YOLOv5-FPN: A Robust Framework for Multi-Sized Cell Counting in Fluorescence Images
Abstract
:1. Introduction
- We utilized YOLOv5, a state-of-the-art object detection algorithm, for cell counting in fluorescence microscopy images.
- We employed the FPN as a feature extractor to handle cells of different sizes in the images.
- We annotated the cell images with bounding boxes using a labeling tool for training the YOLOv5 model.
- We augmented the original dataset of 283 images to 600 images with rotation, scaling, and flipping to improve the model’s performance.
- We evaluated the performance of the YOLOv5 model with an FPN on the cell counting task and compared it to other YOLOv5 model versions.
2. Literature Review
3. Baseline Architecture
3.1. Overview of YOLOv5 Architecture
3.2. Overview of FPN Architecture and Implementation
3.3. Combining YOLOv5 and FPN
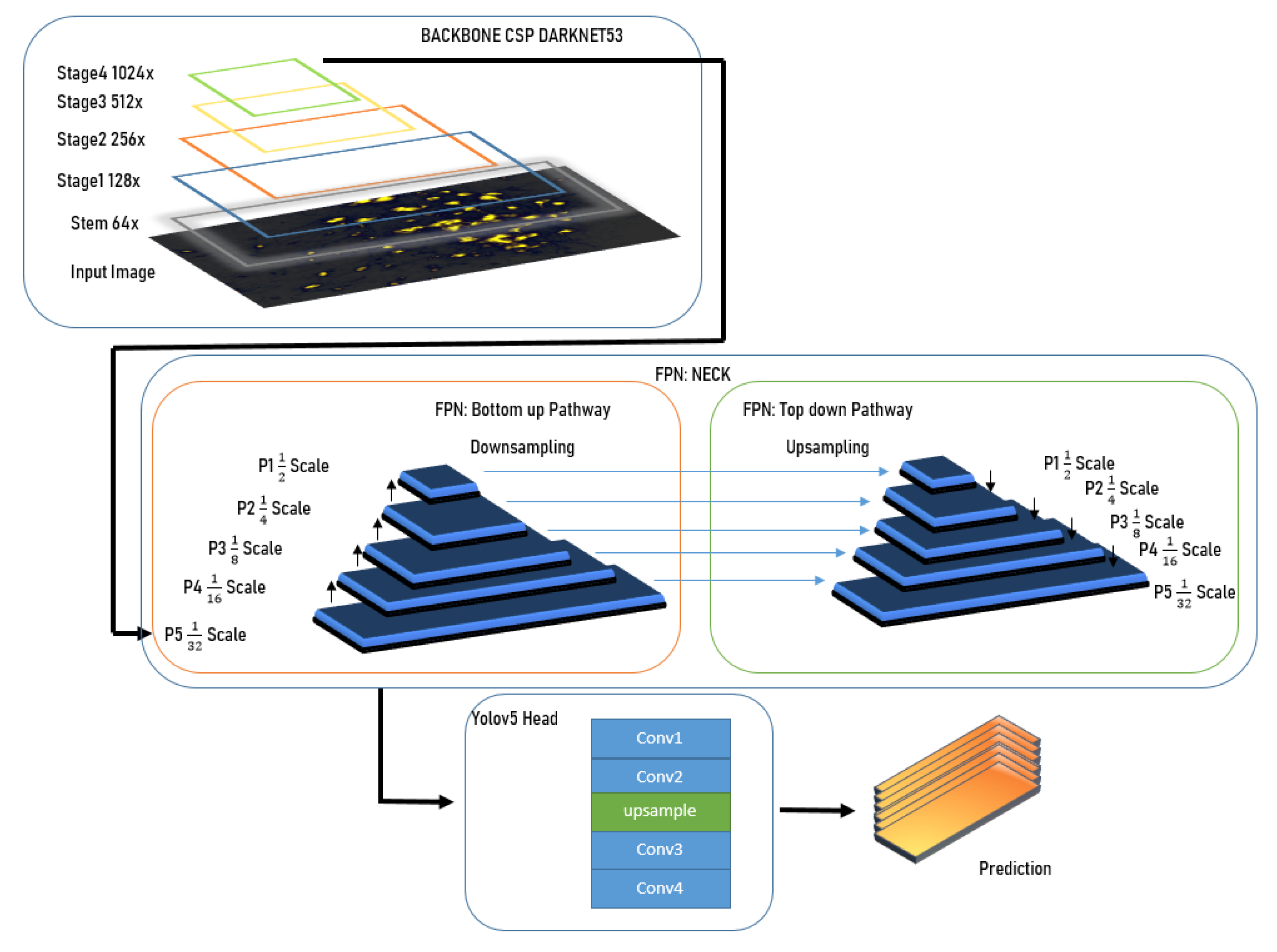
3.3.1. Backbone
3.3.2. Neck
3.3.3. Head
4. Methodology
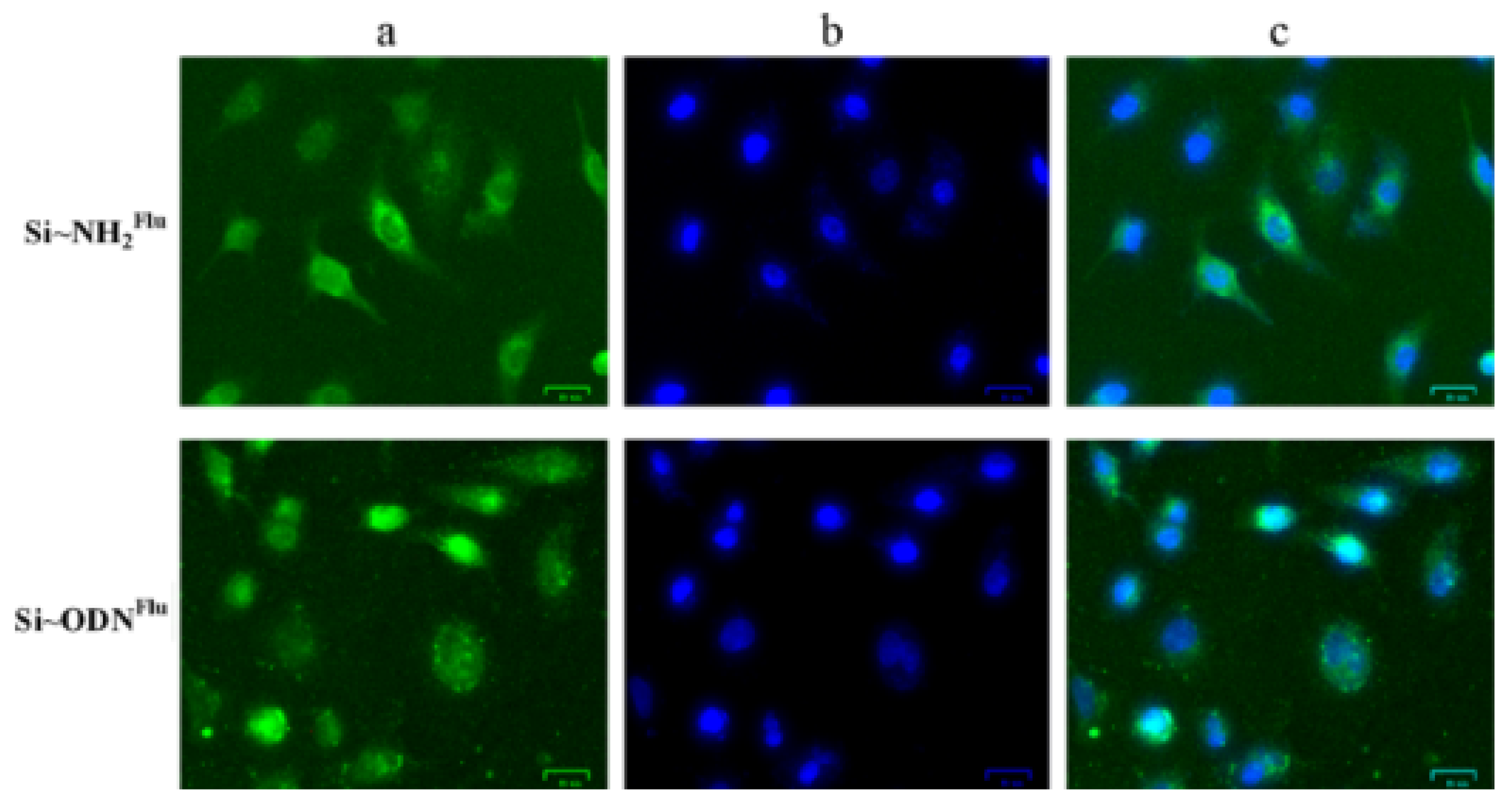
4.1. Dataset Preparation
4.2. Dataset Augmentation
- Horizontal flipping: The images were horizontally flipped to generate new images.
- Rotation: The images were rotated at different angles to create variations in the cell positions and orientations.
- Brightness and contrast adjustment: The brightness and contrast of the images were adjusted within a range of −40 to +40 to simulate different lighting conditions and highlight the dim and dull cells.
4.3. Customizing YOLOv5
| Algorithm 1 Customized YOLOv5 Model with FPN. |
|
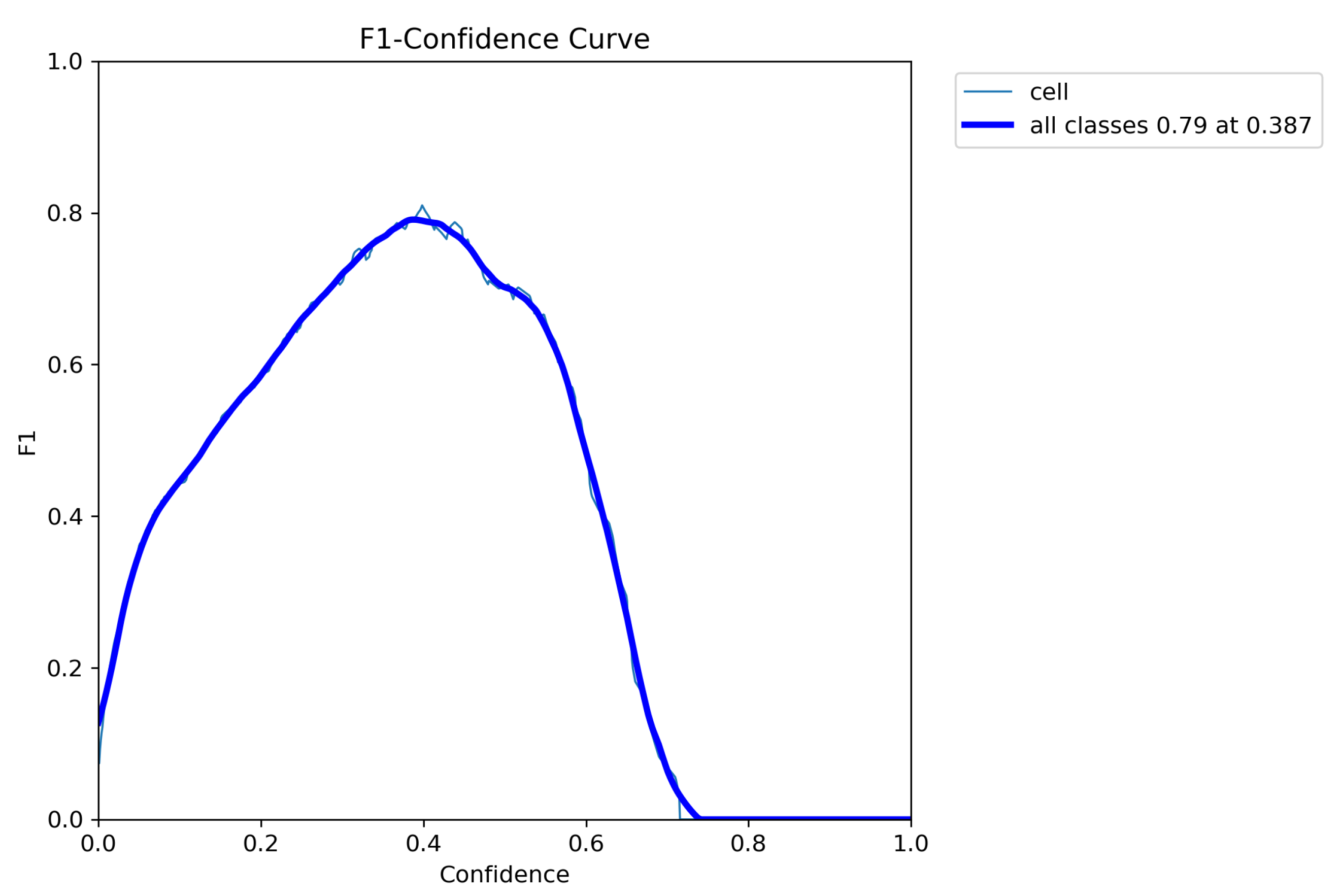
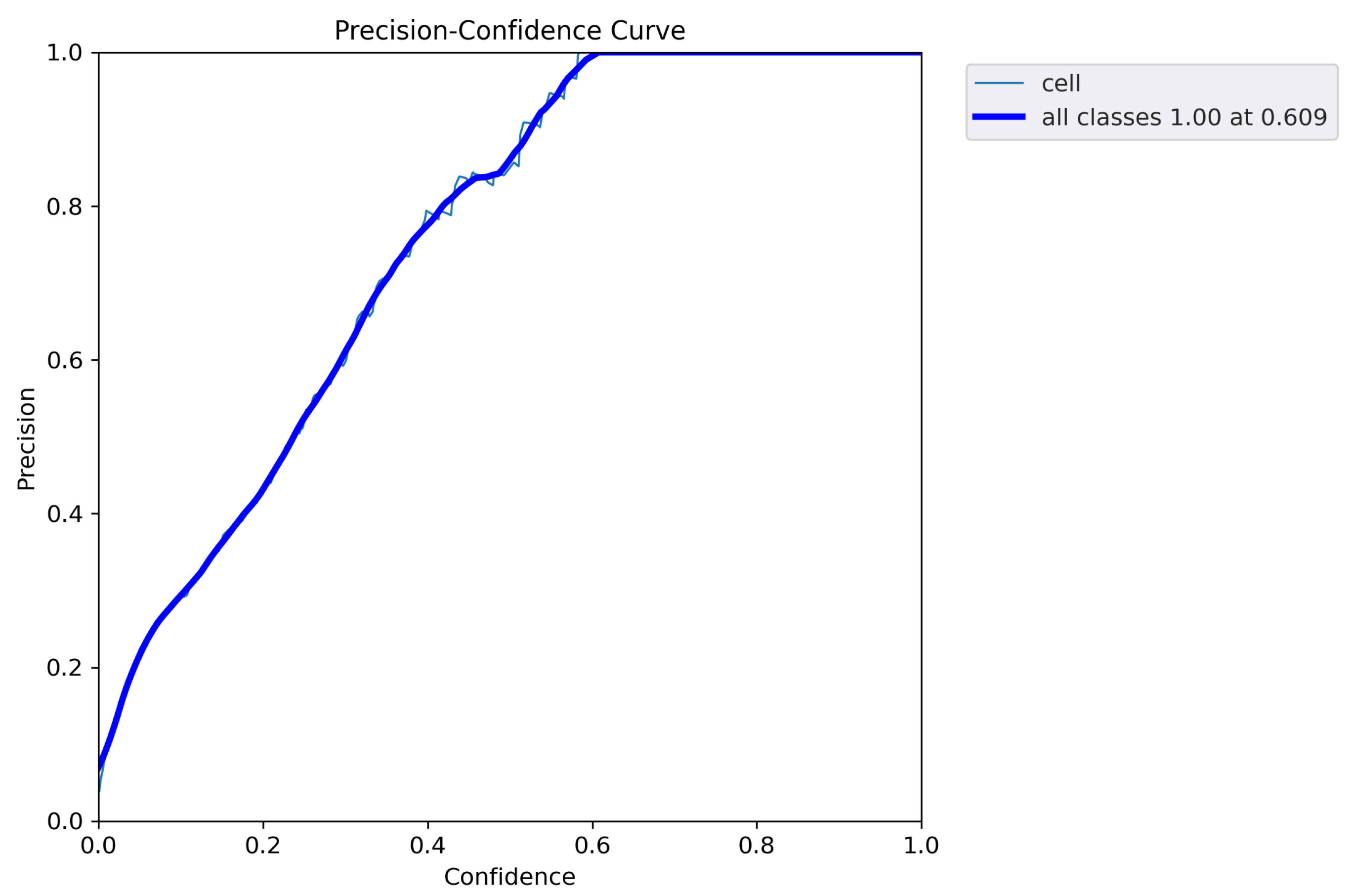
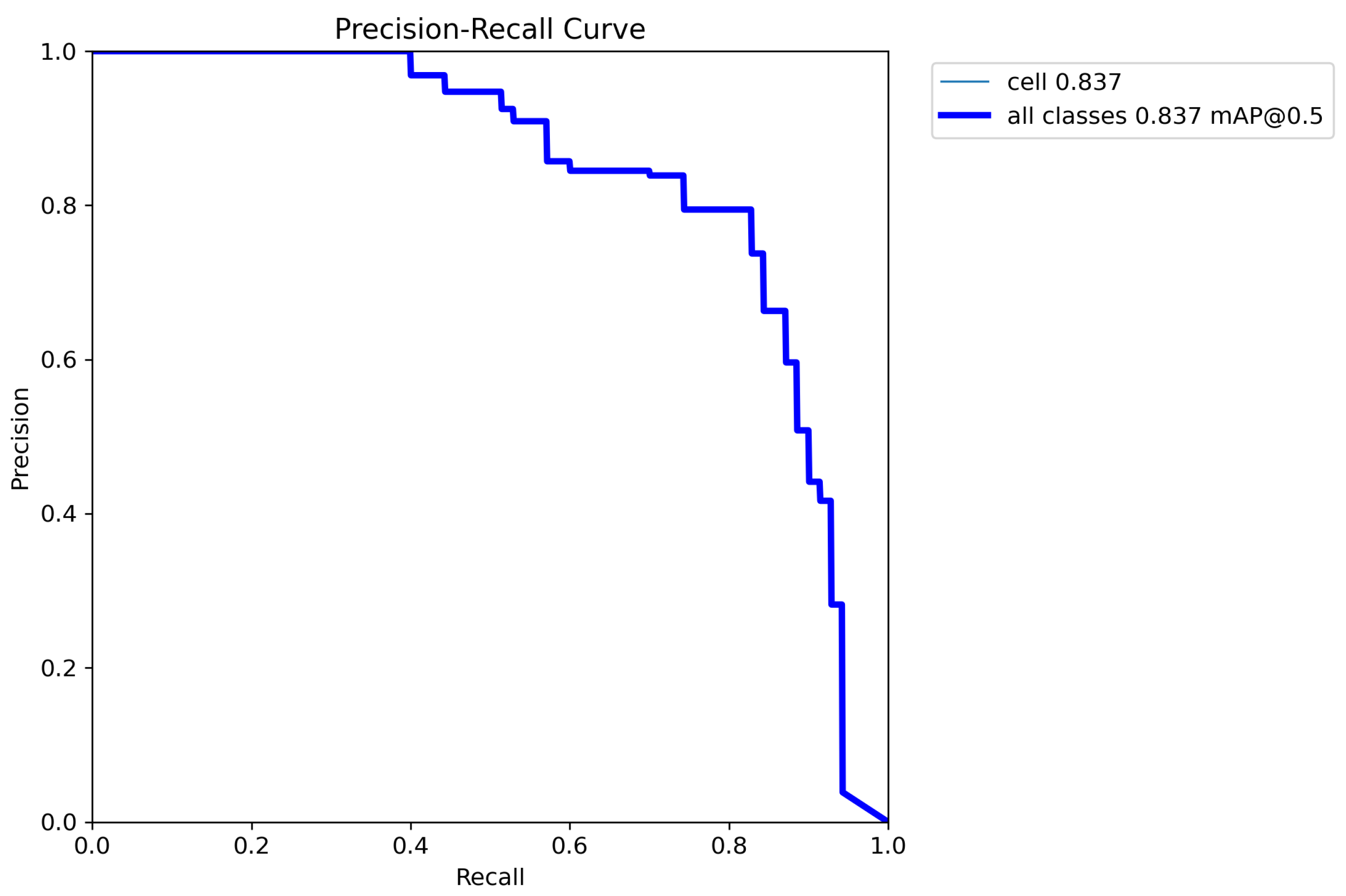
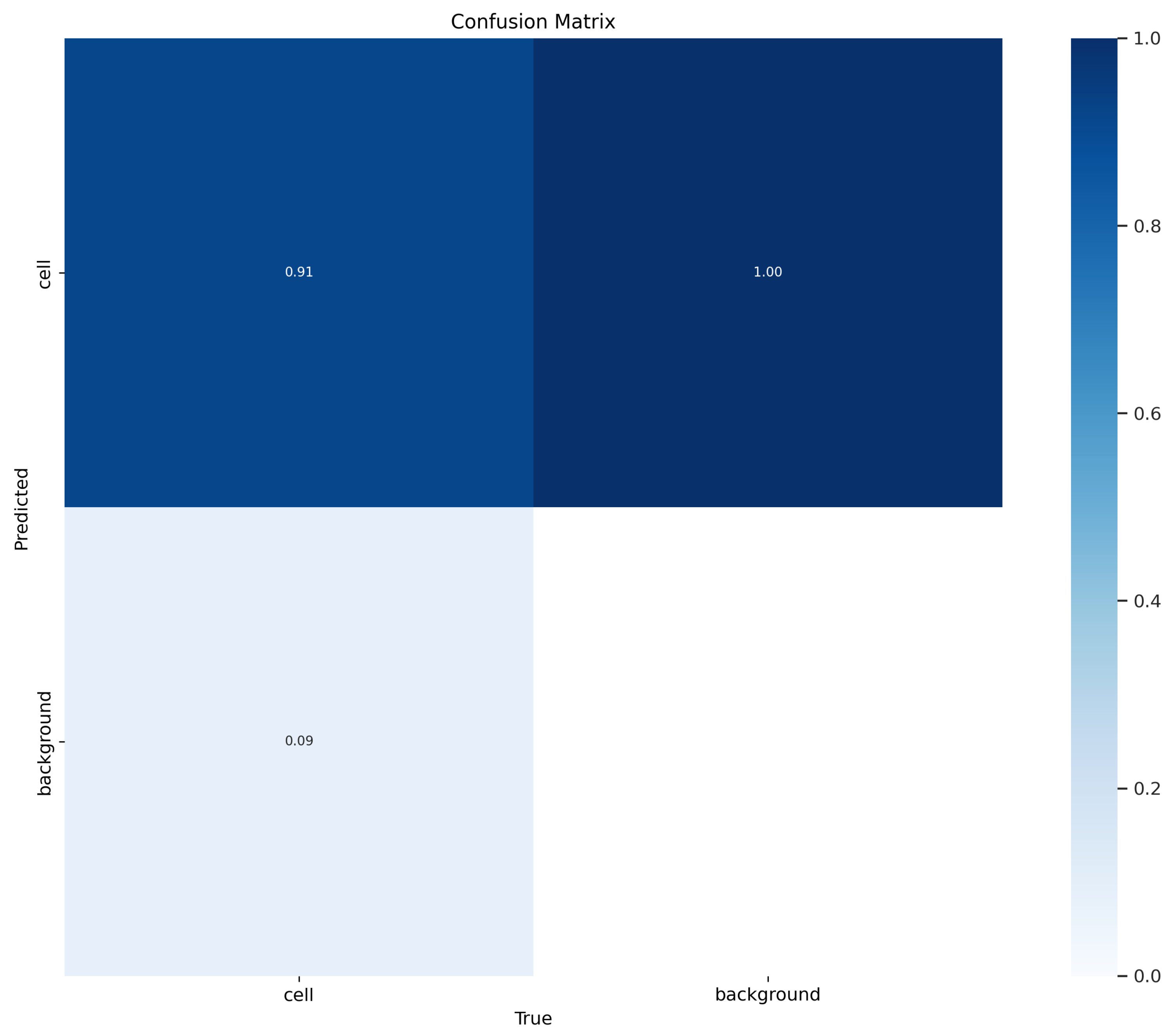
5. Results
6. Discussion
7. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| 2D | Two Dimensional |
| 3D | Three Dimensional |
| CNN | Convolution Neural Network |
| CSP | Cross-Stage Partial |
| DCNN | Deep Convolution Neural Network |
| FCN | Fully Convolution Network |
| FPN | Feature Pyramid Network |
| GAN | Generative Adversarial Network |
| IoU | Intersection Over Union |
| PAN | Path Aggregation Network |
| mAP | Mean Average Precision |
| Mtb | Mycobacterium tuberculosis |
| SSD | Single-Shot Detector |
| UNET | U-Shaped Convolutional Network |
| YOLO | You Only Look Once |
References
- Piccinini, F.; Tesei, A.; Arienti, C.; Bevilacqua, A. Cell counting and viability assessment of 2D and 3D cell cultures: Expected reliability of the trypan blue assay. Biol. Proced. Online 2017, 19, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Ongena, K.; Das, C.; Smith, J.L.; Gil, S.; Johnston, G. Determining cell number during cell culture using the Scepter cell counter. JoVE (J. Vis. Exp.) 2010, e2204. [Google Scholar] [CrossRef]
- Barbosa, M.A.; Xavier, C.P.; Pereira, R.F.; Petrikaitė, V.; Vasconcelos, M.H. 3D cell culture models as recapitulators of the tumor microenvironment for the screening of anti-cancer drugs. Cancers 2022, 14, 190. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Lee, A.M.; Jena, S.; Huang, Y.; Ho, Y.; Tietz, K.T.; Miller, C.R.; Su, M.C.; Mentzer, J.; Ling, A.L.; et al. Computational drug discovery for castration-resistant prostate cancers through in vitro drug response modeling. Proc. Natl. Acad. Sci. USA 2023, 120, e2218522120. [Google Scholar] [CrossRef] [PubMed]
- Briggs, C. Quality counts: New parameters in blood cell counting. Int. J. Lab. Hematol. 2009, 31, 277–297. [Google Scholar] [CrossRef]
- Almufareh, M.F.; Tehsin, S.; Humayun, M.; Kausar, S. A Transfer Learning Approach for Clinical Detection Support of Monkeypox Skin Lesions. Diagnostics 2023, 13, 1503. [Google Scholar] [CrossRef]
- Zou, L.h.; Chen, J.; Zhang, J.; Garcia, N. Malaria cell counting diagnosis within large field of view. In Proceedings of the 2010 International Conference on Digital Image Computing: Techniques and Applications, Sydney, Australia, 1–3 December 2010; pp. 172–177. [Google Scholar]
- Guo, J.; Ge, J.; Guo, Y. Recent advances in methods for the diagnosis of Corona Virus Disease 2019. J. Clin. Lab. Anal. 2022, 36, e24178. [Google Scholar] [CrossRef]
- Pillai, S.; Kunze, E.; Sordillo, L.; Jayarao, B. Application of differential inflammatory cell count as a tool to monitor udder health. J. Dairy Sci. 2001, 84, 1413–1420. [Google Scholar] [CrossRef]
- Phelan, M.C.; Lawler, G. Cell counting. Curr. Protoc. Cytom. 1997, A.3A.1–A.3A.4. [Google Scholar] [CrossRef]
- Hedreen, J.C. What was wrong with the Abercrombie and empirical cell counting methods? A review. Anat. Rec. Off. Publ. Am. Assoc. Anat. 1998, 250, 373–380. [Google Scholar] [CrossRef]
- Kickler, T.S. Clinical analyzers. Advances in automated cell counting. Anal. Chem. 1999, 71, 363–365. [Google Scholar] [CrossRef] [PubMed]
- Alcaide Martín, M.J.; Altimira Queral, L.; Sahuquillo Frías, L.; Valiña Amado, L.; Merino, A.; García de Guadiana-Romualdo, L. Automated cell count in body fluids: A review. Adv. Lab. Med. Med. Lab. 2021, 2, 149–161. [Google Scholar] [CrossRef] [PubMed]
- Cadena-Herrera, D.; Esparza-De Lara, J.E.; Ramírez-Ibañez, N.D.; López-Morales, C.A.; Pérez, N.O.; Flores-Ortiz, L.F.; Medina-Rivero, E. Validation of three viable-cell counting methods: Manual, semi-automated, and automated. Biotechnol. Rep. 2015, 7, 9–16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jindal, D.; Singh, M. Counting of Cells. In Animal Cell Culture: Principles and Practice; Springer: Cham, Switzerland, 2023; pp. 131–145. [Google Scholar]
- Green, M.R.; Sambrook, J. Estimation of cell number by hemocytometry counting. Cold Spring Harb. Protoc. 2019, 2019, pdb-prot097980. [Google Scholar] [CrossRef] [PubMed]
- Pioli, P.D. Protocol: Hemocytometer Cell Counting, 2019. Available online: https://med.wmich.edu/sites/default/files/Hemacytometer_Cell_Counting.pdf (accessed on 8 May 2023).
- Vembadi, A.; Menachery, A.; Qasaimeh, M.A. Cell cytometry: Review and perspective on biotechnological advances. Front. Bioeng. Biotechnol. 2019, 7, 147. [Google Scholar] [CrossRef]
- Khan, S.; Khan, A.; Khattak, F.S.; Naseem, A. An accurate and cost effective approach to blood cell count. Int. J. Comput. Appl. 2012, 50, 18–24. [Google Scholar] [CrossRef] [Green Version]
- Mattern, C.F.; Brackett, F.S.; Olson, B.J. Determination of number and size of particles by electrical gating: Blood cells. J. Appl. Physiol. 1957, 10, 56–70. [Google Scholar] [CrossRef]
- Avci, M.B.; Yasar, S.D.; Cetin, A.E. An Optofluidic Platform for Cell-Counting Applications. Anal. Methods 2023, 15, 2244–2252. [Google Scholar] [CrossRef]
- Philipson, L. Adenovirus assay by the fluorescent cell-counting procedure. Virology 1961, 15, 263–268. [Google Scholar] [CrossRef]
- Chiu, R.J.; Black, L. Assay of wound tumor virus by the fluorescent cell counting technique. Virology 1969, 37, 667–677. [Google Scholar] [CrossRef]
- Levina, A.; Repkova, M.; Shikina, N.; Ismagilov, Z.; Yashnik, S.; Semenov, D.; Savinovskaya, Y.; Mazurkova, N.; Pyshnaya, I.; Zarytova, V. Non-agglomerated silicon-organic nanoparticles and their nanocomplexes with oligonucleotides: Synthesis and properties. Beilstein J. Nanotechnol. 2018, 9, 2516–2525. [Google Scholar] [CrossRef] [Green Version]
- Lo, C.H.; Schaaf, T.M.; Thomas, D.D.; Sachs, J.N. Fluorescence-based TNFR1 biosensor for monitoring receptor structural and conformational dynamics and discovery of small molecule modulators. In The TNF Superfamily: Methods and Protocols; Humana: New York, NY, USA, 2021; pp. 121–137. [Google Scholar]
- Sciences, C.L. Introduction to Fluorescence Cell Counting. 2021. Available online: https://www.corning.com/catalog/cls/documents/application-notes/CLS-AN-495.pdf (accessed on 30 April 2023).
- Parvaiz, A.; Khalid, M.A.; Zafar, R.; Ameer, H.; Ali, M.; Fraz, M.M. Vision transformers in medical computer vision—A contemplative retrospection. Eng. Appl. Artif. Intell. 2023, 122, 106126. [Google Scholar] [CrossRef]
- Greener, J.G.; Kandathil, S.M.; Moffat, L.; Jones, D.T. A guide to machine learning for biologists. Nat. Rev. Mol. Cell Biol. 2022, 23, 40–55. [Google Scholar] [CrossRef] [PubMed]
- Bereciartua-Pérez, A.; Gómez, L.; Picón, A.; Navarra-Mestre, R.; Klukas, C.; Eggers, T. Insect counting through deep learning-based density maps estimation. Comput. Electron. Agric. 2022, 197, 106933. [Google Scholar] [CrossRef]
- Asha, S.; Gopakumar, G.; Subrahmanyam, G.R.S. Saliency and ballness driven deep learning framework for cell segmentation in bright field microscopic images. Eng. Appl. Artif. Intell. 2023, 118, 105704. [Google Scholar] [CrossRef]
- Chen, X.; Kandel, M.E.; He, S.; Hu, C.; Lee, Y.J.; Sullivan, K.; Tracy, G.; Chung, H.J.; Kong, H.J.; Anastasio, M.; et al. Artificial confocal microscopy for deep label-free imaging. Nat. Photonics 2023, 17, 250–258. [Google Scholar] [CrossRef]
- Flórez, S.L.; González-Briones, A.; Hernández, G.; de la Prieta, F. Automated Counting via Multicolumn Network and CytoSMART Exact FL Microscope. In Ambient Intelligence—Software and Applications—13th International Symposium on Ambient Intelligence; Springer: Cham, Switzerland, 2023; pp. 207–218. [Google Scholar]
- Venkatapathiah, S.K.; Selvan, S.S.; Nanda, P.; Shetty, M.; Swamy, V.M.; Awasthi, K. Deep learning based object detection in nailfold capillary images. IAES Int. J. Artif. Intell. 2023, 12, 931. [Google Scholar] [CrossRef]
- Kothari, S.; Chaudry, Q.; Wang, M.D. Automated cell counting and cluster segmentation using concavity detection and ellipse fitting techniques. In Proceedings of the 2009 IEEE International Symposium on Biomedical Imaging: From Nano to Macro, Boston, MA, USA, 28 June–1 July 2009; pp. 795–798. [Google Scholar]
- Mohapatra, S.; Patra, D. Automated cell nucleus segmentation and acute leukemia detection in blood microscopic images. In Proceedings of the 2010 International Conference on Systems in Medicine and Biology, Kharagpur, India, 16–18 December 2010; pp. 49–54. [Google Scholar]
- Fenistein, D.; Lenseigne, B.; Christophe, T.; Brodin, P.; Genovesio, A. A fast, fully automated cell segmentation algorithm for high-throughput and high-content screening. Cytom. Part A J. Int. Soc. Anal. Cytol. 2008, 73, 958–964. [Google Scholar] [CrossRef]
- Kesler, B.; Li, G.; Thiemicke, A.; Venkat, R.; Neuert, G. Automated cell boundary and 3D nuclear segmentation of cells in suspension. Sci. Rep. 2019, 9, 10237. [Google Scholar] [CrossRef] [Green Version]
- Mandal, S.; Uhlmann, V. Splinedist: Automated cell segmentation with spline curves. In Proceedings of the 2021 IEEE 18th International Symposium on Biomedical Imaging (ISBI), Nice, France, 13–16 April 2021; pp. 1082–1086. [Google Scholar]
- Jiang, P.; Ergu, D.; Liu, F.; Cai, Y.; Ma, B. A Review of Yolo algorithm developments. Procedia Comput. Sci. 2022, 199, 1066–1073. [Google Scholar] [CrossRef]
- Redmon, J.; Divvala, S.; Girshick, R.; Farhadi, A. You only look once: Unified, real-time object detection. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016; pp. 779–788. [Google Scholar]
- Redmon, J.; Farhadi, A. YOLO9000: Better, faster, stronger. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21–26 July 2017; pp. 7263–7271. [Google Scholar]
- Humayun, M.; Ashfaq, F.; Jhanjhi, N.Z.; Alsadun, M.K. Traffic management: Multi-scale vehicle detection in varying weather conditions using yolov4 and spatial pyramid pooling network. Electronics 2022, 11, 2748. [Google Scholar] [CrossRef]
- Alwakid, G.; Gouda, W.; Humayun, M.; Jhanjhi, N.Z. Diagnosing Melanomas in Dermoscopy Images Using Deep Learning. Diagnostics 2023, 13, 1815. [Google Scholar] [CrossRef] [PubMed]
- Aldughayfiq, B.; Ashfaq, F.; Jhanjhi, N.; Humayun, M. YOLO-Based Deep Learning Model for Pressure Ulcer Detection and Classification. Healthcare 2023, 11, 1222. [Google Scholar] [CrossRef] [PubMed]
- Alwakid, G.; Gouda, W.; Humayun, M. Deep Learning-based prediction of Diabetic Retinopathy using CLAHE and ESRGAN for Enhancemen. Healthcare 2023, 11, 863. [Google Scholar] [CrossRef]
- Aldughayfiq, B.; Ashfaq, F.; Jhanjhi, N.; Humayun, M. Explainable AI for Retinoblastoma Diagnosis: Interpreting Deep Learning Models with LIME and SHAP. Diagnostics 2023, 13, 1932. [Google Scholar] [CrossRef]
- Ashfaq, F.; Ghoniem, R.; Jhanjhi, N.; Khan, N.; Algarni, A. Using Dual Attention BiLSTM to Predict Vehicle Lane Changing Maneuvers on Highway Dataset. Systems 2023, 11, 196. [Google Scholar] [CrossRef]
- Peng, F.; Miao, Z.; Li, F.; Li, Z. S-FPN: A shortcut feature pyramid network for sea cucumber detection in underwater images. Expert Syst. Appl. 2021, 182, 115306. [Google Scholar] [CrossRef]
- Blay, V.; Tolani, B.; Ho, S.P.; Arkin, M.R. High-throughput screening: Today’s biochemical and cell-based approaches. Drug Discov. Today 2020, 25, 1807–1821. [Google Scholar] [CrossRef]
- Kuusanmäki, H.; Leppä, A.M.; Pölönen, P.; Kontro, M.; Dufva, O.; Deb, D.; Yadav, B.; Brück, O.; Kumar, A.; Everaus, H.; et al. Phenotype-based drug screening reveals association between venetoclax response and differentiation stage in acute myeloid leukemia. Haematologica 2020, 105, 708. [Google Scholar] [CrossRef]
- Costa, A.F.; Campos, D.; Reis, C.A.; Gomes, C. Targeting glycosylation: A new road for cancer drug discovery. Trends Cancer 2020, 6, 757–766. [Google Scholar] [CrossRef]
- Alam, M.M.; Islam, M.T. Machine learning approach of automatic identification and counting of blood cells. Healthc. Technol. Lett. 2019, 6, 103–108. [Google Scholar] [CrossRef] [PubMed]
- Schüffler, P.J.; Fuchs, T.J.; Ong, C.S.; Wild, P.J.; Rupp, N.J.; Buhmann, J.M. TMARKER: A free software toolkit for histopathological cell counting and staining estimation. J. Pathol. Informatics 2013, 4, 2. [Google Scholar] [CrossRef] [PubMed]
- Guo, X.; Yu, F. A method of automatic cell counting based on microscopic image. In Proceedings of the 2013 5th International Conference on Intelligent Human-Machine Systems and Cybernetics, Hangzhou, China, 26–27 August 2013; Volume 1, pp. 293–296. [Google Scholar]
- O’Brien, J.; Hayder, H.; Peng, C. Automated quantification and analysis of cell counting procedures using ImageJ plugins. JoVE (J. Vis. Exp.) 2016, e54719. [Google Scholar] [CrossRef]
- Santen, R.J. Automated estimation of diploid and tetraploid nuclei with an electronic particle counter. Exp. Cell Res. 1965, 40, 413–420. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ertürk, İ.F. Automatic Cell Counting from Microchannel Images. Master’s Thesis, Middle East Technical University, Ankara, Turkey, 2022. [Google Scholar]
- Akkoyun, F.; Öcçelik, A. Rapid characterization of cell and bacteria counts using computer vision. Türk Doğa Ve Fen Dergisi 2021, 10, 269–274. [Google Scholar] [CrossRef]
- Biswas, S.; Ghoshal, D. Blood cell detection using thresholding estimation based watershed transformation with Sobel filter in frequency domain. Procedia Comput. Sci. 2016, 89, 651–657. [Google Scholar] [CrossRef] [Green Version]
- Pandit, A.; Kolhar, S.; Patil, P. Survey on automatic rbc detection and counting. Int. J. Adv. Res. Electr. Electron. Instrum. Eng. 2015, 4, 128–131. [Google Scholar]
- Li, C.; Ma, X.; Deng, J.; Li, J.; Liu, Y.; Zhu, X.; Liu, J.; Zhang, P. Machine learning-based automated fungal cell counting under a complicated background with ilastik and ImageJ. Eng. Life Sci. 2021, 21, 769–777. [Google Scholar] [CrossRef]
- Hagen, G.M.; Bendesky, J.; Machado, R.; Nguyen, T.A.; Kumar, T.; Ventura, J. Fluorescence microscopy datasets for training deep neural networks. GigaScience 2021, 10, giab032. [Google Scholar] [CrossRef]
- Zhang, Q.; Yun, K.K.; Wang, H.; Yoon, S.W.; Lu, F.; Won, D. Automatic cell counting from stimulated Raman imaging using deep learning. PLoS ONE 2021, 16, e0254586. [Google Scholar] [CrossRef]
- Caicedo, J.C.; Roth, J.; Goodman, A.; Becker, T.; Karhohs, K.W.; Broisin, M.; Molnar, C.; McQuin, C.; Singh, S.; Theis, F.J.; et al. Evaluation of deep learning strategies for nucleus segmentation in fluorescence images. Cytom. Part A 2019, 95, 952–965. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, J.; Tárnok, A.; Su, X. Deep learning-based single-cell optical image studies. Cytom. Part A 2020, 97, 226–240. [Google Scholar] [CrossRef]
- Dvanesh, V.D.; Lakshmi, P.S.; Reddy, K.; Vasavi, A.S. Blood cell count using digital image processing. In Proceedings of the 2018 International Conference on Current Trends towards Converging Technologies (ICCTCT), Coimbatore, India, 1–3 March 2018; pp. 1–7. [Google Scholar]
- Grishagin, I.V. Automatic cell counting with ImageJ. Anal. Biochem. 2015, 473, 63–65. [Google Scholar] [CrossRef]
- Huang, X.; Jiang, Y.; Liu, X.; Xu, H.; Han, Z.; Rong, H.; Yang, H.; Yan, M.; Yu, H. Machine learning based single-frame super-resolution processing for lensless blood cell counting. Sensors 2016, 16, 1836. [Google Scholar] [CrossRef] [Green Version]
- Falk, T.; Mai, D.; Bensch, R.; Çiçek, Ö.; Abdulkadir, A.; Marrakchi, Y.; Böhm, A.; Deubner, J.; Jäckel, Z.; Seiwald, K.; et al. U-Net: Deep learning for cell counting, detection, and morphometry. Nat. Methods 2019, 16, 67–70. [Google Scholar] [CrossRef] [PubMed]
- Morelli, R.; Clissa, L.; Amici, R.; Cerri, M.; Hitrec, T.; Luppi, M.; Rinaldi, L.; Squarcio, F.; Zoccoli, A. Automating cell counting in fluorescent microscopy through deep learning with c-ResUnet. Sci. Rep. 2021, 11, 22920. [Google Scholar] [CrossRef] [PubMed]
- Dhieb, N.; Ghazzai, H.; Besbes, H.; Massoud, Y. An automated blood cells counting and classification framework using mask R-CNN deep learning model. In Proceedings of the 2019 31st International Conference on Microelectronics (ICM), Cairo, Egypt, 15–18 December 2019; pp. 300–303. [Google Scholar]
- Kayasandik, C.B.; Ru, W.; Labate, D. A multistep deep learning framework for the automated detection and segmentation of astrocytes in fluorescent images of brain tissue. Sci. Rep. 2020, 10, 5137. [Google Scholar] [CrossRef] [Green Version]
- Tsuzuki, Y.; Sanami, S.; Sugimoto, K.; Fujita, S. Pseudo-nuclear staining of cells by deep learning improves the accuracy of automated cell counting in a label-free cellular population. J. Biosci. Bioeng. 2021, 131, 213–218. [Google Scholar] [CrossRef]
- Zachariou, M.; Arandjelović, O.; Sabiiti, W.; Mtafya, B.; Sloan, D. Tuberculosis bacteria detection and counting in fluorescence microscopy images using a multi-stage deep learning pipeline. Information 2022, 13, 96. [Google Scholar] [CrossRef]
- Khan, A.; Gould, S.; Salzmann, M. Deep convolutional neural networks for human embryonic cell counting. In Proceedings of the Computer Vision–ECCV 2016 Workshops, Amsterdam, The Netherlands, 8–10 and 15–16 October 2016; Proceedings, Part I 14. Springer: Cham, Switzerland, 2016; pp. 339–348. [Google Scholar]
- He, S.; Minn, K.T.; Solnica-Krezel, L.; Anastasio, M.A.; Li, H. Deeply-supervised density regression for automatic cell counting in microscopy images. Med Image Anal. 2021, 68, 101892. [Google Scholar] [CrossRef]
- Lugagne, J.B.; Lin, H.; Dunlop, M.J. DeLTA: Automated cell segmentation, tracking, and lineage reconstruction using deep learning. PLoS Comput. Biol. 2020, 16, e1007673. [Google Scholar] [CrossRef] [Green Version]
- Jeong, E.J.; Choi, D.; Lee, D.W. U-Net Deep-Learning-Based 3D Cell Counter for the Quality Control of 3D Cell-Based Assays through Seed Cell Measurement. SLAS Technol. Transl. Life Sci. Innov. 2021, 26, 468–476. [Google Scholar] [CrossRef]
- Korfhage, N.; Mühling, M.; Ringshandl, S.; Becker, A.; Schmeck, B.; Freisleben, B. Detection and segmentation of morphologically complex eukaryotic cells in fluorescence microscopy images via feature pyramid fusion. PLoS Comput. Biol. 2020, 16, e1008179. [Google Scholar] [CrossRef] [PubMed]
- Morelli, R.; Clissa, L.; Dalla, M.; Luppi, M.; Rinaldi, L.; Zoccoli, A. Automatic cell counting in flourescent microscopy using deep learning. arXiv 2021, arXiv:2103.01141. [Google Scholar]
- Bie, M.; Liu, Y.; Li, G.; Hong, J.; Li, J. Real-time vehicle detection algorithm based on a lightweight You-Only-Look-Once (YOLOv5n-L) approach. Expert Syst. Appl. 2023, 213, 119108. [Google Scholar] [CrossRef]
- Kumar, A. SEAT-YOLO: A Squeeze-Excite and Spatial Attentive You Only Look Once Architecture for Shadow Detection. Optik 2023, 273, 170513. [Google Scholar] [CrossRef]
- Gallo, I.; Rehman, A.U.; Dehkordi, R.H.; Landro, N.; La Grassa, R.; Boschetti, M. Deep Object Detection of Crop Weeds: Performance of YOLOv7 on a Real Case Dataset from UAV Images. Remote Sens. 2023, 15, 539. [Google Scholar] [CrossRef]
- Lin, T.Y.; Dollár, P.; Girshick, R.; He, K.; Hariharan, B.; Belongie, S. Feature pyramid networks for object detection. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21–26 July 2017; pp. 2117–2125. [Google Scholar]
- Min, K.; Lee, G.H.; Lee, S.W. Attentional feature pyramid network for small object detection. Neural Netw. 2022, 155, 439–450. [Google Scholar] [CrossRef] [PubMed]
- Zhu, L.; Lee, F.; Cai, J.; Yu, H.; Chen, Q. An improved feature pyramid network for object detection. Neurocomputing 2022, 483, 127–139. [Google Scholar] [CrossRef]
- Yang, Q.; Zhang, C.; Wang, H.; He, Q.; Huo, L. SV-FPN: Small Object Feature Enhancement and Variance-Guided RoI Fusion for Feature Pyramid Networks. Electronics 2022, 11, 2028. [Google Scholar] [CrossRef]






| Methods | Advantages | Limitations | Applied to Cell Detection and Counting |
|---|---|---|---|
| Image Processing based | Simple and computationally efficient | Limited accuracy and robustness | [54,66,67] |
| No need for large datasets or complex algorithms | Struggle with complex cell morphologies and low SNR | ||
| Easy to implement and interpret | |||
| Machine Learning based | Can handle complex cell morphologies and low SNR | Requires labeled training data | [52,61,68] |
| More accurate and robust than image processing based | Sensitive to variability in data and imaging protocol | ||
| Can be adapted to different imaging modalities | Requires feature engineering, which can be time-consuming | ||
| Deep Learning based | State-of-the-art accuracy for cell detection and counting | Highly dependent on the quality and quantity of training data | [69,70,71] |
| Highly robust to variability in data and imaging protocol | Can be computationally expensive | ||
| Does not require feature engineering, saving time and effort | May be less interpretable than traditional methods |
| 416 × 416 | 640 × 640 | 840 × 840 | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Precision | Recall | mAP | Precision | Recall | mAP | Precision | Recall | mAP | |
| YOLOv5s | 0.741 | 0.701 | 0.732 | 0.787 | 0.744 | 0.764 | 0.756 | 0.723 | 0.741 |
| YOLOv5n | 0.738 | 0.661 | 0.681 | 0.779 | 0.695 | 0.73 | 0.759 | 0.734 | 0.749 |
| YOLOv5fpn | 0.796 | 0.741 | 0.799 | 0.758 | 0.740 | 0.748 | 0.748 | 0.708 | 0.732 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Aldughayfiq, B.; Ashfaq, F.; Jhanjhi, N.Z.; Humayun, M. YOLOv5-FPN: A Robust Framework for Multi-Sized Cell Counting in Fluorescence Images. Diagnostics 2023, 13, 2280. https://doi.org/10.3390/diagnostics13132280
Aldughayfiq B, Ashfaq F, Jhanjhi NZ, Humayun M. YOLOv5-FPN: A Robust Framework for Multi-Sized Cell Counting in Fluorescence Images. Diagnostics. 2023; 13(13):2280. https://doi.org/10.3390/diagnostics13132280
Chicago/Turabian StyleAldughayfiq, Bader, Farzeen Ashfaq, N. Z. Jhanjhi, and Mamoona Humayun. 2023. "YOLOv5-FPN: A Robust Framework for Multi-Sized Cell Counting in Fluorescence Images" Diagnostics 13, no. 13: 2280. https://doi.org/10.3390/diagnostics13132280
APA StyleAldughayfiq, B., Ashfaq, F., Jhanjhi, N. Z., & Humayun, M. (2023). YOLOv5-FPN: A Robust Framework for Multi-Sized Cell Counting in Fluorescence Images. Diagnostics, 13(13), 2280. https://doi.org/10.3390/diagnostics13132280








