Tubular Structure Segmentation via Multi-Scale Reverse Attention Sparse Convolution
Abstract
1. Introduction
- (a)
- In this paper, a multi-scale feature aggregation method is proposed and validated, which can fully extract and fuse the cerebrovascular and airway features with different thicknesses at different scales. The proposed method effectively solves the problem of differences in feature expression at the same scale, thus improving the segmentation accuracy.
- (b)
- Our paper introduces a novel reverse attention module combined with sparse convolution to guide the network effectively. By leveraging reverse attention mechanisms, this module enhances foreground detection by emphasizing the background and excluding areas of prediction. Moreover, it allocates reverse attention weights to extracted features, thereby improving the representation of micro-airways, micro-vessels, and image edges. The utilization of sparse convolution further improves overall feature representation and segmentation accuracy.
- (c)
- Through extensive experimental validation, we investigate the impact of sliding window sequencing and input image dimensions on the segmentation of tubular structures, including cerebral blood vessels and airways. The insights gained from this study contribute to the advancement of artificial intelligence techniques in medical image analysis, specifically focusing on enhancing the segmentation of tubular structures.
2. Related Work
2.1. Tubular Structure Segmentation
2.2. Multi-Scale Feature Fusion and Attention Mechaism
3. Materials and Methods
3.1. Materials
3.1.1. Datasets
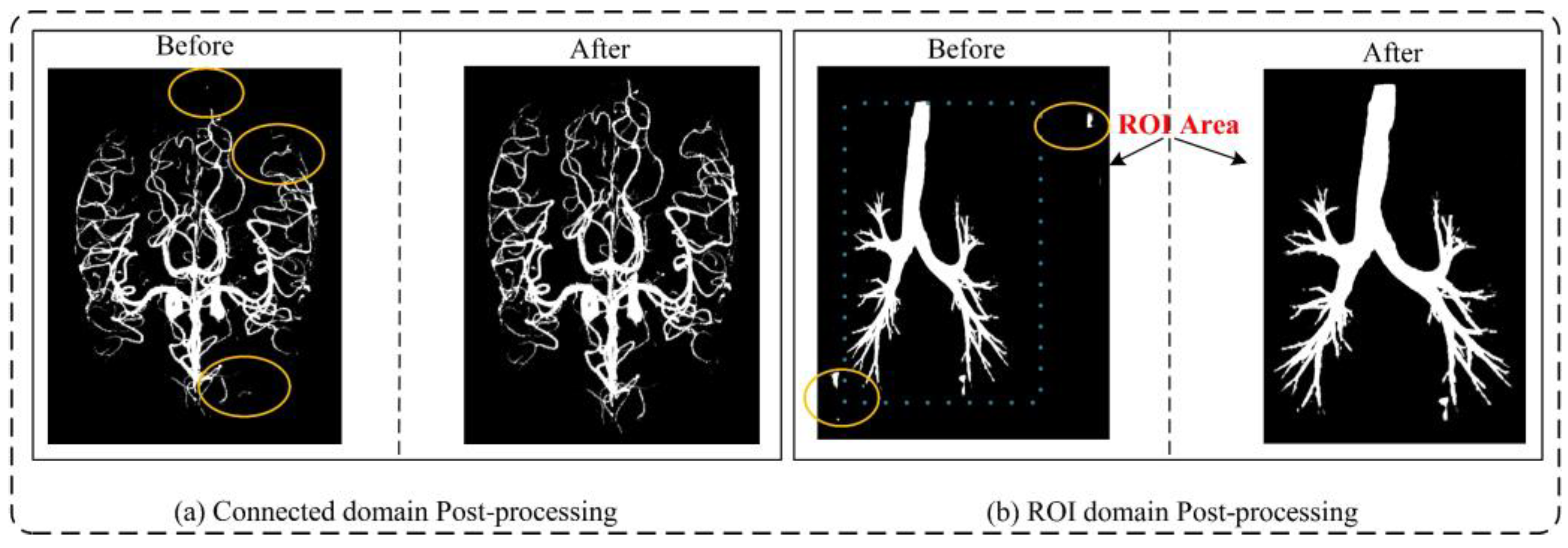
3.1.2. Data Pre-Processing and Sample Cropping
3.2. Methods
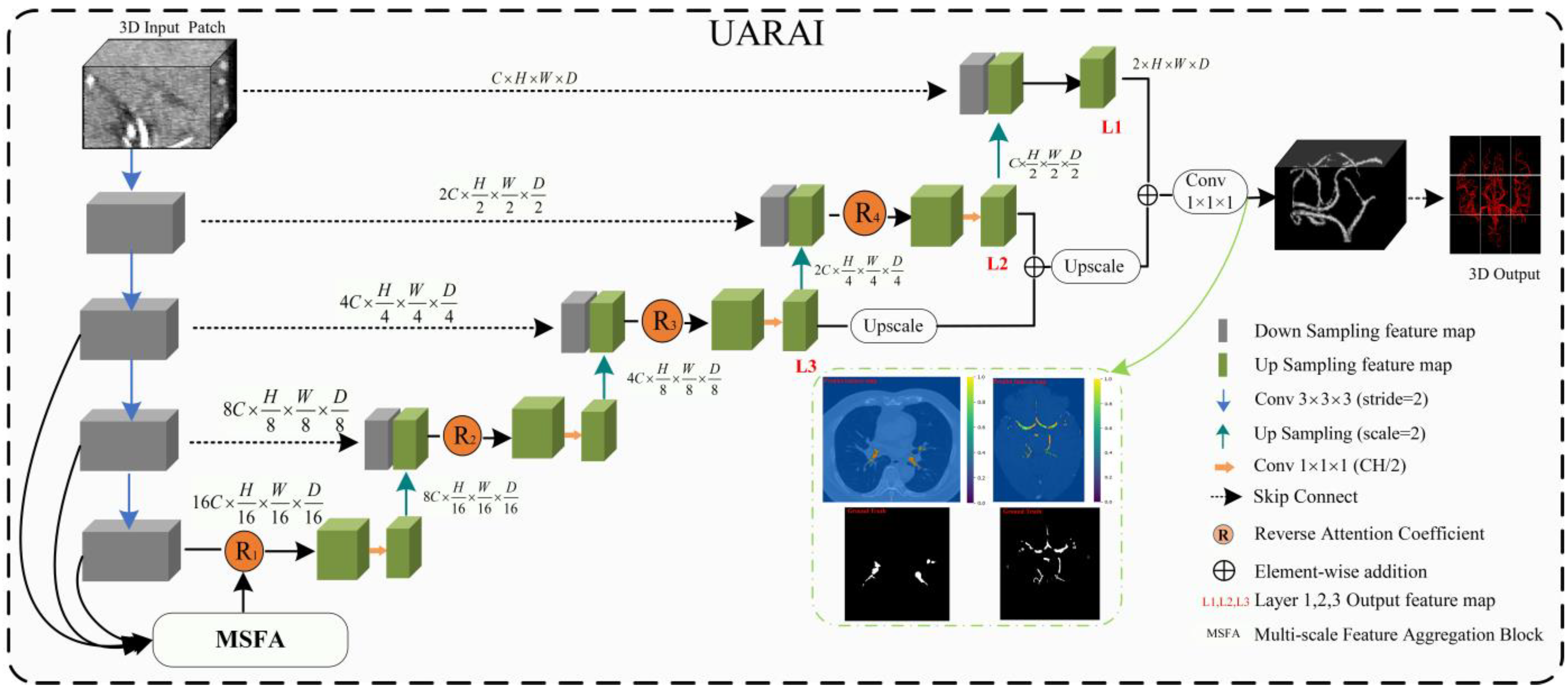
UARAI Overall Framework
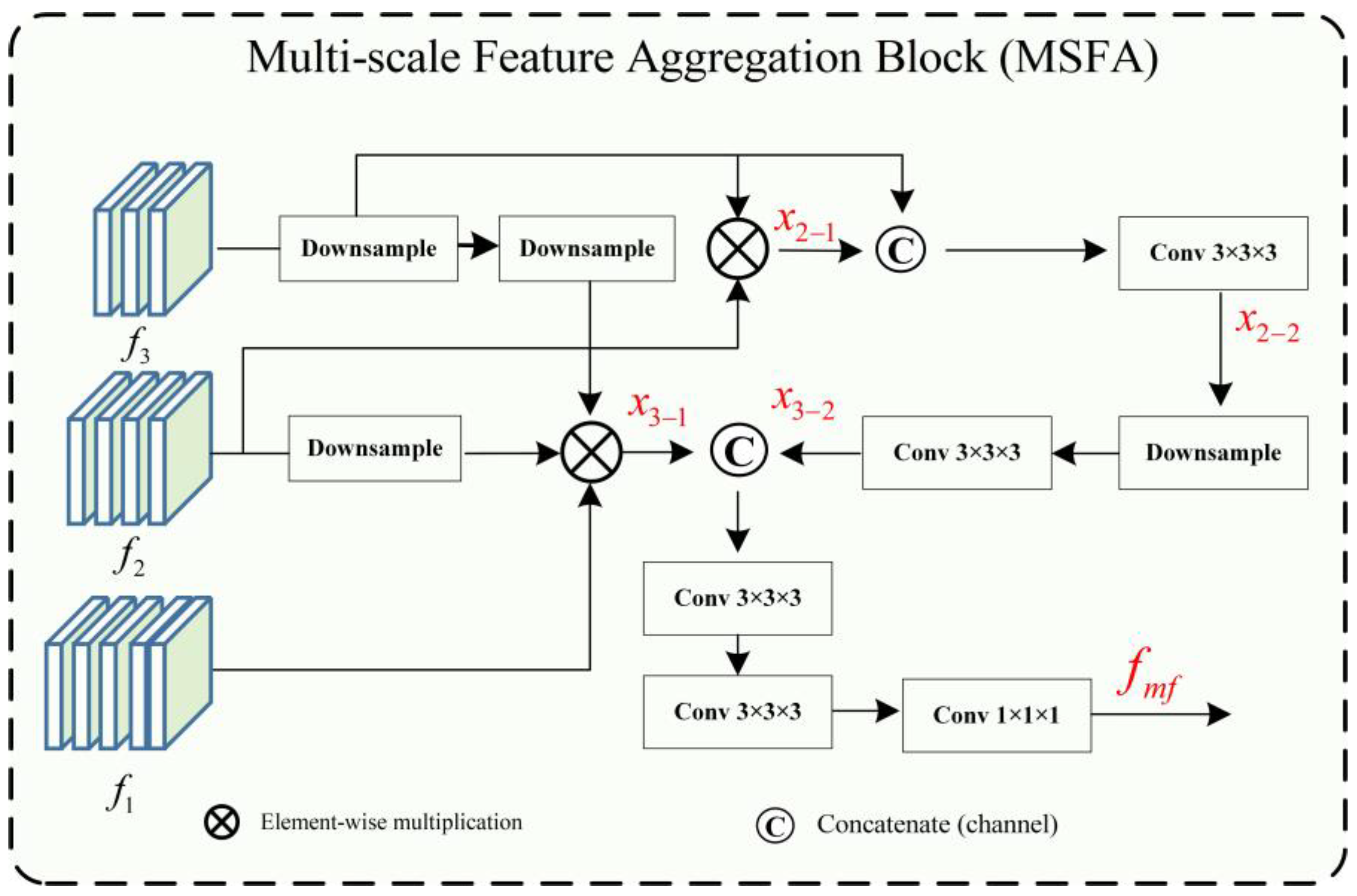
- (A) Multi-Scale Feature Aggregation
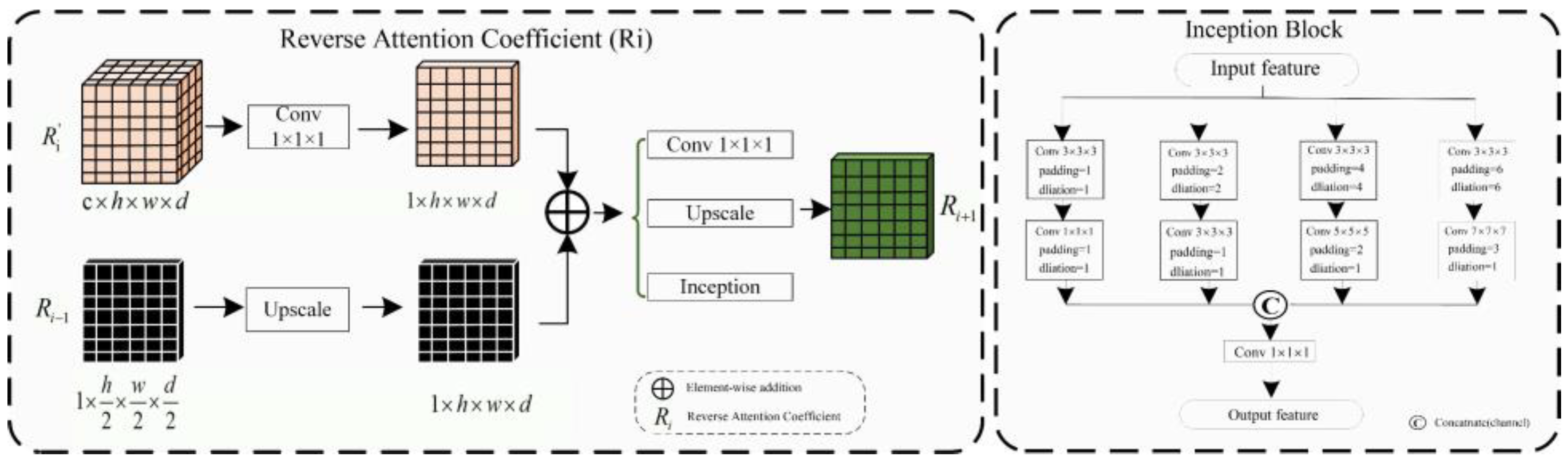
- (B) Reverse Attention Block
- (C) Inception Block
- (D) Loss function
4. Experimental Design
4.1. Experimental and Parameter Settings
4.2. Comparative Experiment
- (a)
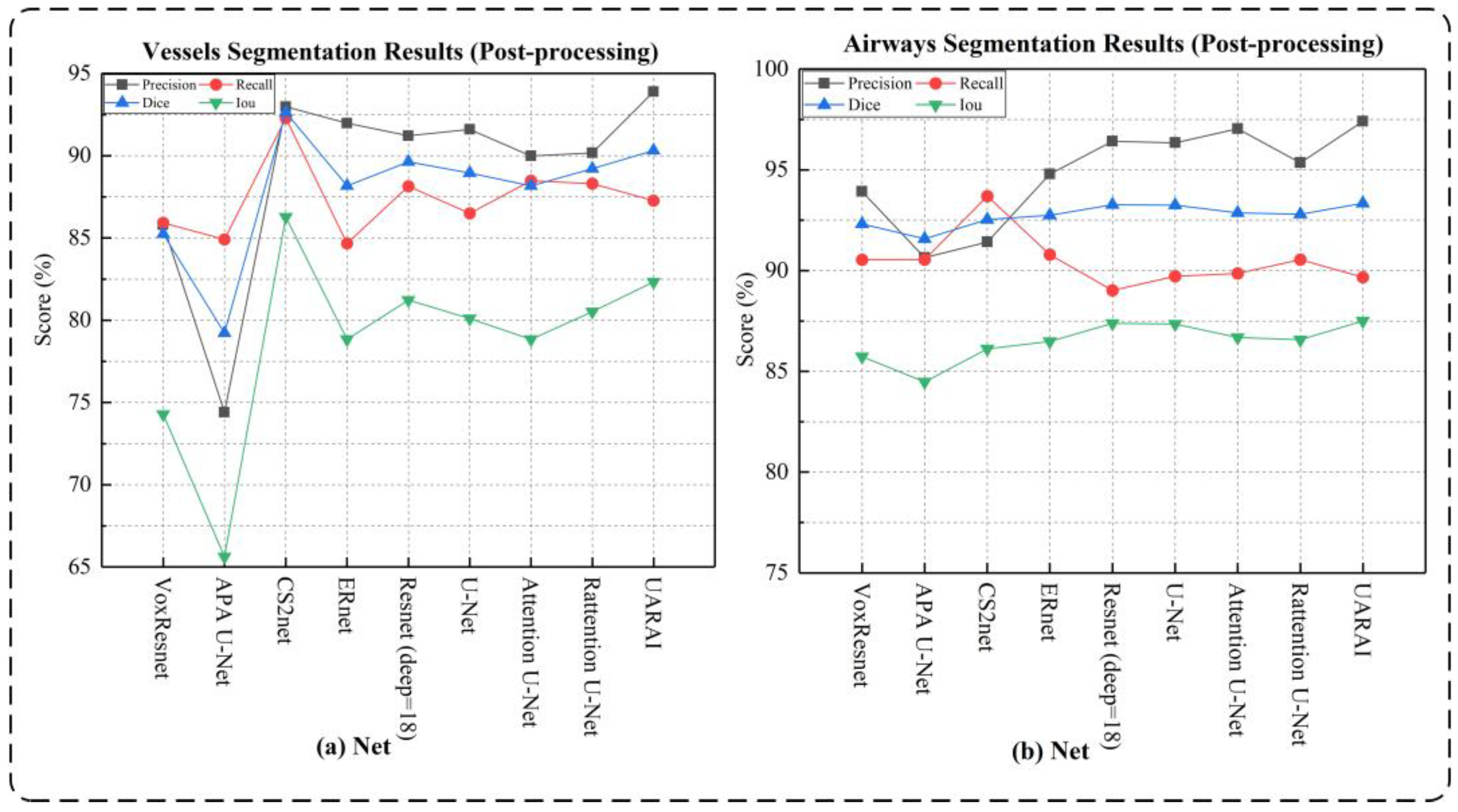
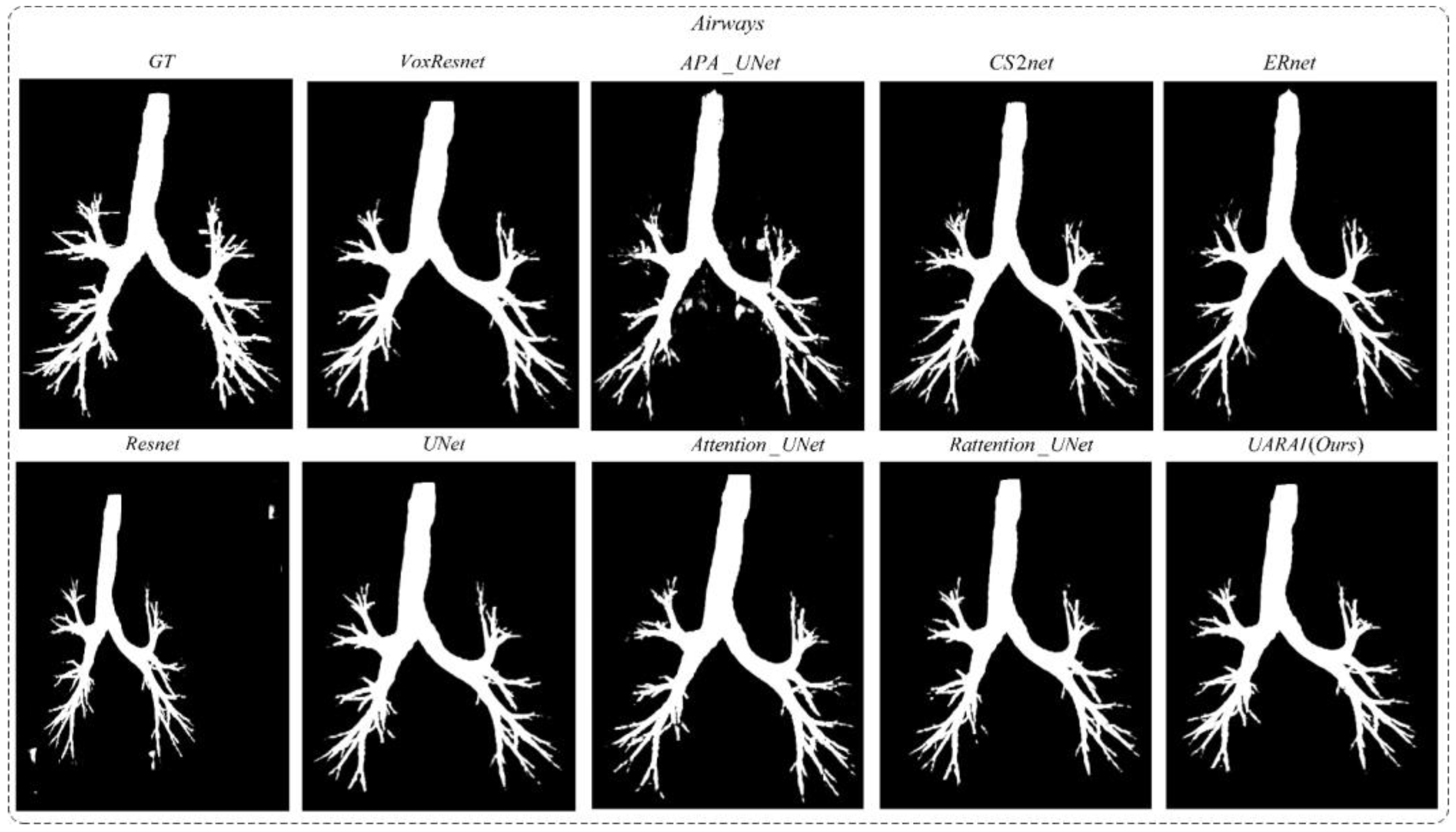
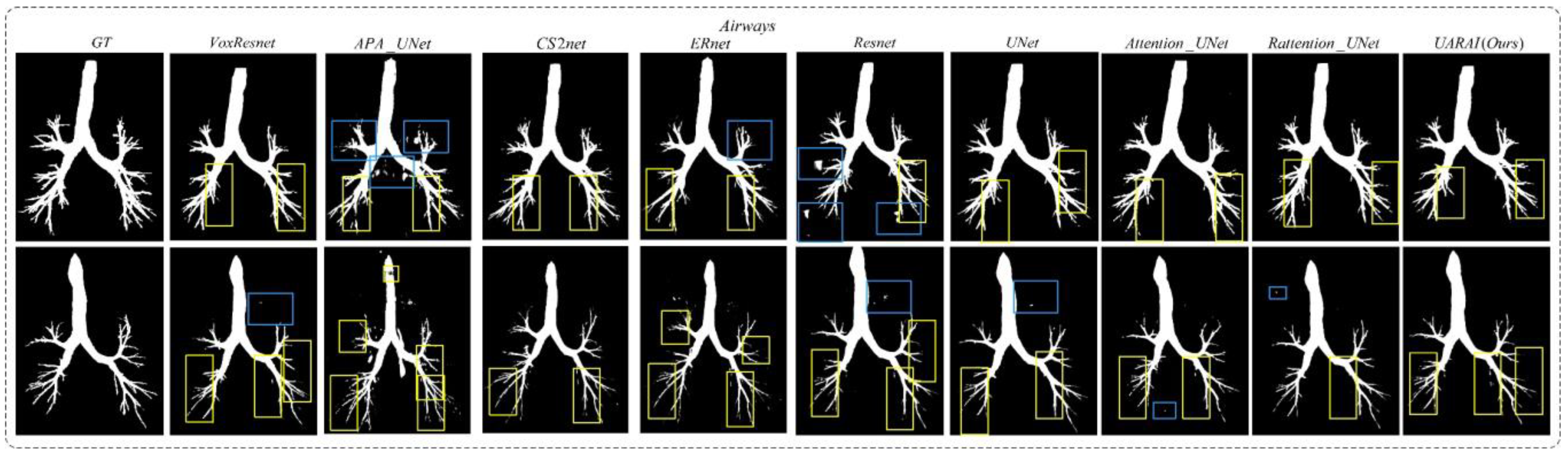
- Network dimension-based comparative experiments: Based on commonly used medical image segmentation networks, this experiment compared and analyzed the performance of VoxResnet [59], Resnet [60], 3D U-Net [13], Attention U-Net [17], Rattention U-Net [50], CS2-Net [35], ER-Net [36], APA U-Net [39], and the UARAI network proposed in this study. Vessel and airway segmentation are evaluated to thoroughly validate the proposed model’s segmentation effect.
- (b)
- Patch-cropping method-based comparative experiments: In order to verify the influence of different patch acquisition methods on model performance, two comparative experiments were designed in this paper. One method is random patch cropping, and the other combines sequential sliding window cropping and random patch cropping. For random patch cropping, the cropping condition was set as the block threshold greater than 0.01 (as shown in Equation (11)), and a total of 150 patches were cropped for each image. This patch type mainly includes coarse tubular structures with fewer vessels and airways in peripheral areas. The other combination method is to sequentially crop samples with a window size of 64 × 64 × 32 and a step size of 32. Then, 30 samples were randomly cropped from each image, and the threshold was set to 0.001 (no need to set a strict threshold). This strategy can obtain all the feature information of the image quickly and increase sample diversity.
- (c)
- Patch-size-based comparative experiments: Cerebrovascular structures are distributed very sparsely in the brain, and the volume fraction of physiological brain arterial vessels is 1.5%. The voxel resolution of arterial vessels that TOF-MRA can detect can be as low as 0.3% of all voxels in the brain [8]. In addition, cerebrovascular and airway structures are complex, and many tubular structures are of different thicknesses. Samples of different sizes cover different features. Smaller patch sizes contain less context information and focus more on detailed features. In comparison, larger patch sizes contain more global features but have a lower training efficiency and require more dimensionality reduction for obtaining high-level features during down-sampling. Therefore, this experiment designed comparative experiments at different patch sizes of 16 × 16 × 32, 32 × 32 × 32, 64 × 64 × 32, 96 × 96 × 32, and 128 × 128 × 32 to explore the performance differences of the network model under different patch sizes.
4.3. Evaluation Metrics
5. Results
5.1. Cerebrovascular and Airway Segmentation Results
5.2. Ablation Studies
6. Discussion
7. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Longde, W. Summary of 2016 Report on Prevention and Treatment of Stroke in China. Chin. J. Cerebrovasc. Dis. 2017, 14, 217–224. [Google Scholar]
- Wang, C.; Xu, J.; Yang, L.; Xu, Y.; Zhang, X.; Bai, C.; Kang, J.; Ran, P.; Shen, H.; Wen, F.; et al. Prevalence and risk factors of chronic obstructive pulmonary disease in China (the China Pulmonary Health [CPH] study): A national cross-sectional study. Lancet N. Am. Ed. 2018, 391, 1706–1717. [Google Scholar] [CrossRef] [PubMed]
- Palágyi, K.; Tschirren, J.; Hoffman, E.A.; Sonka, M. Quantitative analysis of pulmonary airway tree structures. Comput. Biol. Med. 2006, 36, 974–996. [Google Scholar] [CrossRef] [PubMed]
- Society, N.; Chinese Medical Doctor Association. Expert consensus on the clinical practice of neonatal brain magnetic resonance imaging. Chin. J. Contemp. Pediatr. 2022, 24, 14–25. [Google Scholar]
- Sanchesa, P.; Meyer, C.; Vigon, V.; Naegel, B. Cerebrovascular network segmentation of MRA images with deep learning. In Proceedings of the 2019 IEEE 16th International Symposium on Biomedical Imaging (ISBI 2019), Venice, Italy, 8–11 April 2019; IEEE: Piscataway, NJ, USA, 2019; pp. 768–771. [Google Scholar]
- Fan, S.; Bian, Y.; Chen, H.; Kang, Y.; Yang, Q.; Tan, T. Unsupervised cerebrovascular segmentation of TOF-MRA images based on deep neural network and hidden markov random field model. Front. Neuroinformatics 2020, 13, 77. [Google Scholar] [CrossRef]
- Zhao, F.; Chen, Y.; Hou, Y.; He, X. Segmentation of blood vessels using rule-based and machine-learning-based methods: A review. Multimed. Syst. 2019, 25, 109–118. [Google Scholar] [CrossRef]
- Hilbert, A.; Madai, V.; Akay, E.; Aydin, O.; Behland, J.; Sobesky, J.; Galinovic, I.; Khalil, A.; Taha, A.; Wuerfel, J.; et al. BRAVE-NET: Fully automated arterial brain vessel segmentation in patients with cerebrovascular disease. Front. Artif. Intell. 2020, 3, 552258. [Google Scholar] [CrossRef]
- Kuo, W.; de Bruijne, M.; Petersen, J.; Nasserinejad, K.; Ozturk, H.; Chen, Y.; Perez-Rovira, A.; Tiddens, H.A. Diagnosis of bronchiectasis and airway wall thickening in children with cystic fibrosis: Objective airway-artery quantification. Eur. Radiol. 2017, 27, 4680–4689. [Google Scholar] [CrossRef]
- Tschirren, J.; Yavarna, T.; Reinhardt, J. Airway segmentation framework for clinical environments. In Proceedings of the 2nd International Workshop Pulmonary Image Analysis, London, UK, 20 September 2009; pp. 227–238. [Google Scholar]
- Guo, X.; Xiao, R.; Lu, Y.; Chen, C.; Yan, F.; Zhou, K.; He, W.; Wang, Z. Cerebrovascular segmentation from TOF-MRA based on multiple-U-net with focal loss function. Comput. Methods Programs Biomed. 2021, 202, 105998. [Google Scholar] [CrossRef]
- Ronneberger, O.; Fischer, P.; Brox, T. U-net: Convolutional networks for biomedical image segmentation. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Munich, Germany, 5–9 October 2015; pp. 234–241. [Google Scholar]
- Çiçek, Ö.; Abdulkadir, A.; Lienkamp, S.; Brox, T.; Ronneberger, O. 3D U-Net: Learning dense volumetric segmentation from sparse annotation. In Proceedings of the Medical Image Computing and Computer-Assisted Intervention–MICCAI 2016: 19th International Conference, Athens, Greece, 17–21 October 2016; Springer International Publishing: Berlin/Heidelberg, Germany, 2016; pp. 424–432. [Google Scholar]
- Tetteh, G.; Efremov, V.; Forkert, N.; Schneider, M.; Kirschke, J.; Weber, B.; Zimmer, C.; Piraud, M.; Menze, B. Deepvesselnet: Vessel segmentation, centerline prediction, and bifurcation detection in 3-d angiographic volumes. Front. Neurosci. 2020, 14, 592352. [Google Scholar] [CrossRef]
- Livne, M.; Rieger, J.; Aydin, O.; Taha, A.; Akay, E.; Kossen, T.; Sobesky, J.; Kelleher, J.; Hildebrand, K.; Frey, D.; et al. A U-Net deep learning framework for high performance vessel segmentation in patients with cerebrovascular disease. Front. Neurosci. 2019, 13, 97. [Google Scholar] [CrossRef]
- Lee, K.; Sunwoo, L.; Kim, T.; Lee, K. Spider U-Net: Incorporating inter-slice connectivity using LSTM for 3D blood vessel segmentation. Appl. Sci. 2021, 11, 2014. [Google Scholar] [CrossRef]
- Oktay, O.; Schlemper, J.; Folgoc, L.; Lee, M.L.; Heinrich, M.; Misawa, K.; Mori, K.; McDonagh, S.; Hammerla, Y.; Kainz, B.; et al. Attention u-net: Learning where to look for the pancreas. arXiv 2018, arXiv:1804.03999. [Google Scholar]
- Lo, P.; Van Ginneken, B.; Reinhardt, J.M.; Yavarna, T.; de Jong, P.A.; Irving, B.; Fetita, C.; Ortner, M.; Pinho, R.; Sijbers, J.; et al. Extraction of airways from CT (EXACT’09). IEEE Trans. Med. Imaging 2012, 31, 2093–2107. [Google Scholar] [CrossRef] [PubMed]
- Park, J.W. Connectivity-based local adaptive thresholding for carotid artery segmentation using MRA images. Image Vis. Comput. 2005, 23, 1277–1287. [Google Scholar]
- Wang, R.; Li, C.; Wang, J.; Wei, X.; Li, Y.; Zhu, Y.; Zhang, S. Threshold segmentation algorithm for automatic extraction of cerebral vessels from brain magnetic resonance angiography images. J. Neurosci. Methods 2015, 241, 30–36. [Google Scholar] [CrossRef]
- Chen, P.; Zou, T.; Chen, J.Y.; Gao, Z.; Xiong, J. The application of improved pso algorithm in pmmw image ostu threshold segmentation. In Applied Mechanics and Materials; Trans Tech Publications Ltd.: Bäch, Switzerland, 2015; Volume 721, pp. 779–782. [Google Scholar]
- Zhu, Q.; Jing, L.; Bi, R. Exploration and improvement of Ostu threshold segmentation algorithm. In Proceedings of the 2010 8th World Congress on Intelligent Control and Automation, Jinan, China, 7–9 July 2010; IEEE: Piscataway, NJ, USA, 2010; pp. 6183–6188. [Google Scholar]
- Neumann, J.O.; Campos, B.; Younes, B.; Jakobs, A.; Unterberg, A.; Kiening, K.; Hubert, A. Evaluation of three automatic brain vessel segmentation methods for stereotactical trajectory planning. Comput. Methods Programs Biomed. 2019, 182, 105037. [Google Scholar] [CrossRef] [PubMed]
- Rad, A.E.; Mohd Rahim, M.S.; Kolivand, H.; Amin, I. Morphological region-based initial contour algorithm for level set methods in image segmentation. Multimed. Tools Appl. 2017, 76, 2185–2201. [Google Scholar] [CrossRef]
- Frangi, A.F.; Niessen, W.; Vincken, K.; Viergever, M. Multi-scale vessel enhancement filtering. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention; Springer: Berlin/Heidelberg, Germany, 1998; pp. 130–137. [Google Scholar]
- Mori, K.; Hasegawa, J.; Toriwaki, J.; Anno, H.; Katada, K. Recognition of bronchus in three-dimensional X-ray CT images with application to virtualized bronchoscopy system. In Proceedings of the 13th International Conference on Pattern Recognition, Vienna, Austria, 25–29 August 1996; pp. 528–532. [Google Scholar]
- Sonka, M.; Park, W.; Hoffman, E. Rule-based detection of intrathoracic airway trees. IEEE Trans. Med. Imaging 1996, 15, 314–326. [Google Scholar] [CrossRef]
- Tschirren, J.; Hoffman, E.; McLennan, G.; Sonka, M. Intrathoracic airway trees: Segmentation and airway morphology analysis from low-dose CT scans. IEEE Trans. Med. Imaging 2005, 24, 1529–1539. [Google Scholar] [CrossRef]
- Duan, H.H.; Gong, J.; Sun, X.W.; Nie, S. Region growing algorithm combined with morphology and skeleton analysis for segmenting airway tree in CT images. J. X-Ray Sci. Technol. 2020, 28, 311–331. [Google Scholar] [CrossRef]
- Bhargavi, K.; Jyothi, S. A survey on threshold based segmentation technique in image processing. Int. J. Innov. Res. Dev. 2014, 3, 234–239. [Google Scholar]
- Litjens, G.; Kooi, T.; Bejnordi, B.E.; Setio, A.; Ciompi, F.; Ghafoorian, M.; Laak, J.; Ginneken, B.; Sanchez, C.I. A survey on deep learning in medical image analysis. Med. Image Anal. 2017, 42, 60–88. [Google Scholar] [CrossRef]
- Ke, Q.; Zhang, J.; Wei, W.; Polap, D.; Wozniak, M.; Kosmider, L.; Damasevicius, R. A neuro-heuristic approach for recognition of lung diseases from X-ray images. Expert Syst. Appl. 2019, 126, 218–232. [Google Scholar] [CrossRef]
- Jaszcz, A.; Połap, D.; Damaševičius, R. Lung x-ray image segmentation using heuristic red fox optimization algorithm. Sci. Program. 2022, 2022, 4494139. [Google Scholar] [CrossRef]
- Min, Y.; Nie, S. Automatic Segmentation of Cerebrovascular Based on Deep Learning. In Proceedings of the 2021 3rd International Conference on Artificial Intelligence and Advanced Manufacture, Manchester, UK, 23–25 October 2021; pp. 94–98. [Google Scholar]
- Mou, L.; Zhao, Y.; Fu, H.; Liu, Y.; Cheng, J.; Zheng, Y.; Su, P.; Yang, J.; Chen, L.; Frangi, A.F.; et al. CS2-Net: Deep learning segmentation of curvilinear structures in medical imaging. Med. Image Anal. 2021, 67, 101874. [Google Scholar] [CrossRef] [PubMed]
- Xia, L.; Zhang, H.; Wu, Y.; Song, R.; Ma, Y.; Mou, L.; Liu, J.; Xie, Y.; Ma, M.; Zhao, Y. 3D vessel-like structure segmentation in medical images by an edge-reinforced network. Med. Image Anal. 2022, 82, 102581. [Google Scholar] [CrossRef]
- Chen, Y.; Jin, D.; Guo, B.; Bai, X. Attention-Assisted Adversarial Model for Cerebrovascular Segmentation in 3D TOF-MRA Volumes. IEEE Trans. Med. Imaging 2022, 41, 3520–3532. [Google Scholar] [CrossRef] [PubMed]
- Banerjee, S.; Toumpanakis, D.; Dhara, A.K.; Wikstrom, J.; Strand, R. Topology-Aware Learning for Volumetric Cerebrovascular Segmentation. In Proceedings of the 2022 IEEE 19th International Symposium on Biomedical Imaging (ISBI), Kolkata, India, 28–31 March 2022; IEEE: Piscataway, NJ, USA, 2022; pp. 1–4. [Google Scholar]
- Jiang, Y.; Zhang, Z.; Qin, S.; Guo, Y.; Li, Z.; Cui, S. APAUNet: Axis Projection Attention UNet for Small Target in 3D Medical Segmentation. In Proceedings of the Asian Conference on Computer Vision, Macau, China, 4–8 December 2022; pp. 283–298. [Google Scholar]
- Meng, Q.; Roth, H.R.; Kitasaka, T.; Oda, M.; Ueno, J.; Mori, K. Tracking and segmentation of the airways in chest CT using a fully convolutional network. In Proceedings of the Medical Image Computing and Computer-Assisted Intervention—MICCAI 2017: 20th International Conference, Quebec City, QC, Canada, 11–13 September 2017; Springer International Publishing: Berlin/Heidelberg, Germany, 2017; pp. 198–207. [Google Scholar]
- Garcia-Uceda Juarez, A.; Tiddens, H.; de Bruijne, M. Automatic airway segmentation in chest CT using convolutional neural networks. In Proceedings of the Image Analysis for Moving Organ, Breast, and Thoracic Images: Third International Workshop, RAMBO 2018, Granada, Spain, 16–20 September 2018; pp. 238–250. [Google Scholar]
- Garcia-Uceda Juarez, A.; Selvan, R.; Saghir, Z.; de Bruijne, M. A joint 3D UNet-graph neural network-based method for airway segmentation from chest CTs. In Proceedings of the Machine Learning in Medical Imaging: 10th International Workshop, MLMI 2019, Held in Conjunction with MICCAI 2019, Shenzhen, China, 13 October 2019; pp. 583–591. [Google Scholar]
- Wang, C.; Hayashi, Y.; Oda, M.; Itoh, H.; Kitasaka, T.; Frangi, A.F.; Mori, K. Tubular structure segmentation using spatial fully connected network with radial distance loss for 3D medical images. In Proceedings of the Medical Image Computing and Computer Assisted Intervention–MICCAI 2019: 22nd International Conference, Shenzhen, China, 13–17 October 2019; pp. 348–356. [Google Scholar]
- Tan, W.; Liu, P.; Li, X.; Xu, S.; Chen, Y.; Yang, J. Segmentation of lung airways based on deep learning methods. IET Image Process. 2022, 16, 1444–1456. [Google Scholar] [CrossRef]
- Lin, T.Y.; Dollár, P.; Girshick, R.; He, K.; Hariharan, B.; Belongie, S. Feature pyramid networks for object detection. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21–26 July 2017; pp. 2117–2125. [Google Scholar]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Spatial pyramid pooling in deep convolutional networks for visual recognition. IEEE Trans. Pattern Anal. Mach. Intell. 2015, 37, 1904–1916. [Google Scholar] [CrossRef]
- Zhao, H.; Shi, J.; Qi, X.; Wang, X.; Jia, J. Pyramid scene parsing network. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21–26 July 2017; pp. 2881–2890. [Google Scholar]
- Chen, L.C.; Papandreou, G.; Kokkinos, I.; Murphy, K.; Yuille, A. Deeplab: Semantic image segmentation with deep convolutional nets, atrous convolution, and fully connected crfs. IEEE Trans. Pattern Anal. Mach. Intell. 2017, 40, 834–848. [Google Scholar] [CrossRef]
- Hu, J.; Shen, L.; Sun, G. Squeeze-and-excitation networks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Salt Lake City, UT, USA, 18–22 June 2018; pp. 7132–7141. [Google Scholar]
- Fan, D.P.; Ji, G.P.; Zhou, T.; Chen, G.; Fu, H.; Shen, J.; Shao, L. Pranet: Parallel reverse attention network for polyp segmentation. In Proceedings of the Medical Image Computing and Computer Assisted Intervention–MICCAI 2020: 23rd International Conference, Lima, Peru, 4–8 October 2020; Springer International Publishing: Berlin/Heidelberg, Germany, 2020; pp. 263–273. [Google Scholar]
- Bullitt, E.; Zeng, D.; Gerig, G.; Aylward, S.; Joshi, S.; Smith, J.K.; Lin, W.L.; Ewend, M.G. Vessel tortuosity and brain tumor malignancy: A blinded study1. Acad. Radiol. 2005, 12, 1232–1240. [Google Scholar] [CrossRef] [PubMed]
- Palumbo, O.; Dera, D.; Bouaynaya, N.C.; Fathallah-Shaykh, H. Inverted cone convolutional neural network for deboning MRIs. In Proceedings of the 2018 International Joint Conference on Neural Networks (IJCNN), Rio de Janeiro, Brazil, 8–13 July 2018; IEEE: Piscataway, NJ, USA, 2018; pp. 1–6. [Google Scholar]
- Shorten, C.; Khoshgoftaar, T.M. A survey on image data augmentation for deep learning. J. Big Data 2019, 6, 60. [Google Scholar] [CrossRef]
- Hesamian, M.H.; Jia, W.; He, X.; Kennedy, P. Deep learning techniques for medical image segmentation: Achievements and challenges. J. Digit. Imaging 2019, 32, 582–596. [Google Scholar] [CrossRef] [PubMed]
- Biswas, M.; Kuppili, V.; Saba, L.; Edla, D.R.; Suri, H.S.; Cuadrado-Godia, E.; Laird, J.R.; Marinhoe, R.T.; Sanches, J.M.; Nicolaides, A.; et al. State-of-the-art review on deep learning in medical imaging. Front. Biosci.—Landmark 2019, 24, 380–406. [Google Scholar]
- Cao, H.; Wang, Y.; Chen, J.; Jiang, D.; Zhang, X.; Tian, Q.; Wang, M. Swin-unet: Unet-like pure transformer for medical image segmentation. In Proceedings of the Computer Vision–ECCV 2022 Workshops, Tel Aviv, Israel, 23–27 October 2022; Springer International Publishing: Berlin/Heidelberg, Germany, 2022; pp. 205–218. [Google Scholar]
- Wang, X.; Zhanshan, L.; Yingda, L. Medical image segmentation based on multi-scale context-aware and semantic adaptor. J. Jilin Univ. (Eng. Technol. Ed.) 2022, 52, 640–647. [Google Scholar]
- Liu, S.; Qi, L.; Qin, H.; Shi, J.; Jia, J. Path aggregation network for instance segmentation. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Salt Lake City, UT, USA, 18–22 June 2018; pp. 8759–8768. [Google Scholar]
- Chen, H.; Dou, Q.; Yu, L.; Qin, J.; Heng, P. VoxResNet: Deep voxelwise residual networks for brain segmentation from 3D MR images. NeuroImage 2018, 170, 446–455. [Google Scholar] [CrossRef]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep residual learning for image recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016; pp. 770–778. [Google Scholar]
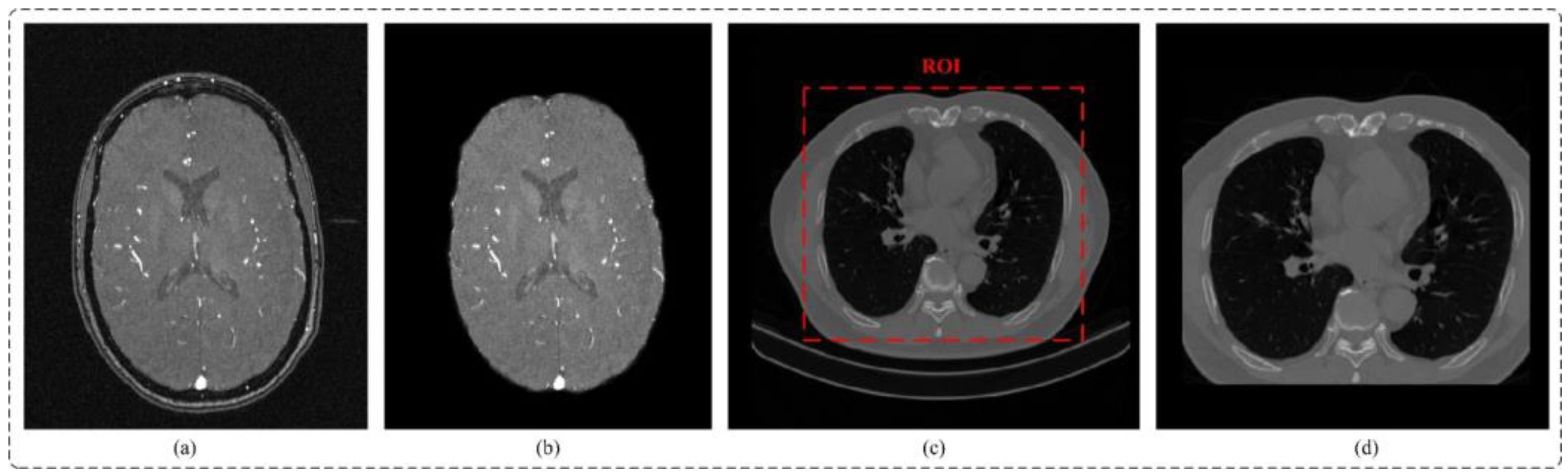








| Dataset | Image Size | Patch Size | Number of Training | Number of Validation | Number of Test | Number of Training Patches |
|---|---|---|---|---|---|---|
| Cerebrovascular (MIDAS) | 448 × 448 × 128 | 64 × 64 × 32 | 76 | 11 | 22 | 46,056 |
| Airway (GMU) | 512 × 512 × 320 | 64 × 64 × 32 | 265 | 38 | 77 | 98,852 |
| Vessels Dataset | Airways Dataset | |||||||
|---|---|---|---|---|---|---|---|---|
| Network | Pre (%) | Re (%) | Di (%) | IoU (%) | Pre (%) | Re (%) | Di (%) | IoU (%) |
| VoxResnet [59] | 85.80 ± 1.92 | 85.92 ± 1.78 | 85.24 ± 1.11 | 74.28 ± 1.64 | 93.92 ± 2.13 | 90.54 ± 3.12 | 92.32 ± 2.03 | 85.74 ± 2.74 |
| APA U-Net [39] | 74.41 ± 3.86 | 84.91 ± 1.63 | 79.22 ± 1.67 | 65.62 ± 2.26 | 90.65 ± 7.71 | 90.55 ± 7.98 | 91.58 ± 6.87 | 84.48 ± 8.57 |
| CS2-Net [35] | 93.15 ± 1.25 | 83.23 ± 1.20 | 87.90 ± 0.67 | 78.42 ± 1.06 | 91.42 ± 1.95 | 93.70 ± 2.01 | 92.54 ± 2.00 | 86.12 ± 2.79 |
| ER-Net [36] | 91.98 ± 1.44 | 84.67 ± 1.22 | 88.17 ± 0.57 | 78.84 ± 1.44 | 94.79 ± 2.48 | 90.80 ± 5.14 | 92.75 ± 3.53 | 86.49 ± 5.46 |
| Resnet (deep = 18) [60] | 91.21 ± 1.18 | 88.14 ± 1.66 | 89.63 ± 0.81 | 81.22 ± 1.33 | 96.42 ± 5.79 | 89.02 ± 3.27 | 93.27 ± 2.54 | 87.39 ± 3.22 |
| U-Net [13] | 91.60 ± 1.92 | 86.49 ± 1.03 | 88.95 ± 0.89 | 80.10 ± 1.44 | 96.34 ± 0.65 | 89.72 ± 3.15 | 93.25 ± 1.84 | 87.35 ± 3.15 |
| Attention U-Net [17] | 89.98 ± 1.35 | 88.48 ± 1.52 | 88.17 ± 0.45 | 78.84 ± 1.05 | 97.04 ± 0.61 | 89.86 ± 3.72 | 92.87 ± 2.25 | 86.69 ± 3.68 |
| Rattention U-Net [50] | 90.17 ± 1.39 | 88.30 ± 1.23 | 89.21 ± 0.60 | 80.52 ± 0.99 | 95.35 ± 5.21 | 90.55 ± 2.97 | 92.80 ± 3.51 | 86.57 ± 5.50 |
| UARAI (Ours) | 93.89 ± 1.22 | 87.27 ± 2.15 | 90.31 ± 0.82 | 82.33 ± 1.37 | 97.41 ± 0.56 | 89.67 ± 3.37 | 93.34 ± 1.98 | 87.51 ± 3.34 |
| Patch Size | 16 × 16 × 32 | 32 × 32 × 32 | ||||||
|---|---|---|---|---|---|---|---|---|
| Network | Pre (%) | Re (%) | Di (%) | IoU (%) | Pre (%) | Re (%) | Di (%) | IoU (%) |
| VoxResnet [59] | 83.75 ± 1.95 | 85.76 ± 1.40 | 84.73 ± 1.24 | 73.51 ± 1.84 | 83.75 ± 1.95 | 85.76 ± 1.40 | 84.73 ± 1.24 | 73.51 ± 1.84 |
| Resnet (deep = 18) [60] | 82.10 ± 1.78 | 79.40 ± 1.25 | 80.20 ± 1.96 | 66.94 ± 2.01 | 83.40 ± 1.09 | 80.00 ± 1.56 | 81.70 ± 1.21 | 69.06 ± 1.43 |
| U-Net [13] | 86.33 ± 1.54 | 87.17 ± 0.79 | 86.74 ± 0.72 | 76.58 ± 1.11 | 72.27 ± 1.74 | 79.64 ± 1.05 | 75.75 ± 0.56 | 60.97 ± 0.72 |
| Attention U-Net [17] | 83.79 ± 2.22 | 85.85 ± 1.23 | 84.78 ± 0.82 | 73.58 ± 1.23 | 87.39 ± 1.83 | 84.89 ± 1.55 | 84.92 ± 0.66 | 73.79 ± 1.07 |
| Attention U-Net (MSFA) | 88.14 ± 1.75 | 85.86 ± 1.10 | 85.96 ± 0.77 | 75.38 ± 1.22 | 87.98 ± 1.87 | 85.95 ± 1.64 | 85.88 ± 0.64 | 75.25 ± 1.12 |
| Rattention U-Net [50] | 86.41 ± 1.00 | 86.61 ± 0.65 | 87.90 ± 0.52 | 78.41 ± 0.51 | 88.01 ± 2.97 | 86.40 ± 1.81 | 88.02 ± 0.93 | 78.60 ± 1.36 |
| Rattention U-Net (MSFA) | 86.47 ± 2.43 | 87.71 ± 1.22 | 87.97 ± 0.82 | 78.52 ± 1.22 | 89.77 ± 1.59 | 88.04 ± 1.76 | 88.17 ± 0.73 | 78.84 ± 1.18 |
| UARAI (Ours) | 90.82 ± 1.88 | 86.79 ± 0.94 | 88.93 ± 1.11 | 80.07 ± 1.74 | 91.60 ± 1.33 | 87.61 ± 0.95 | 89.03 ± 0.80 | 80.23 ± 1.31 |
| Patch Size | 64 × 64 × 32 | 96 × 96 × 32 | ||||||
| Network | Pre (%) | Re (%) | Di (%) | IoU (%) | Pre (%) | Re (%) | Di (%) | IoU (%) |
| VoxResnet [59] | 83.75 ± 1.95 | 85.76 ± 1.40 | 84.73 ± 1.24 | 73.51 ± 1.84 | 83.75 ± 1.95 | 85.76 ± 1.40 | 84.73 ± 1.24 | 73.51 ± 1.84 |
| Resnet (deep = 18) [60] | 82.10 ± 1.78 | 79.40 ± 1.25 | 80.20 ± 1.96 | 66.94 ± 2.01 | 83.40 ± 1.09 | 80.00 ± 1.56 | 81.70 ± 1.21 | 69.06 ± 1.43 |
| U-Net [13] | 86.33 ± 1.54 | 87.17 ± 0.79 | 86.74 ± 0.72 | 76.58 ± 1.11 | 72.27 ± 1.74 | 79.64 ± 1.05 | 75.75 ± 0.56 | 60.97 ± 0.72 |
| Attention U-Net [17] | 83.79 ± 2.22 | 85.85 ± 1.23 | 84.78 ± 0.82 | 73.58 ± 1.23 | 87.39 ± 1.83 | 84.89 ± 1.55 | 84.92 ± 0.66 | 73.79 ± 1.07 |
| Attention U-Net (MSFA) | 88.14 ± 1.75 | 85.86 ± 1.10 | 85.96 ± 0.77 | 75.38 ± 1.22 | 87.98 ± 1.87 | 85.95 ± 1.64 | 85.88 ± 0.64 | 75.25 ± 1.12 |
| Rattention U-Net [50] | 86.41 ± 1.00 | 86.61 ± 0.65 | 87.90 ± 0.52 | 78.41 ± 0.51 | 88.01 ± 2.97 | 86.40 ± 1.81 | 88.02 ± 0.93 | 78.60 ± 1.36 |
| Rattention U-Net (MSFA) | 86.47 ± 2.43 | 87.71 ± 1.22 | 87.97 ± 0.82 | 78.52 ± 1.22 | 89.77 ± 1.59 | 88.04 ± 1.76 | 88.17 ± 0.73 | 78.84 ± 1.18 |
| UARAI (Ours) | 90.82 ± 1.88 | 86.79 ± 0.94 | 88.93 ± 1.11 | 80.07 ± 1.74 | 91.60 ± 1.33 | 87.61 ± 0.95 | 89.03 ± 0.80 | 80.23 ± 1.31 |
| Patch Size | 128 × 128 × 32 | |||||||
| Network | Pre (%) | Re (%) | Di (%) | IoU (%) | ||||
| VoxResnet [59] | 83.75 ± 1.95 | 85.76 ± 1.40 | 84.73 ± 1.24 | 73.51 ± 1.84 | ||||
| Resnet (deep = 18) [60] | 83.10 ± 1.52 | 74.00 ± 1.12 | 80.90 ± 0.88 | 67.93 ± 0.81 | ||||
| U-Net [13] | 79.50 ± 3.60 | 83.25 ± 3.01 | 81.24 ± 2.11 | 68.41 ± 3.02 | ||||
| Attention U-Net [17] | 76.70 ± 2.01 | 86.42 ± 2.69 | 81.21 ± 1.15 | 68.36 ± 1.69 | ||||
| Attention U-Net (MSFA) | 80.45 ± 1.42 | 86.56 ± 1.23 | 83.45 ± 0.56 | 71.60 ± 1.17 | ||||
| Rattention U-Net [50] | 89.20 ± 1.30 | 85.94 ± 1.70 | 88.93 ± 0.77 | 80.07 ± 1.27 | ||||
| Rattention U-Net (MSFA) | 90.02 ± 1.16 | 86.79 ± 0.97 | 88.82 ± 0.73 | 79.89 ± 1.19 | ||||
| UARAI (Ours) | 90.14 ± 1.36 | 85.40 ± 1.39 | 88.82 ± 0.61 | 79.89 ± 1.00 | ||||
| Patch Size | 16 × 16 × 32 | 32 × 32 × 32 | ||||||
|---|---|---|---|---|---|---|---|---|
| Network | Pre (%) | Re (%) | Di (%) | IoU (%) | Pre (%) | Re (%) | Di (%) | IoU (%) |
| VoxResnet [59] | 93.89 ± 2.03 | 90.65 ± 2.51 | 92.21 ± 1.73 | 85.54 ± 2.99 | 93.89 ± 2.03 | 90.65 ± 2.51 | 92.21 ± 1.73 | 85.54 ± 2.99 |
| Resnet (deep = 18) [60] | 75.90 ± 6.25 | 80.25 ± 5.01 | 79.16 ± 4.98 | 65.51 ± 6.98 | 77.54 ± 5.92 | 85.36 ± 5.21 | 82.31 ± 5.37 | 69.94 ± 6.02 |
| U-Net [13] | 81.20 ± 7.52 | 87.17 ± 5.45 | 83.83 ± 5.10 | 72.16 ± 7.42 | 93.81 ± 1.94 | 89.00 ± 3.13 | 90.12 ± 2.01 | 82.02 ± 3.44 |
| Attention U-Net [17] | 81.39 ± 7.97 | 85.88 ± 5.54 | 80.90 ± 5.46 | 67.93 ± 7.55 | 94.44 ± 5.28 | 90.77 ± 2.65 | 92.50 ± 3.47 | 86.05 ± 5.50 |
| Attention U-Net (MSFA) | 81.40 ± 6.88 | 86.23 ± 5.39 | 84.83 ± 4.33 | 73.66 ± 6.51 | 96.86 ± 1.49 | 89.78 ± 3.14 | 92.86 ± 1.93 | 86.67 ± 3.31 |
| Rattention U-Net [50] | 54.88 ± 15.99 | 86.92 ± 5.81 | 66.37 ± 10.59 | 49.67 ± 12.78 | 95.28 ± 3.63 | 90.08 ± 2.75 | 92.55 ± 2.34 | 86.13 ± 3.97 |
| Rattention U-Net (MSFA) | 62.79 ± 12.16 | 85.81 ± 4.80 | 71.08 ± 9.17 | 55.13 ± 10.28 | 96.75 ± 1.36 | 89.77 ± 3.33 | 93.01 ± 1.97 | 86.93 ± 3.35 |
| UARAI (Ours) | 83.22 ± 7.44 | 88.72 ± 4.61 | 85.62 ± 4.54 | 74.86 ± 6.82 | 96.95 ± 4.35 | 89.94 ± 2.30 | 93.04 ± 2.34 | 86.99 ± 3.95 |
| Patch size | 64 × 64 × 32 | 96 × 96 × 32 | ||||||
| Network | Pre (%) | Re (%) | Di (%) | IoU (%) | Pre (%) | Re (%) | Di (%) | IoU (%) |
| VoxResnet [59] | 93.89 ± 2.03 | 90.65 ± 2.51 | 92.21 ± 1.73 | 85.54 ± 2.99 | 93.89 ± 2.03 | 90.65 ± 2.51 | 92.21 ± 1.73 | 85.54 ± 2.99 |
| Resnet (deep = 18) [60] | 82.97 ± 2.21 | 86.25 ± 3.08 | 85.19 ± 2.72 | 74.20 ± 4.21 | 82.91 ± 3.27 | 85.00 ± 4.21 | 84.99 ± 3.10 | 73.90 ± 5.13 |
| U-Net [13] | 95.79 ± 3.83 | 90.00 ± 2.57 | 92.73 ± 2.4 | 86.45 ± 4.04 | 95.64 ± 3.73 | 89.21 ± 2.32 | 92.71 ± 2.65 | 86.41 ± 3.84 |
| Attention U-Net [17] | 96.36 ± 2.42 | 88.99 ± 3.70 | 92.8 ± 2.28 | 86.57 ± 3.72 | 94.12 ± 5.73 | 90.69 ± 2.65 | 92.82 ± 3.71 | 86.60 ± 5.58 |
| Attention U-Net (MSFA) | 96.74 ± 1.10 | 90.11 ± 2.81 | 93.15 ± 2.05 | 87.18 ± 3.50 | 96.27 ± 1.98 | 89.76 ± 3.47 | 93.00 ± 1.37 | 86.92 ± 3.16 |
| Rattention U-Net [50] | 93.61 ± 5.98 | 90.65 ± 2.95 | 91.99 ± 3.92 | 85.17 ± 6.02 | 95.22 ± 3.66 | 90.65 ± 2.21 | 92.09 ± 2.30 | 85.34 ± 3.39 |
| Rattention U-Net (MSFA) | 96.12 ± 1.96 | 89.73 ± 3.36 | 93.14 ± 2.23 | 87.16 ± 3.75 | 96.04 ± 1.53 | 89.71 ± 3.61 | 93.01 ± 1.70 | 86.93 ± 3.49 |
| UARAI (Ours) | 96.90 ± 1.06 | 90.62 ± 5.03 | 93.20 ± 3.29 | 87.27 ± 4.85 | 93.20 ± 4.85 | 91.55 ± 2.98 | 93.05 ± 2.39 | 87.00 ± 3.52 |
| Patch size | 128 × 128 × 32 | |||||||
| Network | Pre (%) | Re (%) | Di (%) | IoU (%) | ||||
| VoxResnet [59] | 93.89 ± 2.03 | 90.65 ± 2.51 | 92.21 ± 1.73 | 85.54 ± 2.99 | ||||
| Resnet (deep = 18) [60] | 81.90 ± 2.13 | 84.27 ± 3.58 | 84.46 ± 3.25 | 73.10 ± 3.24 | ||||
| U-Net [13] | 96.42 ± 1.25 | 88.34 ± 4.20 | 92.63 ± 2.62 | 86.27 ± 4.28 | ||||
| Attention U-Net [17] | 96.92 ± 1.90 | 89.15 ± 3.10 | 92.84 ± 2.01 | 86.64 ± 3.44 | ||||
| Attention U-Net (MSFA) | 97.00 ± 1.37 | 89.54 ± 3.32 | 92.74 ± 2.33 | 86.46 ± 3.87 | ||||
| Rattention U-Net [50] | 97.07 ± 1.22 | 86.40 ± 4.40 | 91.56 ± 2.61 | 84.43 ± 4.30 | ||||
| Rattention U-Net (MSFA) | 97.06 ± 0.58 | 87.79 ± 6.28 | 92.22 ± 3.80 | 85.56 ± 6.02 | ||||
| UARAI (Ours) | 97.07 ± 1.77 | 88.87 ± 3.65 | 93.09 ± 2.04 | 87.07 ± 3.46 | ||||
| Vessels Dataset | Airways Dataset | |||||||
|---|---|---|---|---|---|---|---|---|
| Network | Pre (%) | Re (%) | Di (%) | IoU (%) | Pre (%) | Re (%) | Di (%) | IoU (%) |
| Baseline | 91.60 ± 1.92 | 86.49 ± 1.03 | 88.95 ± 0.89 | 80.10 ± 1.44 | 96.34 ± 0.65 | 89.72 ± 3.15 | 93.25 ± 1.84 | 87.35 ± 3.15 |
| Baseline + MSFA | 91.88 ± 1.74 | 87.01 ± 1.62 | 89.07 ± 0.74 | 80.29 ± 1.22 | 96.68 ± 0.91 | 89.49 ± 3.33 | 93.28 ± 2.03 | 87.41 ± 3.45 |
| Baseline + Att | 89.98 ± 1.35 | 88.48 ± 1.52 | 88.17 ± 0.45 | 78.84 ± 1.05 | 97.04 ± 0.61 | 89.86 ± 3.72 | 92.87 ± 2.25 | 86.69 ± 3.68 |
| Baseline + MSFA + Att | 91.90 ± 1.24 | 86.36 ± 1.47 | 89.03 ± 0.60 | 80.23 ± 0.98 | 97.06 ± 1.37 | 89.99 ± 2.17 | 93.24 ± 1.76 | 87.34 ± 3.03 |
| Baseline + Ra | 90.17 ± 1.39 | 88.30 ± 1.23 | 89.21 ± 0.60 | 80.52 ± 0.99 | 95.35 ± 5.21 | 90.55 ± 2.97 | 92.80 ± 3.51 | 86.57 ± 5.50 |
| Baseline + MSFA + Ra | 92.60 ± 1.32 | 86.82 ± 1.35 | 89.60 ± 0.69 | 81.16 ± 1.14 | 96.34 ± 0.78 | 90.54 ± 5.03 | 93.27 ± 3.31 | 87.38 ± 4.89 |
| Baseline + MSFA + Ra + Icp | 93.89 ± 1.22 | 87.27 ± 2.15 | 90.31 ± 0.82 | 82.33 ± 1.37 | 97.41 ± 0.56 | 89.67 ± 3.37 | 93.34 ± 1.98 | 87.51 ± 3.34 |
| Vessels Dataset | Airways Dataset | |||||||
|---|---|---|---|---|---|---|---|---|
| Network | Pre (%) | Re (%) | Di (%) | IoU (%) | Pre (%) | Re (%) | Di (%) | IoU (%) |
| Baseline + MSFA | 91.88 ± 1.74 | 87.01 ± 1.62 | 89.07 ± 0.74 | 80.29 ± 1.22 | 96.68 ± 0.91 | 89.49 ± 3.33 | 93.28 ± 2.03 | 87.41 ± 3.45 |
| Baseline + MSFA + Att | 91.90 ± 1.24 | 86.36 ± 1.47 | 89.03 ± 0.60 | 80.52 ± 0.98 | 97.06 ± 1.37 | 89.99 ± 2.17 | 93.24 ± 1.76 | 87.34 ± 3.03 |
| Baseline + MSFA + RA + Icp | 93.89 ± 1.22 | 87.27 ± 2.15 | 90.31 ± 0.82 | 82.33 ± 1.37 | 97.41 ± 0.56 | 89.67 ± 3.37 | 93.34 ± 1.98 | 87.51 ± 3.34 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zeng, X.; Guo, Y.; Zaman, A.; Hassan, H.; Lu, J.; Xu, J.; Yang, H.; Miao, X.; Cao, A.; Yang, Y.; et al. Tubular Structure Segmentation via Multi-Scale Reverse Attention Sparse Convolution. Diagnostics 2023, 13, 2161. https://doi.org/10.3390/diagnostics13132161
Zeng X, Guo Y, Zaman A, Hassan H, Lu J, Xu J, Yang H, Miao X, Cao A, Yang Y, et al. Tubular Structure Segmentation via Multi-Scale Reverse Attention Sparse Convolution. Diagnostics. 2023; 13(13):2161. https://doi.org/10.3390/diagnostics13132161
Chicago/Turabian StyleZeng, Xueqiang, Yingwei Guo, Asim Zaman, Haseeb Hassan, Jiaxi Lu, Jiaxuan Xu, Huihui Yang, Xiaoqiang Miao, Anbo Cao, Yingjian Yang, and et al. 2023. "Tubular Structure Segmentation via Multi-Scale Reverse Attention Sparse Convolution" Diagnostics 13, no. 13: 2161. https://doi.org/10.3390/diagnostics13132161
APA StyleZeng, X., Guo, Y., Zaman, A., Hassan, H., Lu, J., Xu, J., Yang, H., Miao, X., Cao, A., Yang, Y., Chen, R., & Kang, Y. (2023). Tubular Structure Segmentation via Multi-Scale Reverse Attention Sparse Convolution. Diagnostics, 13(13), 2161. https://doi.org/10.3390/diagnostics13132161









