Multimodal Spatiotemporal Deep Learning Framework to Predict Response of Breast Cancer to Neoadjuvant Systemic Therapy
Abstract
:1. Introduction
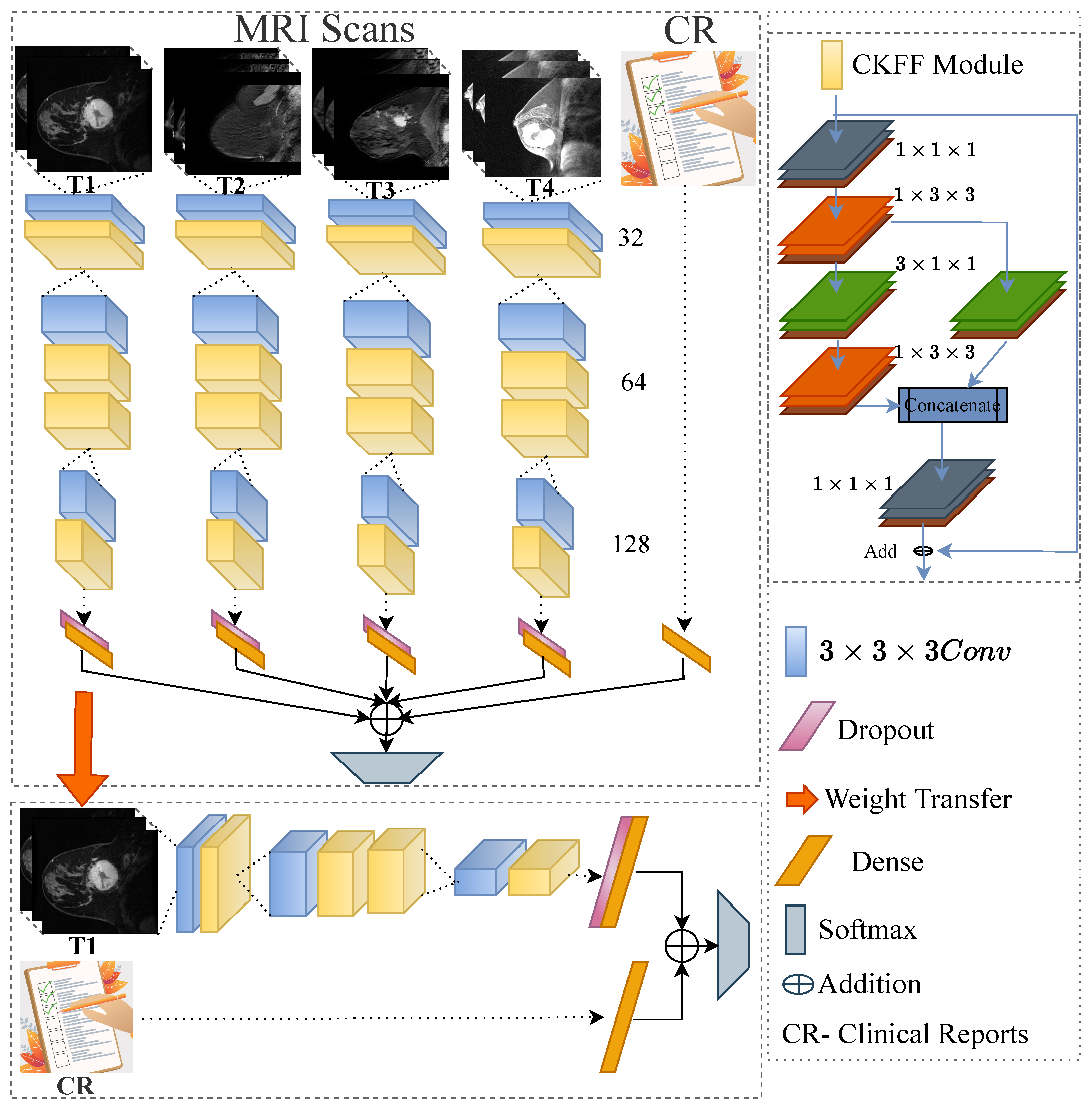
- We develop a multimodal spatiotemporal deep learning by integrating the following multi-modalities: imaging data with N-time stamps (pre-treatment, early treatment, inter-regimen, prior to surgery, etc.), molecular data (ER, PgRPos, HRPos, BilateralCa, Laterality, HER2Pos, HR_HER2_Category, and HR_HER2_Status), and demographical data (age and race). We demonstrate the influence of each time point on the predictions made by the network through ablation experiments.
- We design a novel 3D-CNN-based deep learning framework by introducing a cross-kernel feature fusion (CKFF) module.
- The CKFF module makes the architecture more learnable at a lower computational cost by paying attention to multiple receptive fields to extract the spatiotemporal features.
- The efficacy of the proposed framework is tested on a challenging breast cancer data set, ISPY-1 [15], in terms of accuracy and AUC.
2. Methodology
Proposed Method
3. Experimental Results and Analysis
3.1. Dataset
3.2. Training and Implementation Details
3.3. Experimental Results Analysis
3.3.1. Discussion
3.3.2. Ablation Study
3.3.3. Complexity Analysis
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Tao, Z.; Shi, A.; Lu, C.; Song, T.; Zhang, Z.; Zhao, J. Breast cancer: Epidemiology and etiology. Cell Biochem. Biophys. 2015, 72, 333–338. [Google Scholar] [CrossRef] [PubMed]
- Yankeelov, T.E.; Atuegwu, N.; Hormuth, D.; Weis, J.A.; Barnes, S.L.; Miga, M.I.; Rericha, E.C.; Quaranta, V. Clinically relevant modeling of tumor growth and treatment response. Sci. Transl. Med. 2013, 5, 187ps9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Braman, N.M.; Etesami, M.; Prasanna, P.; Dubchuk, C.; Gilmore, H.; Tiwari, P.; Plecha, D.; Madabhushi, A. Intratumoral and peritumoral radiomics for the pretreatment prediction of pathological complete response to neoadjuvant chemotherapy based on breast DCE-MRI. Breast Cancer Res. 2017, 19, 1–14. [Google Scholar]
- Hylton, N.M.; Blume, J.D.; Bernreuter, W.K.; Pisano, E.D.; Rosen, M.A.; Morris, E.A.; Weatherall, P.T.; Lehman, C.D.; Newstead, G.M.; Polin, S.; et al. Locally advanced breast cancer: MR imaging for prediction of response to neoadjuvant chemotherapy—Results from ACRIN 6657/I-SPY TRIAL. Radiology 2012, 263, 663–672. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hylton, N.M.; Gatsonis, C.A.; Rosen, M.A.; Lehman, C.D.; Newitt, D.C.; Partridge, S.C.; Bernreuter, W.K.; Pisano, E.D.; Morris, E.A.; Weatherall, P.T.; et al. Neoadjuvant chemotherapy for breast cancer: Functional tumor volume by MR imaging predicts recurrence-free survival—Results from the ACRIN 6657/CALGB 150007 I-SPY 1 TRIAL. Radiology 2016, 279, 44–55. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, W.; Arasu, V.; Newitt, D.C.; Jones, E.F.; Wilmes, L.; Gibbs, J.; Kornak, J.; Joe, B.N.; Esserman, L.J.; Hylton, N.M. Effect of MR imaging contrast thresholds on prediction of neoadjuvant chemotherapy response in breast cancer subtypes: A subgroup analysis of the ACRIN 6657/I-SPY 1 TRIAL. Tomography 2016, 2, 378–387. [Google Scholar] [CrossRef] [PubMed]
- Ravichandran, K.; Braman, N.; Janowczyk, A.; Madabhushi, A. A deep learning classifier for prediction of pathological complete response to neoadjuvant chemotherapy from baseline breast DCE-MRI. In Medical Imaging 2018: Computer-Aided Diagnosis; SPIE: Bellingham, WA, USA, 2018; Volume 10575, pp. 79–88. [Google Scholar]
- Ha, R.; Chin, C.; Karcich, J.; Liu, M.Z.; Chang, P.; Mutasa, S.; Pascual Van Sant, E.; Wynn, R.T.; Connolly, E.; Jambawalikar, S. Prior to initiation of chemotherapy, can we predict breast tumor response? Deep learning convolutional neural networks approach using a breast MRI tumor dataset. J. Digit. Imaging 2019, 32, 693–701. [Google Scholar] [CrossRef] [PubMed]
- Braman, N.; Adoui, M.E.; Vulchi, M.; Turk, P.; Etesami, M.; Fu, P.; Bera, K.; Drisis, S.; Varadan, V.; Plecha, D.; et al. Deep learning-based prediction of response to HER2-targeted neoadjuvant chemotherapy from pre-treatment dynamic breast MRI: A multi-institutional validation study. arXiv 2020, arXiv:2001.08570. [Google Scholar]
- Marinovich, M.L.; Macaskill, P.; Irwig, L.; Sardanelli, F.; Mamounas, E.; von Minckwitz, G.; Guarneri, V.; Partridge, S.C.; Wright, F.C.; Choi, J.H.; et al. Agreement between MRI and pathologic breast tumor size after neoadjuvant chemotherapy, and comparison with alternative tests: Individual patient data meta-analysis. BMC Cancer 2015, 15, 662. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lobbes, M.; Prevos, R.; Smidt, M.; Tjan-Heijnen, V.; Van Goethem, M.; Schipper, R.; Beets-Tan, R.; Wildberger, J. The role of magnetic resonance imaging in assessing residual disease and pathologic complete response in breast cancer patients receiving neoadjuvant chemotherapy: A systematic review. Insights Imaging 2013, 4, 163–175. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Duanmu, H.; Huang, P.B.; Brahmavar, S.; Lin, S.; Ren, T.; Kong, J.; Wang, F.; Duong, T.Q. Prediction of pathological complete response to neoadjuvant chemotherapy in breast cancer using deep learning with integrative imaging, molecular and demographic data. In Medical Image Computing and Computer Assisted Intervention–MICCAI 2020, Proceedings of the 23rd International Conference, Lima, Peru, 4–8 October 2020; Springer: Berlin/Heidelberg, Germany, 2020; pp. 242–252. [Google Scholar]
- Bouzón, A.; Acea, B.; Soler, R.; Iglesias, Á.; Santiago, P.; Mosquera, J.; Calvo, L.; Seoane-Pillado, T.; García, A. Diagnostic accuracy of MRI to evaluate tumour response and residual tumour size after neoadjuvant chemotherapy in breast cancer patients. Radiol. Oncol. 2016, 50, 73–79. [Google Scholar] [CrossRef] [PubMed]
- Qu, Y.H.; Zhu, H.T.; Cao, K.; Li, X.T.; Ye, M.; Sun, Y.S. Prediction of pathological complete response to neoadjuvant chemotherapy in breast cancer using a deep learning (DL) method. Thorac. Cancer 2020, 11, 651–658. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Newitt, D.; Hylton, N. Multi-center breast DCE-MRI data and segmentations from patients in the I-SPY 1/ACRIN 6657 trials. Cancer Imaging Arch. 2016, 10. [Google Scholar]
- Krizhevsky, A.; Sutskever, I.; Hinton, G.E. Imagenet classification with deep convolutional neural networks. Adv. Neural Inf. Process. Syst. 2012, 25, 84–90. [Google Scholar] [CrossRef] [Green Version]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep residual learning for image recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016; pp. 770–778. [Google Scholar]
- Verma, M.; Vipparthi, S.K.; Singh, G.; Murala, S. LEARNet: Dynamic imaging network for micro expression recognition. IEEE Trans. Image Process. 2019, 29, 1618–1627. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schindler, A.; Lidy, T.; Rauber, A. Comparing Shallow versus Deep Neural Network Architectures for Automatic Music Genre Classification. In Proceedings of the FMT, Pölten, Austria, 23–24 November 2016; pp. 17–21. [Google Scholar]
- Verma, M.; Vipparthi, S.K.; Singh, G. Hinet: Hybrid inherited feature learning network for facial expression recognition. IEEE Lett. Comput. Soc. 2019, 2, 36–39. [Google Scholar] [CrossRef]
- Huynh, B.Q.; Antropova, N.; Giger, M.L. Comparison of breast DCE-MRI contrast time points for predicting response to neoadjuvant chemotherapy using deep convolutional neural network features with transfer learning. In Medical Imaging 2017: Computer-Aided Diagnosis; SPIE: Bellingham, WA, USA, 2017; Volume 10134, pp. 207–213. [Google Scholar]
- Verma, M.; Reddy, M.S.K.; Meedimale, Y.R.; Mandal, M.; Vipparthi, S.K. Automer: Spatiotemporal neural architecture search for microexpression recognition. IEEE Transactions on Neural Networks and Learning Systems 2021, 33, 6116–6128. [Google Scholar] [CrossRef] [PubMed]
- Lin, T.Y.; Goyal, P.; Girshick, R.; He, K.; Dollár, P. Focal loss for dense object detection. In Proceedings of the IEEE International Conference on Computer Vision, Venice, Italy, 22–29 October 2017; pp. 2980–2988. [Google Scholar]
- Joo, S.; Ko, E.S.; Kwon, S.; Jeon, E.; Jung, H.; Kim, J.Y.; Chung, M.J.; Im, Y.H. Multimodal deep learning models for the prediction of pathologic response to neoadjuvant chemotherapy in breast cancer. Sci. Rep. 2021, 11, 18800. [Google Scholar] [CrossRef] [PubMed]
- Szegedy, C.; Liu, W.; Jia, Y.; Sermanet, P.; Reed, S.; Anguelov, D.; Erhan, D.; Vanhoucke, V.; Rabinovich, A. Going deeper with convolutions. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Boston, MA, USA, 7–12 June 2015; pp. 1–9. [Google Scholar]
- Szegedy, C.; Vanhoucke, V.; Ioffe, S.; Shlens, J.; Wojna, Z. Rethinking the inception architecture for computer vision. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 26 June–1 July 2016; pp. 2818–2826. [Google Scholar]




| Method | AUC | ACC |
|---|---|---|
| Volume [4] | 0.73 | N/A |
| FTV [5] | 0.73 | N/A |
| FTV and Varying PER and SER [6] | 0.90 | N/A |
| CNN and Feature Convolution [12] | 0.80 | N/A |
| CNN pre-post contrast [7] | 0.85 | N/A |
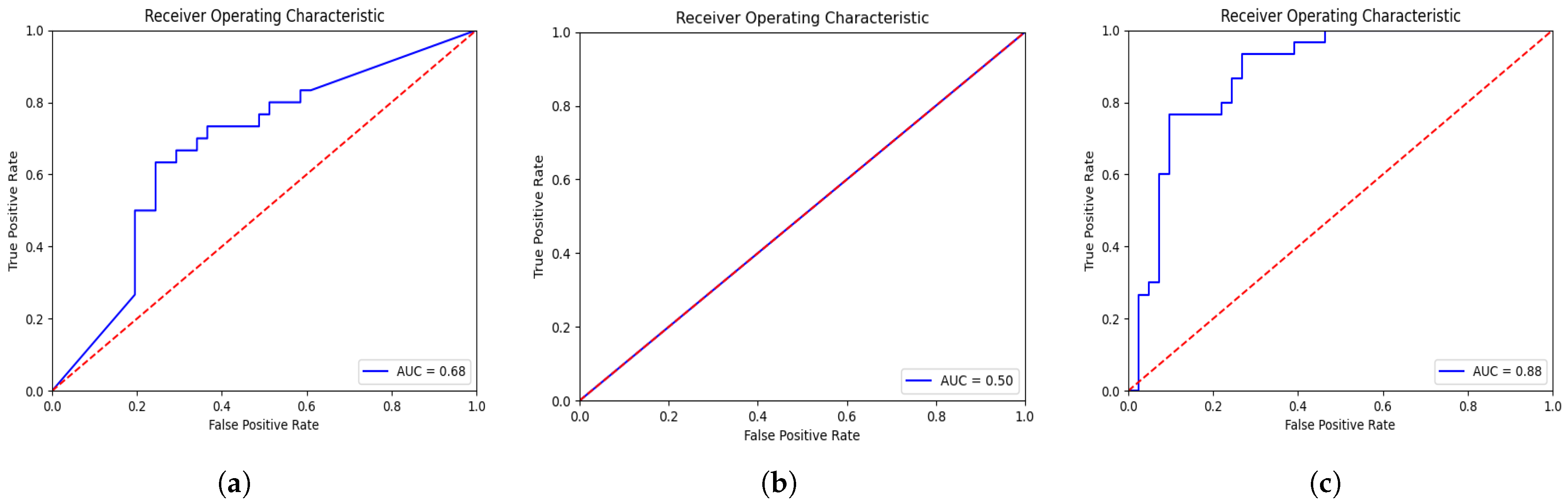
| 3D-VGGNet * [16] | 0.68 | 0.68 |
| 3D-ResNet * [17] | 0.50 | 0.42 |
| Deep-NST+Focal | 0.88 | 0.79 |
| Deep-NST+3DCNN | 0.60 | 0.58 |
| Deep-NST+3DInception | 0.61 | 0.58 |
| UniModal ST | 0.84 | 0.72 |
| Deep-NST | 0.88 | 0.85 |
| Input | AUC | ACC |
|---|---|---|
| MRI Scans | 0.87 | 0.77 |
| MRI Scans + Clinical Data | 0.85 | 0.82 |
| + MRI Scans + Clinical Data | 0.86 | 0.73 |
| + + MRI Scans + Clinical Data | 0.87 | 0.77 |
| + + + MRI Scans + Clinical Data | 0.88 | 0.85 |
| Method | #Param. | #Mem | #FLOPS |
|---|---|---|---|
| 3D-VGGNet [16] | 225 MB | 1.8 GB | G |
| 3D-ResNet [17] | 311.0 MB | 2.5 GB | G |
| Deep-NST+Focal | 4 MB | 37.0 MB | 30.5 G |
| Deep-NST+3DCNN | 3.8 MB | 31.3 MB | 2.85 G |
| Deep-NST+3DInception | 171.0 MB | 1.4 GB | G |
| Unimodal ST | 4 MB | 36.6 MB | 30.5 G |
| Deep-NST | 4 MB | 37.0 MB | 30.5 G |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Verma, M.; Abdelrahman, L.; Collado-Mesa, F.; Abdel-Mottaleb, M. Multimodal Spatiotemporal Deep Learning Framework to Predict Response of Breast Cancer to Neoadjuvant Systemic Therapy. Diagnostics 2023, 13, 2251. https://doi.org/10.3390/diagnostics13132251
Verma M, Abdelrahman L, Collado-Mesa F, Abdel-Mottaleb M. Multimodal Spatiotemporal Deep Learning Framework to Predict Response of Breast Cancer to Neoadjuvant Systemic Therapy. Diagnostics. 2023; 13(13):2251. https://doi.org/10.3390/diagnostics13132251
Chicago/Turabian StyleVerma, Monu, Leila Abdelrahman, Fernando Collado-Mesa, and Mohamed Abdel-Mottaleb. 2023. "Multimodal Spatiotemporal Deep Learning Framework to Predict Response of Breast Cancer to Neoadjuvant Systemic Therapy" Diagnostics 13, no. 13: 2251. https://doi.org/10.3390/diagnostics13132251
APA StyleVerma, M., Abdelrahman, L., Collado-Mesa, F., & Abdel-Mottaleb, M. (2023). Multimodal Spatiotemporal Deep Learning Framework to Predict Response of Breast Cancer to Neoadjuvant Systemic Therapy. Diagnostics, 13(13), 2251. https://doi.org/10.3390/diagnostics13132251






