopy: Detection | Using “Cystonet” a CNN to discriminate malignant from benign images | SENS: 91% SPEC: 99% |
| Zheng et al., 2019 [66] | T2w MRI images Training: 130 pt Validation: 69 pt | LASSO, LR | Histopathology: pT stage | Building a nomogram with mpMRI radiomic features | AUC: 0.88 |
| Wang et al., 2019 [71] | T2w MRI images Training: 70 pt Validation: 30 pt | LR | Histopathology: Grading | Utilizing MRI radiomics features to discriminate low and high-grade BCa | AUC: 88.2 |
| Zhang et al., 2019 [76] | Histopathology digital images Training: 620 Validation: 193 | CNN | Histopathology: pT stage | Utilizing CNN to analyze bladder Ca WSI compared to expert histopathologists | AUC: 0.97 |
| Sanghvi et al., 2019 [46] | Urine cytology Training: 2405 urine sample Prospective Validation | CNN | Cistoscopy: Detection | Artificial Intelligence Algorithm for Reporting Urine Cytopathology | AUC: 0.88 |
| Kouznetsova et al., 2019 [51] | Urine metabolomes | ANN, LR | Histopathology: pT stage | Recognition of Early and Late Stages of Bladder Cancer Using Metabolites and Machine Learning | ACC: 0.82 |
| Ma et al., 2019 [57] | CT Urography Training: 81 pt Validation: 92 pt | U-net DCNN | Segmentation | Deep Learning Bladder Segmentation in CT Urography | JSC: 0.85 |
| Xu et al., 2019 [79] | T2w and DWI MRI images Training: 54 pt | SVM | Histopathology: pT stage | BCa staging with MRI Radiomics Analysis | AUC:0.97 |
| Ikeda et al., 2020 [36] | 2102 Cystoscopy images | CNN | Cistoscopy: Detection | Development of a Support System for Cystoscopic Diagnosis of BCa | AUC: 0.98 |
| Lorencin et al., 2020 [34] | 2983 Cystoscopy images | ANN | Cistoscopy: Detection | Development of a Support System for Cystoscopic Diagnosis of BCa | AUC: 0.99 |
| Li et al., 2020 [55] | MRI 1092 pt | U-net | Segmentation | Deep Learning Bladder Segmentation in MRI images | DSC: 0.85 |
| Niazi et al., 2020 [56] | Histopathology digital images of pT1 pt | U-net | Segmentation | Deep Learning for bladder layers identification on Pathology images | ACC: 0.90 |
| Yin et al., 2020 [80] | Histopathology digital images of pTa and pT1 pt | SVM, LR, RF, ANN | Histopathology: pT stage | Histopathological staging of BCa using different ML Approaches | ACC: 0.96 |
| Jansen et al., 2020 [72] | Histopathology digital images | U-net | Histopathology: Grading | Detection and grading of BCa | ACC: 0.76 |
| Lorencin et al., 2021 [35] | 2983 Cystoscopy images | CNN | Cystoscopy: Detection | Development of a Support System for Cystoscopic Diagnosis of BCa | AUC:0.99 |
| Nojima et al., 2021 [43] | Urine cytology | 16-layer Visual Geometry Group CNN | Detection and Grading | DL diagnosis and grading of BCa using urine Cytology | AUC: 0.98, F1 score: 0.90 (Presence/Absence) AUC: 0.86, F1 score: 0.82 (Invasive/non invasive) AUC: 0.86, F1 score: 0.82 (low-grade/high-grade |
| Yang et al., 2021 [37] | Cystoscopy images | CNN | Cystoscopy: Detection | Comparisons of a Support Systems for Cystoscopic Diagnosis of BCa | ACC: 0.97 |
| Awan et al., 2021 [44] | Urine cytology | CNN | Detection | Identification of atypic cells | AUC: 0.99 |
| Yang et al. (2021b) [81] | CT Images, 1200 images from 369 pt | CNN | Histopathology: pT stage | DL to differentiate Muscle-Invasive BCa with CT | AUC: 0.99 |
| Lilli et al., 2021 [49] | Urine cytology | CNN | Detection | Identification of Cancer cells | ACC: 89.90% |
| Du et al., 2021 [38] | Cystoscopy images 1736 pt | CNN EasyDL Caffe DL | Cystoscopy: Detection | Comparisons of a Support Systems for Cystoscopic Diagnosis of BCa | ACC = 82.9% (Caffe DL) ACC = 96.9% (EasyDL) |
| Taguchi et al., 2021 [68] | MRI images 68 pt | CNN | Detection | VI-RADS score and DL for BCa detection | AUC: 0.92 |
| Velmahos et al., 2021 [77] | Histopathology digital images | CNN | Histopathology: FGFR alterations and tumor-infiltrating lymphocytes | Deep Learning to Identify Bladder Cancers with FGFR-Activating Mutations | AUC: 0.86 |
| Ali et al., 2021 [41] | Blue light cystoscopy images | CNN | Cystoscopy: Detection Histopathology: Staging | Blue-light cystoscopy and CNN algorithm to detect, grade, and stage BCa | Detect-SENS = 95.77% SPEC = 87.84% Staging-SENS = 88% SPEC = 96.56% |
| Yoo et al., 2022 [42] | Cystoscopy images | SVM | Cystoscopy: Detection | Cystoscopic Diagnosis of BCa using a red-green-blue method. | SENS = 95.0% SPEC = 93.7% DSC = 74.7% |
| Wu et al., 2022 [40] | Cystoscopy images | CNN (ResNet) | Cystoscopy: Detection | Support Systems for Cystoscopic Diagnosis of BCa | ACC = 93.9%, SENS = 95.4% |
| Xu et al. [84] 2022 | CT images 60 pt | CNN YOLO | Histopathology: pT stage | Predicting pT stage at pre-operative CT scan | CR: T1 stage = 50.01% T2a = 91.65%, T2b, T3 and T4 stage = 100.00% |
| Zou et al., 2022 [85] | T2w MRI images Prospective cohort | CNN Inception V3 | Histopathology: pT stage | CNN to extract features and build a model predicting pT stage | ACC = 92.3% SENS = 100% SPEC = 88.5% |
| Zhang et al., 2023 [58] | Cystoscopy images | U-Net | Segmentation | Deep Learning Tumor Segmentation during cystoscopy | Dice = 82.7% MioU = 69% |
| Li et al., 2023 [82] | T2w MRI images | CNN LASSO SVM | Histopathology: pT stage | Accuracy of radiomics, single- and multi-task DL on T2w MRI images for staging | Radiomics-AUC = 0.920 Singletask = AUC = 0.933 Multitask = AUC = 0.932 |
| Sarkar et al., 2023 [69] | CT | Hybrid ML and DL | Histopathology: Detection Staging | Hybrid ML and DL model to automatically detect and stage BCa | Detection: ACC = 86.07% Staging: ACC = 79.72% |
| Li et al., 2023 [83] | T2w MRI images | CNN VI-RADS | Staging | DL-CNN model based on T2w vs. VI-RADS in BCa staging | (CNN) AUC = 0.963 (VIRADS) AUC = 0.84 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ferro, M.; Falagario, U.G.; Barone, B.; Maggi, M.; Crocetto, F.; Busetto, G.M.; Giudice, F.d.; Terracciano, D.; Lucarelli, G.; Lasorsa, F.; et al. Artificial Intelligence in the Advanced Diagnosis of Bladder Cancer-Comprehensive Literature Review and Future Advancement. Diagnostics 2023, 13, 2308. https://doi.org/10.3390/diagnostics13132308
Ferro M, Falagario UG, Barone B, Maggi M, Crocetto F, Busetto GM, Giudice Fd, Terracciano D, Lucarelli G, Lasorsa F, et al. Artificial Intelligence in the Advanced Diagnosis of Bladder Cancer-Comprehensive Literature Review and Future Advancement. Diagnostics. 2023; 13(13):2308. https://doi.org/10.3390/diagnostics13132308
Chicago/Turabian StyleFerro, Matteo, Ugo Giovanni Falagario, Biagio Barone, Martina Maggi, Felice Crocetto, Gian Maria Busetto, Francesco del Giudice, Daniela Terracciano, Giuseppe Lucarelli, Francesco Lasorsa, and et al. 2023. "Artificial Intelligence in the Advanced Diagnosis of Bladder Cancer-Comprehensive Literature Review and Future Advancement" Diagnostics 13, no. 13: 2308. https://doi.org/10.3390/diagnostics13132308
APA StyleFerro, M., Falagario, U. G., Barone, B., Maggi, M., Crocetto, F., Busetto, G. M., Giudice, F. d., Terracciano, D., Lucarelli, G., Lasorsa, F., Catellani, M., Brescia, A., Mistretta, F. A., Luzzago, S., Piccinelli, M. L., Vartolomei, M. D., Jereczek-Fossa, B. A., Musi, G., Montanari, E., ... Tataru, O. S. (2023). Artificial Intelligence in the Advanced Diagnosis of Bladder Cancer-Comprehensive Literature Review and Future Advancement. Diagnostics, 13(13), 2308. https://doi.org/10.3390/diagnostics13132308













