Detection and Characterization of Gastric Cancer Using Cascade Deep Learning Model in Endoscopic Images
Abstract
:1. Introduction
1.1. Background
1.2. Related Works
2. Methods
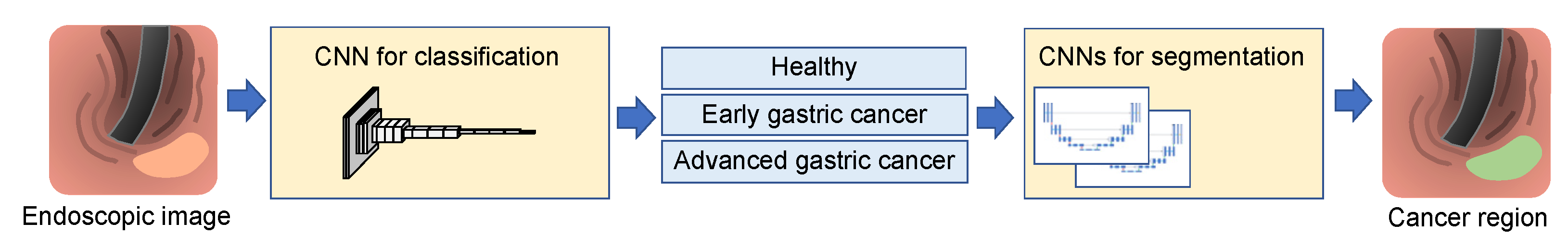
2.1. Proposed Method
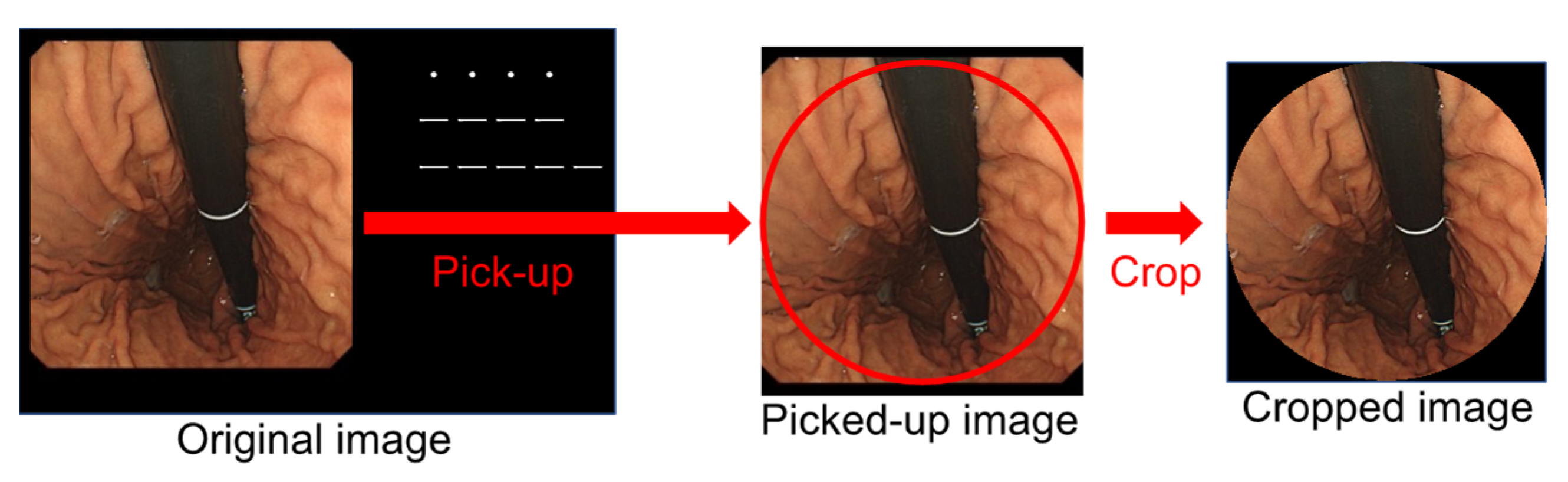
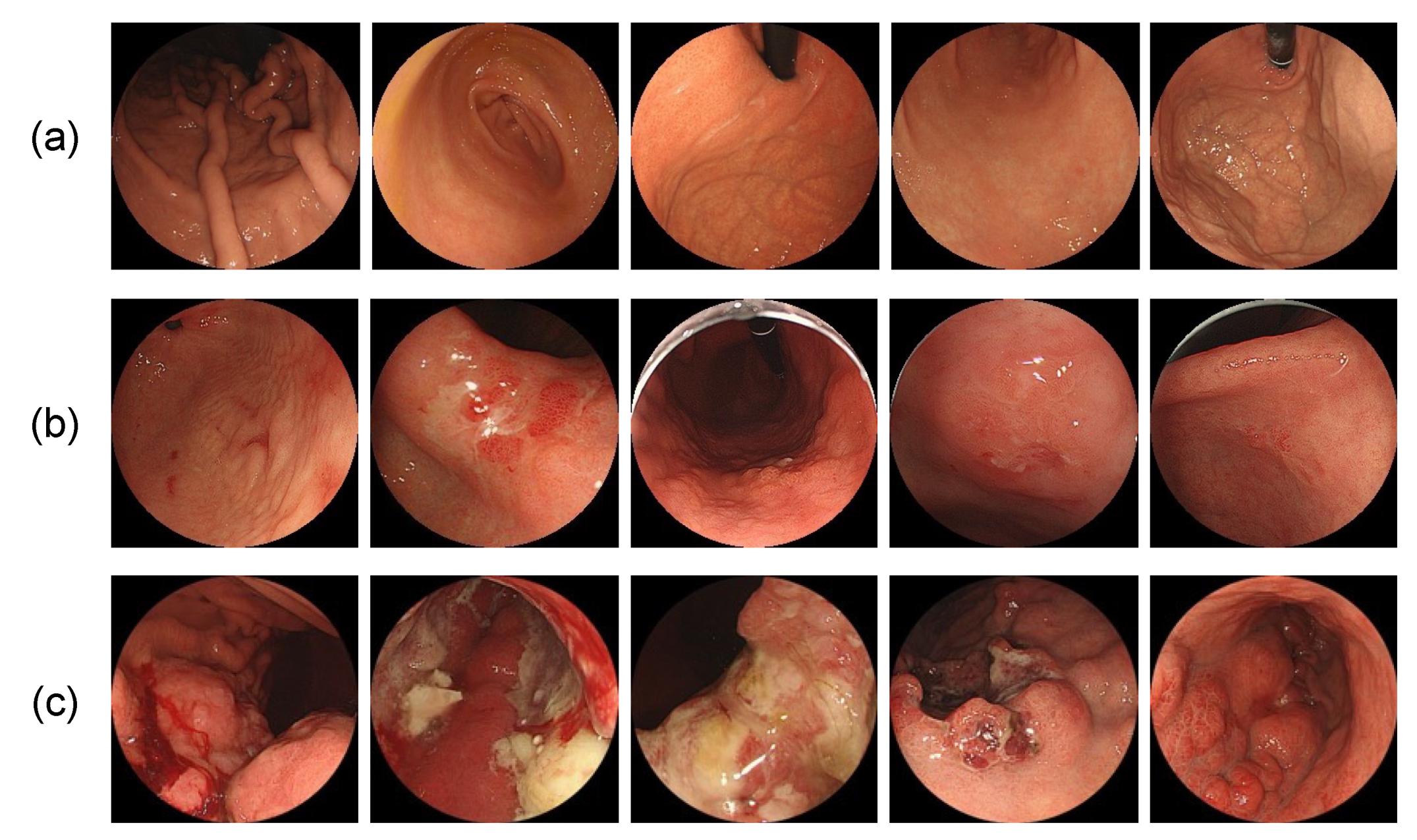
2.2. Image Dataset
2.3. Annotation of Gastric Cancer Region
2.4. Data Augmentation
2.5. Network Architecture for Image Classification
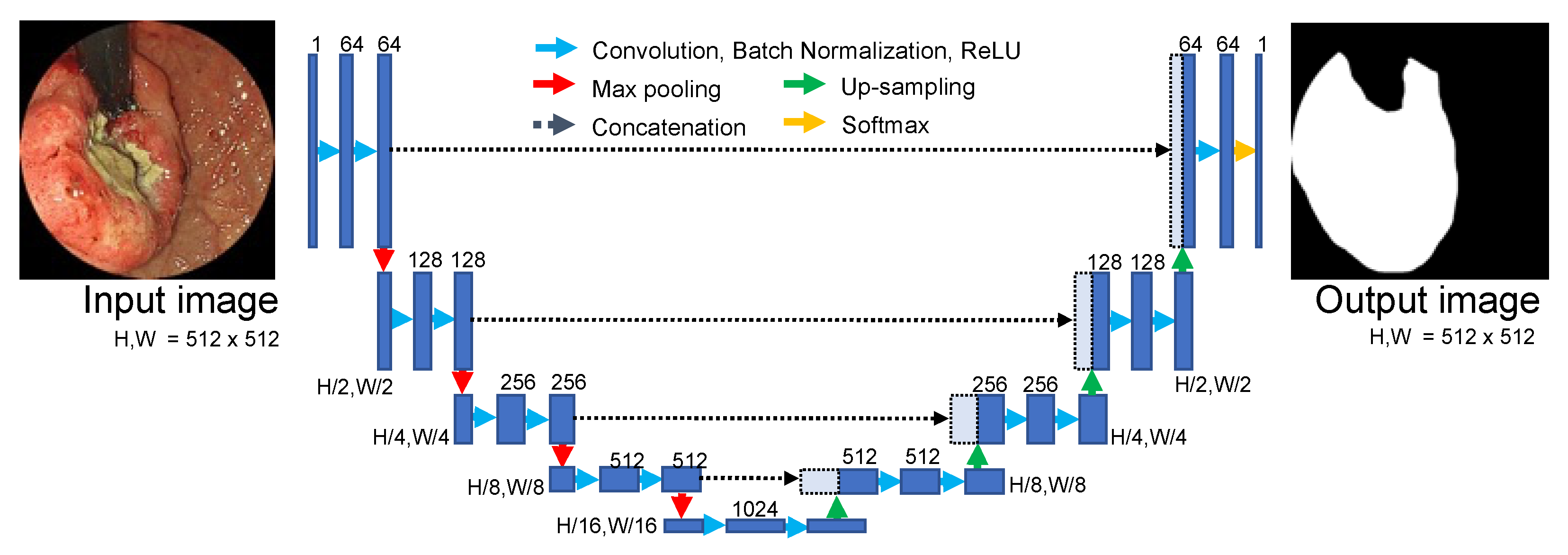
2.6. Network Architecture for Image Segmentation
2.7. Evaluation Metrics
3. Results
Image Classification Result
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Fitzmaurice, C.; Akinyemiju, T.F.; Al Lami, F.H.; Alam, T.; Alizadeh-Navaei, R.; Allen, C.; Alsharif, U.; Alvis-Guzman, N.; Amini, E.; Anderson, B.O.; et al. Global, regional, and national cancer incidence, mortality, years of life lost, years lived with disability, and disability-adjusted life-years for 29 cancer groups, 1990 to 2016: A systematic analysis for the global burden of disease study global burden of disease cancer collaboration. JAMA Oncol. 2018, 4, 1553–1568. [Google Scholar] [PubMed]
- Karger Publishers. GLOBOCAN 2012: Estimated Cancer Incidence, Mortality, and Prevalence Worldwide in 2012. Available online: http://globocan.iarc.fr/Pages/fact_sheets_cancer.aspx (accessed on 31 October 2021).
- Tashiro, A.; Sano, M.; Kinameri, K.; Fujita, K.; Takeuchi, Y. Comparing mass screening techniques for gastric cancer in Japan. World J. Gastroenterol. 2006, 12, 4873–4874. [Google Scholar]
- Krizhevsky, A.; Sutskever, I.; Hinton, G.E. ImageNet classification with deep convolutional neural networks. Adv. Neural Inf. Process. Syst. 2012, 25, 1106–1114. [Google Scholar] [CrossRef]
- LeCun, Y.; Bengio, Y.; Hinton, G.E. Deep learning. Nature 2015, 521, 436–444. [Google Scholar] [CrossRef]
- Teramoto, A.; Fujita, H.; Yamamuro, O.; Tamaki, T. Automated detection of pulmonary nodules in PET/CT images: Ensemble false-positive reduction using a convolutional neural network technique. Med. Phys. 2016, 43, 2821–2827. [Google Scholar] [CrossRef] [PubMed]
- Yan, K.; Cai, J.; Zheng, Y.; Harrison, A.P.; Jin, D.; Tang, Y.B.; Tang, Y.X.; Huang, L.; Xiao, J.; Lu, L. Learning from Multiple Datasets with Heterogeneous and Partial Labels for Universal Lesion Detection in CT. arXiv 2020, arXiv:2009.02577. [Google Scholar] [CrossRef] [PubMed]
- Teramoto, A.; Kiriyama, Y.; Tsukamoto, T.; Sakurai, E.; Michiba, E.; Imaizumi, K.; Saito, K.; Fujita, H. Weakly Supervised Learning for Classification of Lung Cytological Images Using Attention-Based Multiple Instance Learning. Sci. Rep. 2011, 11, 20317. [Google Scholar] [CrossRef]
- Jin, Z.; Gan, T.; Wang, P.; Fu, Z.; Zhang, C.; Yan, Q.; Zheng, X.; Liang, X.; Ye, X. Deep learning for gastroscopic images: Computer-aided techniques for clinicians. BioMed. Eng. OnLine 2022, 21, 12. [Google Scholar] [CrossRef]
- Hirasawa, T.; Aoyama, K.; Tanimoto, T.; Ishihara, S.; Shichijo, S.; Ozawa, T.; Ohnishi, T.; Fujishiro, M.; Matsuo, K.; Fujisaki, J.; et al. Application of artificial intelligence using a convolutional neural network for detecting gastric cancer in endoscopic images. Gastric Cancer 2018, 21, 653–660. [Google Scholar] [CrossRef]
- Sakai, Y.; Takemoto, S.; Hori, K.; Nishimura, M.; Ikematsu, H.; Yano, T.; Yokota, H. Automatic detection of early gastric cancer in endoscopic images using a transferring convolutional neural network. In Proceedings of the 2018 40th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), Honolulu, HI, USA, 18–21 July 2018; pp. 4138–4141. [Google Scholar]
- Shibata, T.; Teramoto, A.; Yamada, H.; Ohmiya, N.; Saito, K.; Fujita, H. Automated detection and segmentation of early gastric cancer from endoscopic images using mask R-CNN. Appl. Sci. 2020, 10, 3842. [Google Scholar] [CrossRef]
- Teramoto, A.; Shibata, T.; Yamada, H.; Hirooka, Y.; Saito, K.; Fujita, H. Automated Detection of Gastric Cancer by Retrospective Endoscopic Image Dataset Using U-Net R-CNN. Appl. Sci. 2021, 11, 11275. [Google Scholar] [CrossRef]
- Zhu, Y.; Wang, Q.C.; Xu, M.D.; Zhang, Z.; Cheng, J.; Zhong, Y.S.; Zhang, Y.-Q.; Chen, W.-F.; Yao, L.-Q.; Zhou, P.-H.; et al. Application of convolutional neural networks in the diagnosis of the invasion depth of gastric cancer based on conventional endoscopy. Gastrointest. Endosc. 2019, 89, 806–815. [Google Scholar] [CrossRef]
- Hamada, K.; Kawahara, Y.; Tanimoto, T.; Ohto, A.; Aida, T.; Yamasaki, Y.; Gotoda, T.; Ogawa, T.; Abe, M.; Okanoue, S.; et al. Application of convolutional neural networks for evaluating the depth of invasion of early gastric cancer based on endoscopic images. Gastroenterol. Hepatol. 2022, 37, 352–357. [Google Scholar] [CrossRef]
- Ungureanu, B.; Sacerdotianu, V.; Turcu-Stiolica, A.; Cazacu, I.; Saftoiu, A. Endoscopic Ultrasound vs. Computed Tomography for Gastric Cancer Staging: A Network Meta-Analysis. Diagnostics 2021, 11, 134. [Google Scholar] [CrossRef]
- Hirai, K.; Kuwahara, T.; Furukawa, K.; Kakushima, N.; Furune, S.; Yamamoto, H.; Marukawa, T.; Asai, H.; Matsui, K.; Sasaki, Y.; et al. Artificial intelligence-based diagnosis of upper gastrointestinal subepithelial lesions on endoscopic ultrasonography images. Gastric Cancer 2022, 25, 382–391. [Google Scholar] [CrossRef] [PubMed]
- Kotowski, K.; Adamski, S.; Malara, W.; Machura, B.; Zarudzki, L.; Nalepa, J. Segmenting Brain Tumors from MRI Using Cascaded 3D U-Nets. Lect. Notes. Comput. Sci. 2021, 12659, 265–277. [Google Scholar]
- Wei, J.; Wu, Z.; Wang, L.; Toan, D.; Qu, L.; Yap, P.-T.; Xia, Y.; Li, G.; Shen, D. A cascaded nested network for 3T brain MR image segmentation guided by 7T labeling. Pattern Recognit. 2022, 124, 108420. [Google Scholar] [CrossRef]
- Krüger, N.; Meyer, A.; Tautz, L.; Hüllebrand, M.; Wamala, I.; Pullig, M.; Kofler, M.; Kempfert, J.; Sündermann, S.; Falk, V.; et al. Cascaded neural network-based CT image processing for aortic root analysis. Int. J. Comput. Assist. Radiol. Surg. 2022, 17, 507–519. [Google Scholar] [CrossRef]
- Simonyan, K.; Zisserman, A. Very deep convolutional networks for large-scale image recognition. arXiv 2015, arXiv:1409.1556. [Google Scholar]
- Szegedy, C.; Liu, W.; Jia, Y.; Sermanet, P.; Reed, S.; Anguelov, D.; Erhan, D.; Vanhoucke, V.; Rabinovich, A. Going Deeper with Convolutions. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Boston, MA, USA, 7–12 June 2015; pp. 1–9. [Google Scholar]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep Residual Learning for Image Recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Las Vegas, NV, USA, 27–30 June 2016; pp. 770–778. [Google Scholar]
- Huang, G.; Liu, Z.; van der Maaten, L.; Weinberger, K.Q. Densely connected convolutional networks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Honolulu, HI, USA, 21–26 July 2017; pp. 4700–4708. [Google Scholar]
- Zamir, A.R.; Sax, A.; Shen, W.; Guibas, L.J.; Malik, J.; Savarese, S. Taskonomy: Disentangling Task Transfer Learning. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Salt Lake City, UT, USA, 18–22 June 2018; pp. 3712–3722. [Google Scholar]
- Ronneberger, O.; Fischer, P.; Brox, T. U-Net: Convolutional networks for biomedical image segmentation. Lect. Notes. Comput. Sci. 2015, 9351, 234–241. [Google Scholar]
- Selvaraju, R.R.; Cogswell, M.; Das, A.; Vedantam, R.; Parikh, D.; Batra, D. Grad-CAM: Visual explanations from deep networks via gradient-based localization. In Proceedings of the International Conference on Computer Vision (ICCV), Venice, Italy, 22–29 October 2017; pp. 618–626. [Google Scholar]
- Efron, B. Estimating the error rate of a prediction rule: Improvement on cross-validation. J. Am. Stat. Assoc. 1983, 78, 316–331. [Google Scholar] [CrossRef]
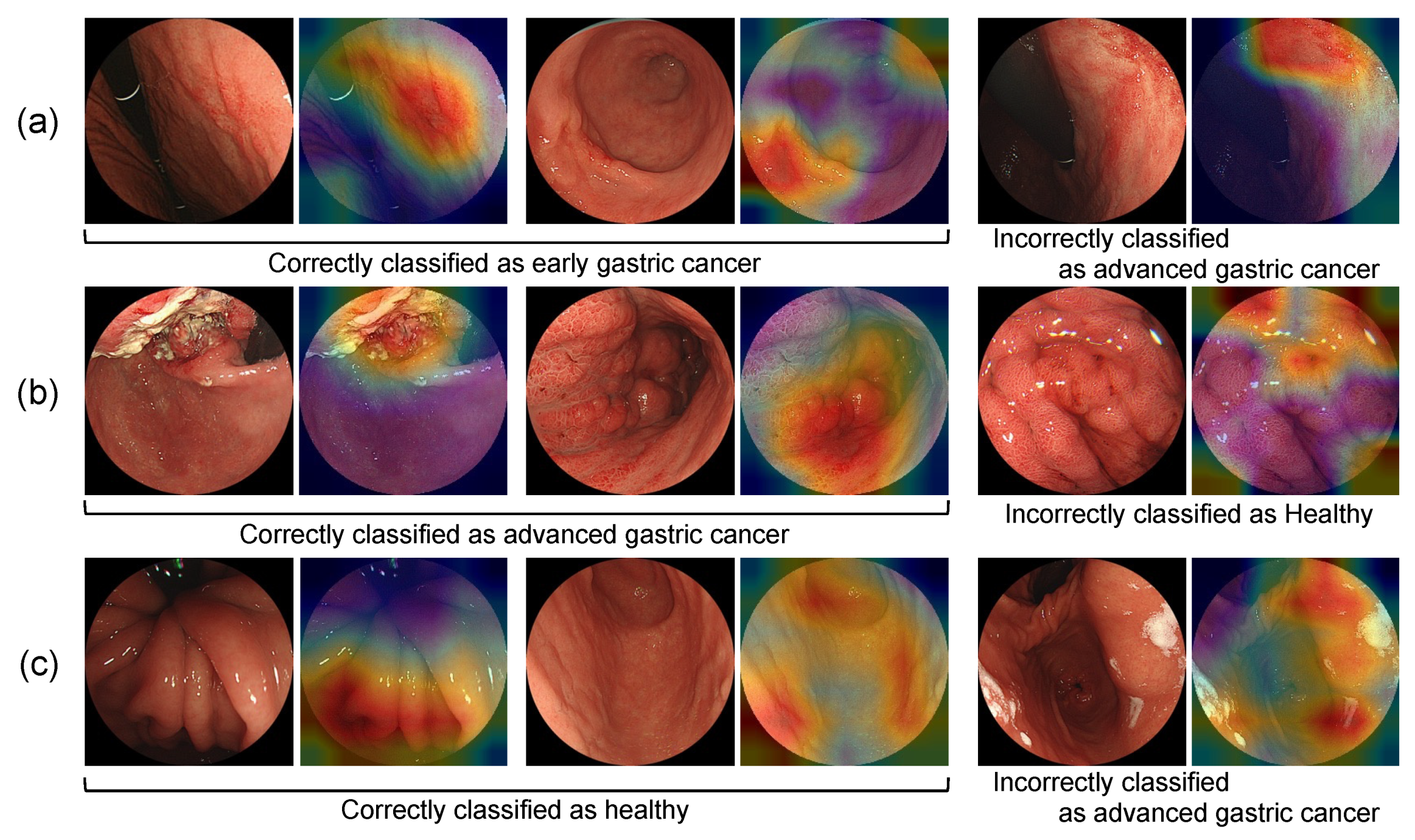
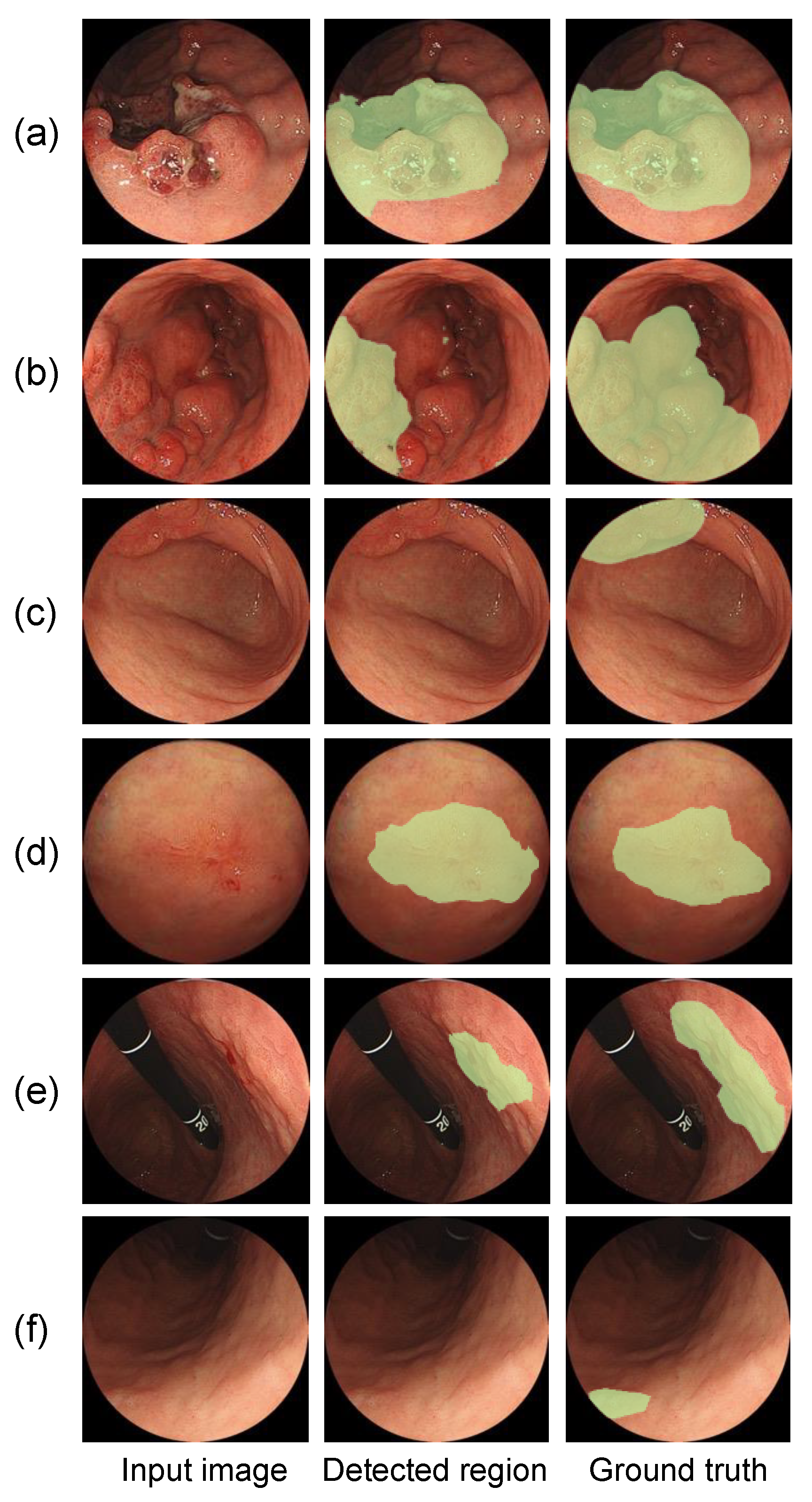
| (a) Image-Based Classification | |||||||
| AccuracyHealthy | AccuracyEGC | AccuracyAGC | Accuracyoverall | AccuracyBalanced | Sensitivity | Specificity | |
| VGG16 | 0.964 | 0.919 | 0.922 | 0.943 | 0.935 | 0.951 | 0.964 |
| VGG19 | 0.994 | 0.947 | 0.904 | 0.960 | 0.949 | 0.939 | 0.994 |
| InceptionV3 | 0.988 | 0.921 | 0.906 | 0.951 | 0.938 | 0.938 | 0.988 |
| DenseNet121 | 0.994 | 0.998 | 0.945 | 0.982 | 0.979 | 0.970 | 0.994 |
| DenseNet169 | 0.995 | 0.998 | 0.903 | 0.971 | 0.965 | 0.948 | 0.995 |
| DenseNet201 | 0.996 | 0.989 | 0.943 | 0.980 | 0.976 | 0.965 | 0.996 |
| ResNet50 | 0.991 | 0.991 | 0.922 | 0.972 | 0.968 | 0.956 | 0.991 |
| ResNet101 | 0.990 | 0.981 | 0.920 | 0.969 | 0.964 | 0.949 | 0.990 |
| ResNet152 | 0.996 | 0.994 | 0.918 | 0.975 | 0.970 | 0.954 | 0.996 |
| (b) Case-based classification | |||||||
| AccuracyHealthy | AccuracyEGC | AccuracyAGC | Accuracyoverall | AccuracyBalanced | Sensitivity | Specificity | |
| VGG16 | 0.976 | 0.937 | 0.960 | 0.952 | 0.958 | 0.979 | 0.976 |
| VGG19 | 1.000 | 0.947 | 0.940 | 0.957 | 0.962 | 0.972 | 1.000 |
| InceptionV3 | 1.000 | 0.958 | 0.980 | 0.973 | 0.979 | 0.986 | 1.000 |
| DenseNet121 | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 |
| DenseNet169 | 1.000 | 1.000 | 0.960 | 0.989 | 0.987 | 0.986 | 1.000 |
| DenseNet201 | 1.000 | 1.000 | 0.980 | 0.995 | 0.993 | 0.993 | 1.000 |
| ResNet50 | 1.000 | 0.989 | 1.000 | 0.995 | 0.996 | 1.000 | 1.000 |
| ResNet101 | 1.000 | 1.000 | 0.980 | 0.995 | 0.993 | 0.993 | 1.000 |
| ResNet152 | 1.000 | 1.000 | 0.980 | 0.995 | 0.993 | 0.993 | 1.000 |
| (a) Image-Based Classification | ||||
| Predicted | ||||
| Healthy | Early Gastric Cancer | Advanced Gastric Cancer | ||
| Actual | Healthy | 1201 | 0 | 7 |
| Early gastric cancer | 0 | 531 | 1 | |
| Advanced gastric cancer | 35 | 0 | 602 | |
| (b) Case-based classification | ||||
| Predicted | ||||
| Healthy | Early Gastric Cancer | Advanced Gastric Cancer | ||
| Actual | Healthy | 42 | 0 | 0 |
| Early gastric cancer | 0 | 95 | 0 | |
| Advanced gastric cancer | 0 | 0 | 50 | |
| Di | Ji | |
|---|---|---|
| Early gastric cancer | 0.555 | 0.427 |
| Advanced gastric cancer | 0.716 | 0.611 |
| Author | Method | Image Dataset | Detection Performance | Segmentation Performance |
|---|---|---|---|---|
| Hirasawa et al. [10] | SSD | Original 15,880 images | Sensitivity = 0.922 Positive predictive value = 0.306 | - |
| Sakai et al. [11] | CNN | Original 926 images | Sensitivity = 0.800 Specificity = 0.948 | - |
| Shibata et al. [12] | Mask R-CNN | Original 1741 images | Sensitivity = 0.96 False positives per image = 0.105 | Dice index = 0.54 |
| Teramoto et al. [13] | U-Net + CNN | Original 1741 images | Sensitivity = 0.98 False positives per image = 0.011 | Dice index = 0.56 |
| Proposed method | Cascade CNN | Original 2378 images | Sensitivity = 1.00 False positives per image = 0.005 | Dice index = 0.56 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Teramoto, A.; Shibata, T.; Yamada, H.; Hirooka, Y.; Saito, K.; Fujita, H. Detection and Characterization of Gastric Cancer Using Cascade Deep Learning Model in Endoscopic Images. Diagnostics 2022, 12, 1996. https://doi.org/10.3390/diagnostics12081996
Teramoto A, Shibata T, Yamada H, Hirooka Y, Saito K, Fujita H. Detection and Characterization of Gastric Cancer Using Cascade Deep Learning Model in Endoscopic Images. Diagnostics. 2022; 12(8):1996. https://doi.org/10.3390/diagnostics12081996
Chicago/Turabian StyleTeramoto, Atsushi, Tomoyuki Shibata, Hyuga Yamada, Yoshiki Hirooka, Kuniaki Saito, and Hiroshi Fujita. 2022. "Detection and Characterization of Gastric Cancer Using Cascade Deep Learning Model in Endoscopic Images" Diagnostics 12, no. 8: 1996. https://doi.org/10.3390/diagnostics12081996
APA StyleTeramoto, A., Shibata, T., Yamada, H., Hirooka, Y., Saito, K., & Fujita, H. (2022). Detection and Characterization of Gastric Cancer Using Cascade Deep Learning Model in Endoscopic Images. Diagnostics, 12(8), 1996. https://doi.org/10.3390/diagnostics12081996






