Core Needle Biopsy Guidance Based on Tissue Morphology Assessment with AI-OCT Imaging
Abstract
:1. Introduction
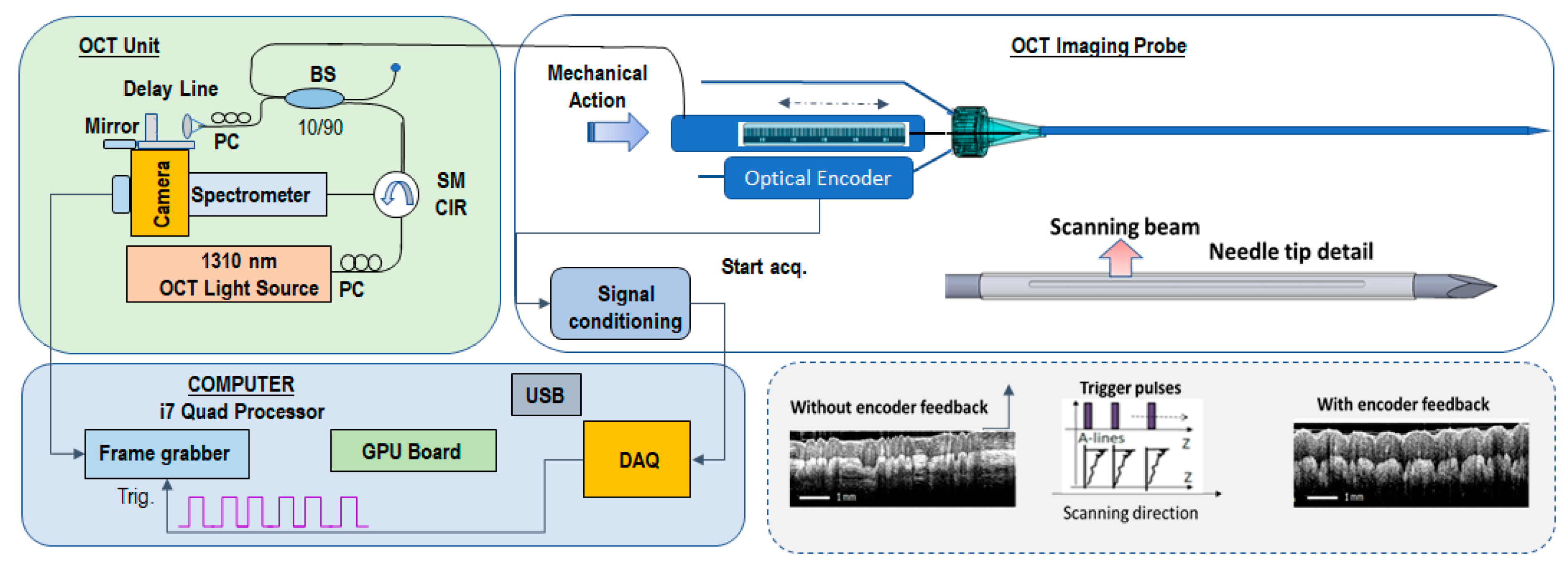
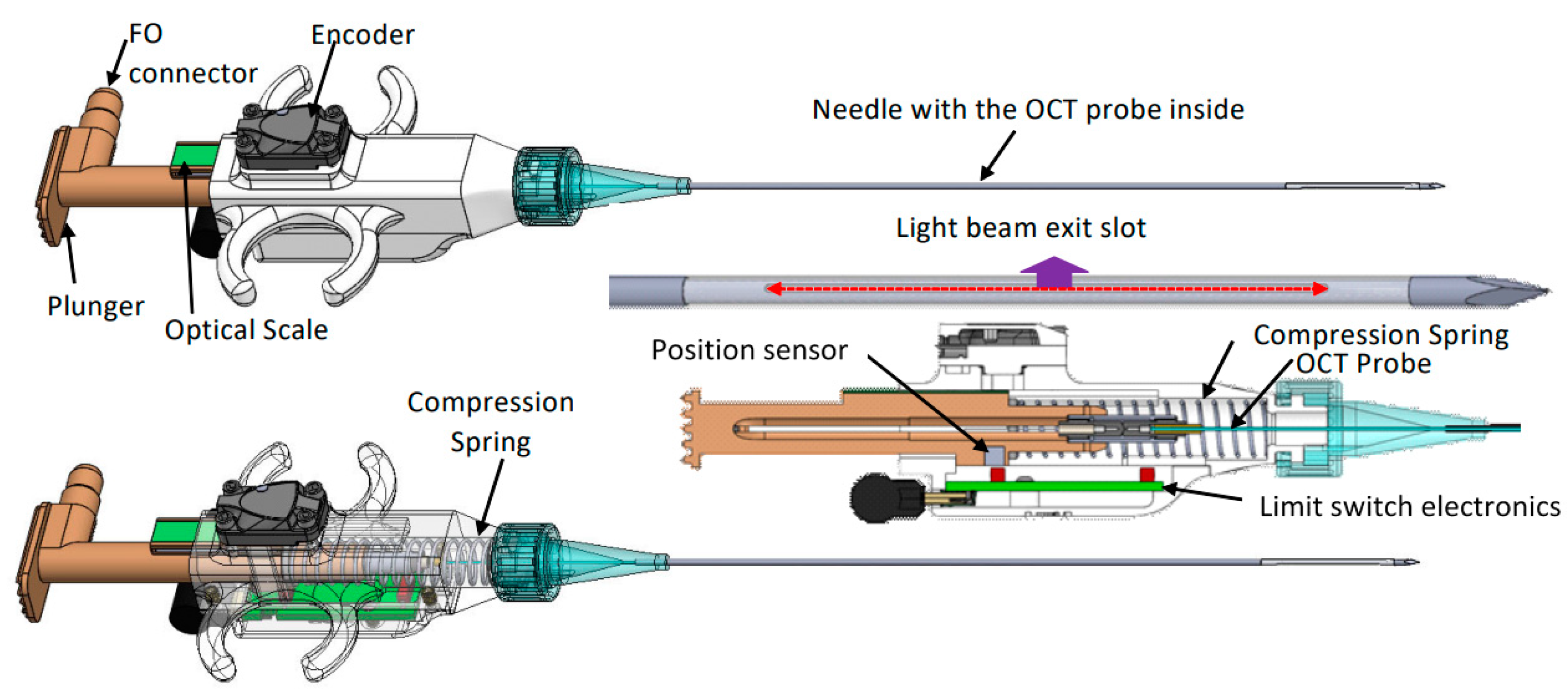
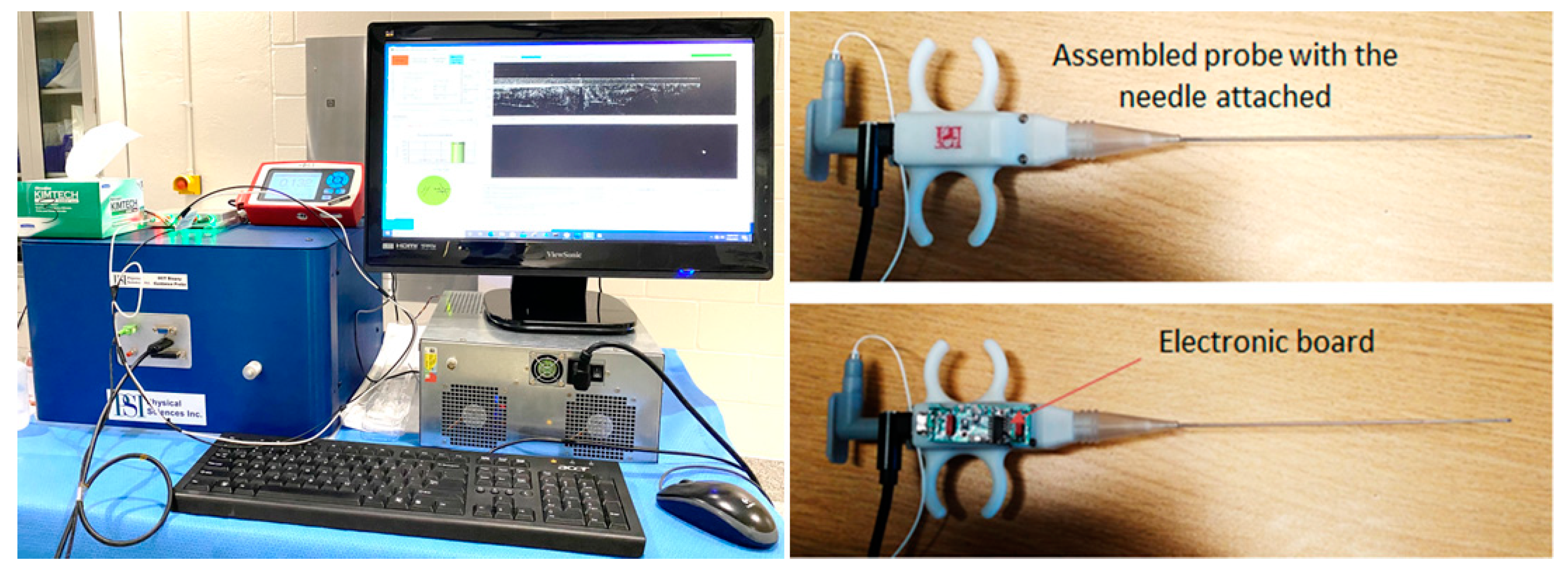
2. Materials and Methods
- (a)
- Percutaneously, intramuscularly inject VX2 tumor in both thighs of each rabbit;
- (b)
- Allow tumor to grow for 10 to 14 days +/− 2 days to reach a size of 1.5 to 2 cm in diameter (appropriate size for use);
- (c)
- Use palpation to verify tumor growth in thighs and determine tumor growth and volume.
- Percutaneously insert a biopsy guidance needle (18 Ga) within the tumor using ultrasound guidance;
- Remove the needle stylet and insert the optical probe into the tumor site through the bore of the guidance needle;
- Perform up to 4 quadrant OCT measurements (4 × 90 deg angular orientations) at each location and collect at least 2 images/quadrant;
- Retract the OCT probe and use an 18 Ga core biopsy gun to collect 1 biopsy core after imaging is performed;
- Reinsert the guidance needle in the tumor-adjacent area and repeat the steps above to collect OCT images of heathy tissue;
- Following the final biopsy, euthanize the animal using Beuthanasia-D (1 mL/10 lb) solution.
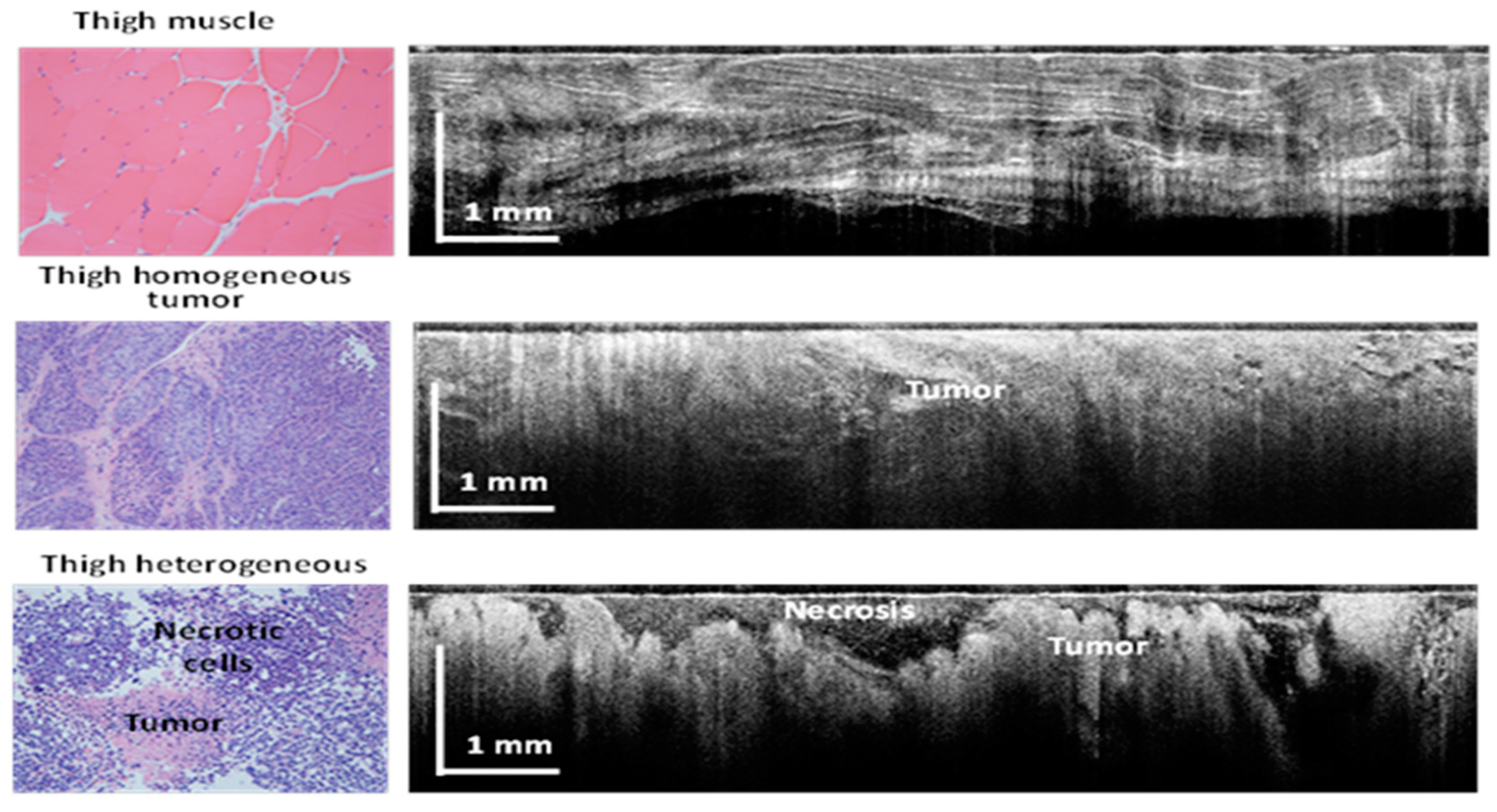
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Basik, M.; Aguilar-Mahecha, A.; Rousseau, C.; Diaz, Z.; Tejpar, S.; Spatz, A.; Greenwood Celia, M.T.; Batist, G. Biopsies: Next-generation biospecimens for tailoring therapy. Nat. Rev. Clin. Oncol. 2013, 10, 437–450. [Google Scholar] [CrossRef]
- Sabir, S.H.; Krishnamurthy, S.; Gupta, S.; Mills, G.B.; Wei, W.; Cortes, A.C.; Mills Shaw, K.R.; Luthra, R.; Wallace, M.J. Characteristics of percutaneous core biopsies adequate for next generation genomic sequencing. PLoS ONE 2017, 12, e0189651. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Swanton, C. Intratumor heterogeneity. Evolution through space and time. Cancer Res. 2012, 72, 4875–4882. [Google Scholar] [CrossRef] [Green Version]
- Marusyk, A.; Almendro, V.; Polyak, K. Intra-tumour heterogeneity: A looking glass for cancer? Nat. Rev. Cancer 2012, 12, 323–334. [Google Scholar] [CrossRef] [PubMed]
- Hatada, T.; Ishii, H.; Ichii, S.; Okada, K.; Fujiwara, Y.; Yamamura, T. Diagnostic value of ultrasound-guided fine-needle aspiration biopsy, core-needle biopsy, and evaluation of combined use in the diagnosis of breast lesions. J. Am. Coll. Surg. 2000, 190, 299–303. [Google Scholar] [CrossRef] [PubMed]
- Mitra, S.; Dey, P. Fine-needle aspiration and core biopsy in the diagnosis of breast lesions: A comparison and review of the literature. Cytojournal 2016, 13, 18. [Google Scholar] [CrossRef]
- Brem, R.F.; Lenihan, M.J.; Lieberman, J.; Torrente, J. Screening breast ultrasound: Past, present, and future. Am. J. Roentgenol. 2015, 204, 234–240. [Google Scholar] [CrossRef] [PubMed]
- Cummins, T.; Yoon, C.; Choi, H.; Eliahoo, P.; Kim, H.H.; Yamashita, M.W.; Hovanessian-Larsen, L.J.; Lang, J.E.; Sener, S.F.; Vallone, J.; et al. High-frequency ultrasound imaging for breast cancer biopsy guidance. J. Med. Imaging 2015, 2, 047001. [Google Scholar] [CrossRef] [Green Version]
- Bui-Mansfield, L.T.; Chen, D.C.; O’Brien, S.D. Accuracy of ultrasound of musculoskeletal soft-tissue tumors. AJR Am. J. Roentgenol. 2015, 204, W218. [Google Scholar] [CrossRef]
- Carra, B.J.; Bui-Mansfield, L.T.; O’Brien, S.D.; Chen, D.C. Sonography of musculoskeletal soft-tissue masses: Techniques, pearls, and pitfalls. AJR Am. J. Roentgenol. 2014, 202, 1281–1290. [Google Scholar] [CrossRef]
- Resnick, M.J.; Lee, D.J.; Magerfleisch, L.; Vanarsdalen, K.N.; Tomaszewski, J.E.; Wein, A.J.; Malkowicz, S.B.; Guzzo, T.J. Repeat prostate biopsy and the incremental risk of clinically insignificant prostate cancer. Urology 2011, 77, 548–552. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.S.; McMahon, C.J.; Lozano-Calderon, S.; Kung, J.W. Utility of Repeat Core Needle Biopsy of Musculoskeletal Lesions With Initially Nondiagnostic Findings. Am. J. Roentgenol. 2017, 208, 609–616. [Google Scholar] [CrossRef]
- Katsis, J.M.; Rickman, O.B.; Maldonado, F.; Lentz, R.J. Bronchoscopic biopsy of peripheral pulmonary lesions in 2020, a review of existing technologies. J. Thorac. Dis. 2020, 12, 3253–3262. [Google Scholar] [CrossRef] [PubMed]
- Chappy, S.L. Women’s experience with breast biopsy. AORN J. 2004, 80, 885–901. [Google Scholar] [CrossRef] [PubMed]
- Silverstein, M.J.; Recht, A.; Lagios, M.D.; Bleiweiss, I.J.; Blumencranz, P.W.; Gizienski, T.; Harms, S.E.; Harness, J.; Jackman, R.J.; Klimberg, V.S.; et al. Special report: Consensus conference III. Image-detected breast cancer: State-of-the-art diagnosis and treatment. J. Am. Coll. Surg. 2009, 209, 504–520. [Google Scholar] [CrossRef] [PubMed]
- Tam, A.L.; Lim, H.J.; Wistuba, I.I.; Tamrazi, A.; Kuo, M.D.; Ziv, E.; Wong, S.; Shih, A.J.; Webster, R.J., 3rd; Fischer, G.S.; et al. Image-Guided Biopsy in the Era of Personalized Cancer Care: Proceedings from the Society of Interventional Radiology Research Consensus Panel. J. Vasc. Interv. Radiol. 2016, 27, 8–19. [Google Scholar] [CrossRef] [Green Version]
- Lee, J.M.; Han, J.J.; Altwerger, G.; Kohn, E.C. Proteomics and biomarkers in clinical trials for drug development. J. Proteom. 2011, 74, 2632–2641. [Google Scholar] [CrossRef] [Green Version]
- Myers, M.B. Targeted therapies with companion diagnostics in the management of breast cancer: Current perspectives. Pharmgenomics Pers. Med. 2016, 22, 7–16. [Google Scholar] [CrossRef] [Green Version]
- Akshulakov, S.; Kerimbayev, T.T.; Biryuchkov, M.Y.; Urunbayev, Y.A.; Farhadi, D.S.; Byvaltsev, V. Current Trends for Improving Safety of Stereotactic Brain Biopsies: Advanced Optical Methods for Vessel Avoidance and Tumor Detection. Front. Oncol. 2019, 9, 947. [Google Scholar] [CrossRef]
- Wilson, R.H.; Vishwanath, K.; Mycek, M.A. Optical methods for quantitative and label-free sensing in living human tissues: Principles, techniques, and applications. Adv. Phys. 2016, 1, 523–543. [Google Scholar] [CrossRef] [Green Version]
- Krishnamurthy, S. Microscopy: A Promising Next-Generation Digital Microscopy Tool for Surgical Pathology Practice. Arch. Pathol. Lab. Med. Sept. 2019, 143, 1058–1068. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Konecky, S.D.; Mazhar, A.; Cuccia, D.; Durkin, A.J.; Schotland, J.C.; Tromberg, B.J. Quantitative optical tomography of sub-surface heterogeneities using spatially modulated structured light. Opt. Express 2009, 17, 14780–14790. [Google Scholar] [CrossRef] [Green Version]
- Iftimia NMujat, M.; Ustun, T.; Ferguson, D.; Vu, D.; Hammer, D. Spectral-domain low coherence interferometry/optical coherence tomography system for fine needle breast biopsy guidance. Rev. Sci. Instrum. 2009, 80, 024302. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Iftimia, N.; Park, J.; Maguluri, G.; Krishnamurthy, S.; McWatters, A.; Sabir, S.H. Investigation of tissue cellularity at the tip of the core biopsy needle with optical coherence tomography. Biomed. Opt. Express 2018, 9, 694–704. [Google Scholar] [CrossRef]
- Quirk, B.C.; McLaughlin, R.A.; Curatolo, A.; Kirk, R.W.; Noble, P.B.; Sampson, D.D. In situ imaging of lung alveoli with an optical coherence tomography needle probe. J. Biomed. Opt. 2011, 16, 036009. [Google Scholar] [CrossRef] [Green Version]
- Liang, C.P.; Wierwille, J.; Moreira, T.; Schwartzbauer, G.; Jafri, M.S.; Tang, C.M.; Chen, Y. A forward-imaging needle-type OCT probe for image guided stereotactic procedures. Opt. Express 2011, 19, 26283–26294. [Google Scholar] [CrossRef] [PubMed]
- Chang, E.W.; Gardecki, J.; Pitman, M.; Wilsterman, E.J.; Patel, A.; Tearney, G.J.; Iftimia, N. Low coherence interferometry approach for aiding fine needle aspiration biopsies. J. Biomed. Opt. 2014, 19, 116005. [Google Scholar] [CrossRef] [Green Version]
- Curatolo, A.; McLaughlin, R.A.; Quirk, B.C.; Kirk, R.W.; Bourke, A.G.; Wood, B.A.; Robbins, P.D.; Saunders, C.M.; Sampson, D.D. Ultrasound-guided optical coherence tomography needle probe for the assessment of breast cancer tumor margins. AJR Am. J. Roentgenol. 2012, 199, W520–W522. [Google Scholar] [CrossRef]
- Wang, J.; Xu, Y.; Boppart, S.A. Review of optical coherence tomography in oncology. J. Biomed. Opt. 2017, 22, 1–23. [Google Scholar] [CrossRef]
- Hesamian, M.H.; Jia, W.; He, X.; Kennedy, P. Deep learning techniques for medical image segmentation: Achievements and challenges. J. Digit. Imaging 2019, 32, 582–596. [Google Scholar] [CrossRef] [Green Version]
- Moorthy, U.; Gandhi, U.D. A survey of big data analytics using machine learning algorithms. In Research Anthology on Big Data Analytics, Architectures, and Applications; IGI Global: Hershey, PA, USA, 2022; pp. 655–677. [Google Scholar]
- Luis, F.; Kumar, I.; Vijayakumar, V.; Singh, K.U.; Kumar, A. Identifying the patterns state of the art of deep learning models and computational models their challenges. Multimed. Syst. 2021, 27, 599–613. [Google Scholar]
- Moorthy, U.; Gandhi, U.D. A novel optimal feature selection for medical data classification using CNN based Deep learning. J. Ambient. Intell. Humaniz. Comput. 2021, 12, 3527–3538. [Google Scholar] [CrossRef]
- Chen, S.X.; Ni, Y.Q.; Zhou, L. A deep learning framework for adaptive compressive sensing of high-speed train vibration responses. Struct. Control Health Monit. 2020, 29, e2979. [Google Scholar] [CrossRef]
- Finck, T.; Singh, S.P.; Wang, L.; Gupta, S.; Goli, H.; Padmanabhan, P.; Gulyás, B. A basic introduction to deep learning for medical image analysis. Sensors 2021, 20, 5097. [Google Scholar]
- Dahrouj, M.; Miller, J.B. Artificial Intelligence (AI) and Retinal Optical Coherence Tomography (OCT). Semin. Ophthalmol. 2021, 36, 341–345. [Google Scholar] [CrossRef]
- Kapoor, R.; Whigham, B.T.; Al-Aswad, L.A. Artificial Intelligence and Optical Coherence Tomography Imaging. Asia Pac. J. Ophthalmol. 2019, 8, 187–194. [Google Scholar]
- Iftimia, N.; Maguluri, G.; Chang, E.W.; Chang, S.; Magill, J.; Brugge, W. Hand scanning optical coherence tomography imaging using encoder feedback. Opt. Lett. 2014, 39, 6807–6810. [Google Scholar] [CrossRef] [Green Version]



| CNN | Definition | Excluded Features | Number of Annotated Images |
|---|---|---|---|
| CNN1: Tissue | All tissue including muscle, fat, vessel, and tumor. | Dark background, catheter, and tissue holes. | 89 |
| CNN2: Tumor |
|
| 94 |
| CNN3: Necrotic Tumor | Focal dark region surrounded by tumor region. | Dark regions away from catheter surface where signal fades. | 72 |
| CNN1: Tissue | CNN2: Tumor | CNN3: Necrotic Tumor | ||
|---|---|---|---|---|
| Training parameters | Weight decay | 0.0001 | 0.0001 | 0.000140 |
| Mini-batches size | 20 | 20 | 20 | |
| Mini-batches per iteration | 40 | 20 | 40 | |
| Iterations without progress | 500 | 500 | 500 | |
| Initial learning rate | 1 | 1 | 1 | |
| Image augmentation | Scale (Min/max) | −1/1.01 | −1/1.01 | −1/1.01 |
| Aspect ratio | 1 | 1 | 1 | |
| Maximum shear | 1 | 1 | 1 | |
| Luminance (min/max) | −1/50 | −1/1 | −1/1 | |
| Contrast (min/max) | −1/50 | −1/50 | −1/50 | |
| Max with balance change | 1 | 1 | 1 | |
| Noise | 0 | 0 | 0 | |
| JPG compression (min/max) | 40/60 | 40/60 | 40/60 | |
| Blur max pixels | 1 | 1 | 1 | |
| JPG compression percentage | 0.5 | 0.5 | 0.5 | |
| Blur percentage | 0.5 | 0.5 | 0.5 | |
| Rotation angle (min/max) | 180/180 | −180/180 | −180/180 | |
| Gain | 1 | 1.5 | 1.3 | |
| Level of detail | Low | Medium | Medium | |
| Parameter | Formula |
|---|---|
| False Positive (FP) (%) | This parameter determines the proportion of pixels incorrectly classified as positive in the verification region. |
| False Negative (FN) (%) | This parameter determines the proportion of pixels incorrectly classified as negative in the verification region. |
| Error (%) | (FP + FN)/All validation area |
| Precision (%) | TP/(TP + FP) |
| Sensitivity (%) | TP/TP + FN |
| F1 Score (%) | 2TP/(2TP + FP + FN) |
| FP % | FN % | Error % | Precision % | Sensitivity % | F1 Score % | |
|---|---|---|---|---|---|---|
| AI vs. Human | 0.86 | 1.81 | 2.67 | 73.17 | 66.98 | 77.36 |
| Human vs. Human | 1.53 | 1.47 | 3.00 | 71.23 | 71.23 | 76.74 |
| F1 score Agreement | 99.38% | |||||
| FP % | FN % | Error % | Precision % | Sensitivity % | F1 Score % | |
|---|---|---|---|---|---|---|
| AI vs. Human | 1.25 | 1.12 | 2.37 | 65.11 | 68.5 | 74.74 |
| Human vs. Human | 1.41 | 1.37 | 2.78 | 69.66 | 69.66 | 71.89 |
| F1 score Agreement | 97.15% | |||||
| FP % | FN % | Error % | Precision % | Sensitivity % | F1 Score % | |
|---|---|---|---|---|---|---|
| AI vs. Human | 1.14 | 1.21 | 2.35 | 77.36 | 73.15 | 76.45 |
| Human vs. Human | 1.39 | 1.26 | 2.64 | 73.96 | 73.96 | 78.48 |
| F1 score Agreement | 97.97% | |||||
| FP % | FN % | Error % | Precision % | Sensitivity % | F1 Score % | |
|---|---|---|---|---|---|---|
| AI vs. Human | 0.58 | 4.23 | 4.82 | 38.9 | 18.27 | 42.11 |
| Human vs. Human | 2.62 | 2.56 | 5.17 | 41.7 | 41.7 | 57.53 |
| F1 score Agreement | 84.58% | |||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Maguluri, G.; Grimble, J.; Caron, A.; Zhu, G.; Krishnamurthy, S.; McWatters, A.; Beamer, G.; Lee, S.-Y.; Iftimia, N. Core Needle Biopsy Guidance Based on Tissue Morphology Assessment with AI-OCT Imaging. Diagnostics 2023, 13, 2276. https://doi.org/10.3390/diagnostics13132276
Maguluri G, Grimble J, Caron A, Zhu G, Krishnamurthy S, McWatters A, Beamer G, Lee S-Y, Iftimia N. Core Needle Biopsy Guidance Based on Tissue Morphology Assessment with AI-OCT Imaging. Diagnostics. 2023; 13(13):2276. https://doi.org/10.3390/diagnostics13132276
Chicago/Turabian StyleMaguluri, Gopi, John Grimble, Aliana Caron, Ge Zhu, Savitri Krishnamurthy, Amanda McWatters, Gillian Beamer, Seung-Yi Lee, and Nicusor Iftimia. 2023. "Core Needle Biopsy Guidance Based on Tissue Morphology Assessment with AI-OCT Imaging" Diagnostics 13, no. 13: 2276. https://doi.org/10.3390/diagnostics13132276
APA StyleMaguluri, G., Grimble, J., Caron, A., Zhu, G., Krishnamurthy, S., McWatters, A., Beamer, G., Lee, S.-Y., & Iftimia, N. (2023). Core Needle Biopsy Guidance Based on Tissue Morphology Assessment with AI-OCT Imaging. Diagnostics, 13(13), 2276. https://doi.org/10.3390/diagnostics13132276






