Automated Identification of Multiple Findings on Brain MRI for Improving Scan Acquisition and Interpretation Workflows: A Systematic Review
Abstract
:1. Introduction
2. Materials and Methods
2.1. Literature Search
2.2. Study Selection
2.3. Data Extraction and Analysis
2.4. Quality Assessment of Included Studies
2.5. Evaluation of Applicability for Workflow Improvements
3. Results
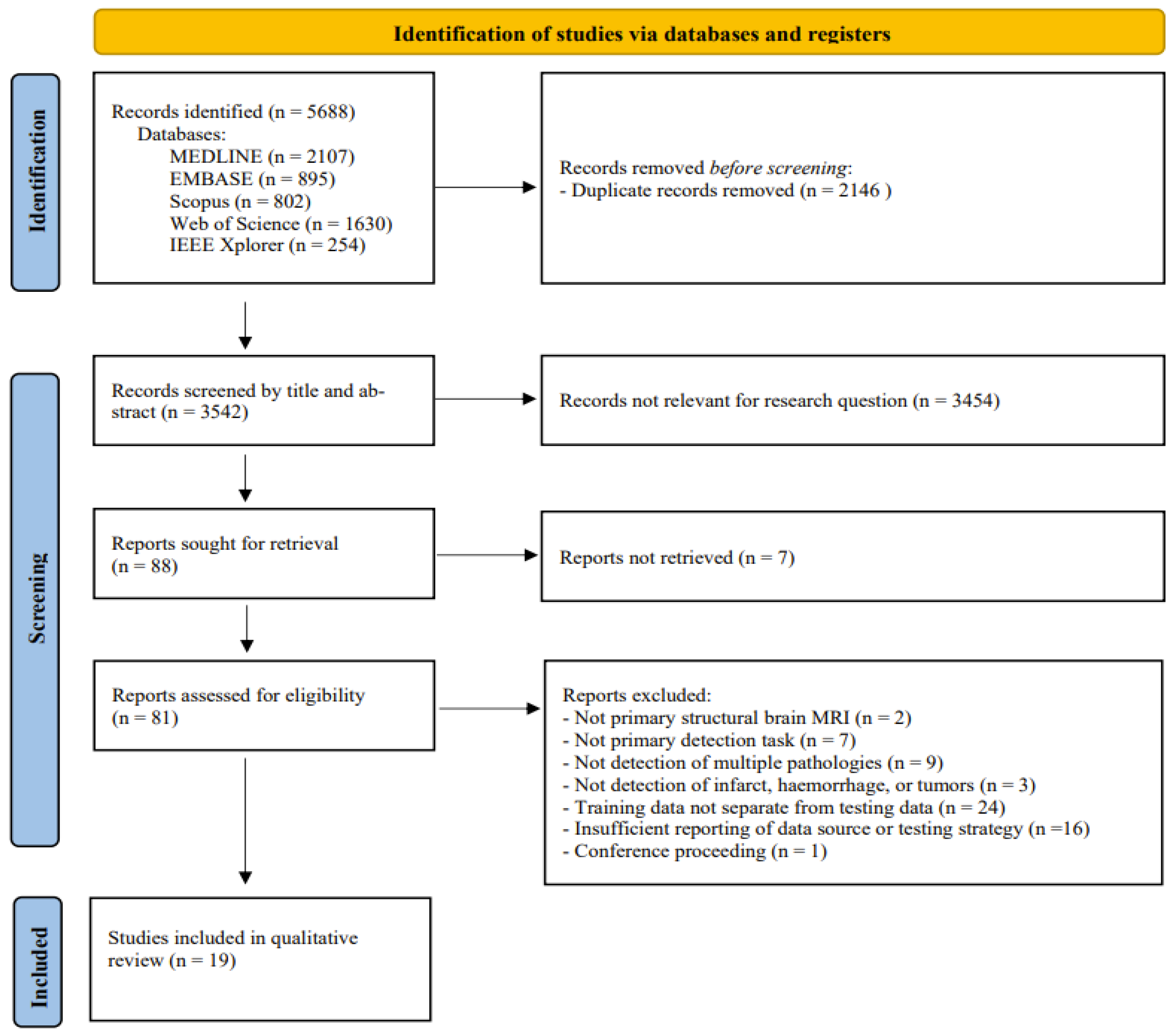
3.1. Study Selection and Data Extraction
3.2. Applicability to Workflow Improvement
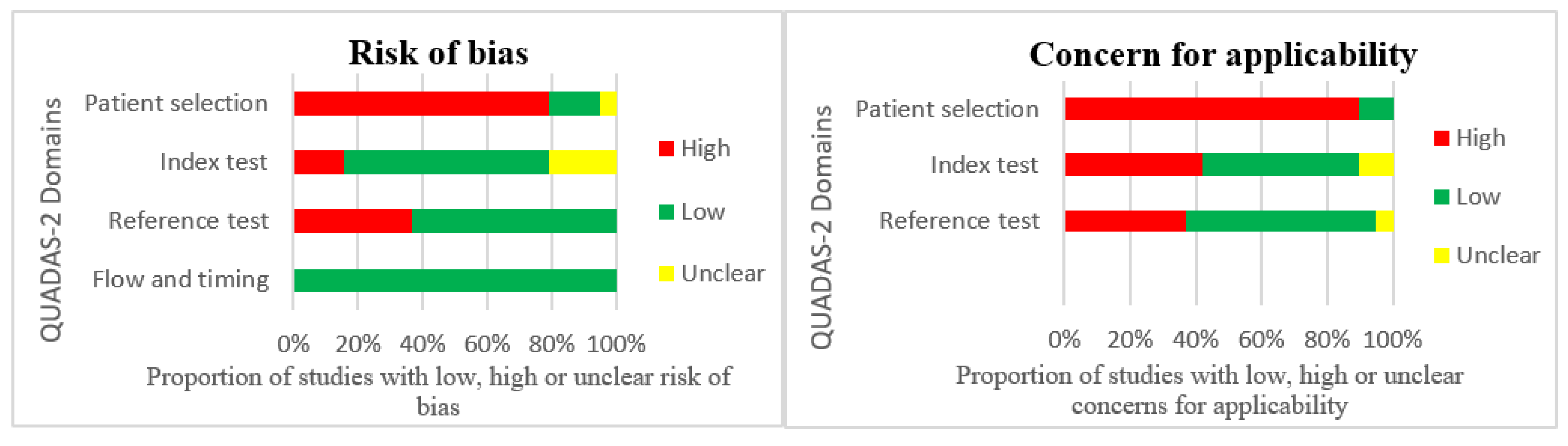
3.3. Quality Assessment
4. Discussion
4.1. Potential Benefits of Integrating ML into Existing Scan- and Interpretation Workflows
4.2. Limitations of Included Studies and Future Directions
4.3. Limitations of This Review
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Appendix A
References
- Radue, E.W.; Weigel, M.; Wiest, R.; Urbach, H. Introduction to Magnetic Resonance Imaging for Neurologists. Contin. Lifelong Learn. Neurol. 2016, 22, 1379–1398. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Luttrull, M.D.; Boulter, D.J.; Kirsch, C.F.E.; Aulino, J.M.; Broder, J.S.; Chakraborty, S.; Choudhri, A.F.; Ducruet, A.F.; Kendi, A.T.; Lee, R.K.; et al. ACR Appropriateness Criteria® Acute Mental Status Change, Delirium, and New Onset Psychosis. J. Am. Coll. Radiol. 2019, 16, S26–S37. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Salmela, M.B.; Mortazavi, S.; Jagadeesan, B.D.; Broderick, D.F.; Burns, J.; Deshmukh, T.K.; Harvey, H.B.; Hoang, J.; Hunt, C.H.; Kennedy, T.A.; et al. ACR Appropriateness Criteria® Cerebrovascular Disease. J. Am. Coll. Radios. 2017, 14, S34–S61. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Policeni, B.; Corey, A.S.; Burns, J.; Conley, D.B.; Crowley, R.W.; Harvey, H.B.; Hoang, J.; Hunt, C.H.; Jagadeesan, B.D.; Juliano, A.F.; et al. ACR Appropriateness Criteria® Cranial Neuropathy. J. Am. Coll. Radiol. 2017, 14, S406–S420. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Harvey, H.B.; Watson, L.C.; Subramaniam, R.M.; Burns, J.; Bykowski, J.; Chakraborty, S.; Ledbetter, L.N.; Lee, R.K.; Pannell, J.S.; Pollock, J.M.; et al. ACR Appropriateness Criteria® Movement Disorders and Neurodegenerative Diseases. J. Am. Coll. Radiol. 2020, 17, S175–S187. [Google Scholar] [CrossRef] [PubMed]
- Murphy, A.; di Muzio, B. Brain Screen Protocol (MRI). Radiopaedia. Org. 2015. [Google Scholar] [CrossRef]
- Subramaniam, R.M.; Kurth, D.A.; Waldrip, C.A.; Rybicki, F.J. American College of Radiology Appropriateness Criteria: Advancing Evidence-Based Imaging Practice. Semin. Nucl. Med. 2019, 49, 161–165. [Google Scholar] [CrossRef]
- Mehan, W.A.; González, R.G.; Buchbinder, B.R.; Chen, J.W.; Copen, W.A.; Gupta, R.; Hirsch, J.A.; Hunter, G.J.; Hunter, S.; Johnson, J.M.; et al. Optimal Brain MRI Protocol for New Neurological Complaint. PLoS ONE 2014, 9, e110803. [Google Scholar] [CrossRef] [Green Version]
- Chetlen, A.L.; Chan, T.L.; Ballard, D.H.; Frigini, L.A.; Hildebrand, A.; Kim, S.; Brian, J.M.; Krupinski, E.A.; Ganeshan, D. Addressing Burnout in Radiologists. Acad. Radiol. 2019, 26, 526. [Google Scholar] [CrossRef] [PubMed]
- Statistics Monthly Diagnostic Waiting Times and Activity. Available online: https://www.england.nhs.uk/statistics/statistical-work-areas/diagnostics-waiting-times-and-activity/monthly-diagnostics-waiting-times-and-activity/ (accessed on 21 May 2022).
- Choy, G.; Khalilzadeh, O.; Michalski, M.; Do, S.; Samir, A.E.; Pianykh, O.S.; Geis, J.R.; Pandharipande, P.V.; Brink, J.A.; Dreyer, K.J. Current Applications and Future Impact of Machine Learning in Radiology. Radiology 2018, 288, 318–328. [Google Scholar] [CrossRef]
- Letourneau-Guillon, L.; Camirand, D.; Guilbert, F.; Forghani, R. Artificial Intelligence Applications for Workflow, Process Optimization and Predictive Analytics. Neuroimaging Clin. 2020, 30, e1–e15. [Google Scholar] [CrossRef] [PubMed]
- Park, S.H.; Han, K. Methodologic Guide for Evaluating Clinical Performance and Effect of Artificial Intelligence Technology for Medical Diagnosis and Prediction. Radiology 2018, 286, 800–809. [Google Scholar] [CrossRef]
- Wichmann, J.L.; Willemink, M.J.; de Cecco, C.N. Artificial Intelligence and Machine Learning in Radiology: Current State and Considerations for Routine Clinical Implementation. Investig. Radiol. 2020, 55, 619–627. [Google Scholar] [CrossRef] [PubMed]
- Paniagua Bravo, A.; Albillos Merino, J.C.; Ibáñez Sanz, L.; Alba de Cáceres, I. Analysis of the Appropriateness of the Clinical Indications for Neuroimaging Studies. Radiología 2013, 55, 37–45. [Google Scholar] [CrossRef] [PubMed]
- Vernooij, M.W.; Ikram, M.A.; Tanghe, H.L.; Vincent, A.J.P.E.; Hofman, A.; Krestin, G.P.; Niessen, W.J.; Breteler, M.M.B.; van der Lugt, A. Incidental Findings on Brain MRI in the General Population. N. Engl. J. Med. 2007, 357, 1821–1828. [Google Scholar] [CrossRef] [PubMed]
- Kelly, C.J.; Karthikesalingam, A.; Suleyman, M.; Corrado, G.; King, D. Key Challenges for Delivering Clinical Impact with Artificial Intelligence. BMC Med. 2019, 17, 195. [Google Scholar] [CrossRef] [Green Version]
- Strohm, L.; Hehakaya, C.; Ranschaert, E.R.; Boon, W.P.C.; Moors, E.H.M. Implementation of Artificial Intelligence (AI) Applications in Radiology: Hindering and Facilitating Factors. Eur. Radiol. 2020, 30, 5525–5532. [Google Scholar] [CrossRef]
- Yao, A.D.; Cheng, D.L.; Pan, I.; Kitamura, F. Deep Learning in Neuroradiology: A Systematic Review of Current Algorithms and Approaches for the New Wave of Imaging Technology. Radiol. Artif. Intell. 2020, 2, e190026. [Google Scholar] [CrossRef] [PubMed]
- McInnes, M.D.F.; Moher, D.; Thombs, B.D.; McGrath, T.A.; Bossuyt, P.M.; Clifford, T.; Cohen, J.F.; Deeks, J.J.; Gatsonis, C.; Hooft, L.; et al. Preferred Reporting Items for a Systematic Review and Meta-Analysis of Diagnostic Test Accuracy Studies: The PRISMA-DTA Statement. JAMA 2018, 319, 388–396. [Google Scholar] [CrossRef]
- Bertels, J.; Eelbode, T.; Berman, M.; Vandermeulen, D.; Maes, F.; Bisschops, R.; Blaschko, M.B. Optimizing the Dice Score and Jaccard Index for Medical Image Segmentation: Theory and Practice. In Medical Image Computing and Computer Assisted Intervention—MICCAI 2019; Lecture Notes in Computer Science; Springer: Cham, Switzerland, 2019; Volume 11765, pp. 92–100. [Google Scholar] [CrossRef]
- QUADAS-2 | Bristol Medical School: Population Health Sciences | University of Bristol. Available online: https://www.bristol.ac.uk/population-health-sciences/projects/quadas/quadas-2/ (accessed on 11 March 2022).
- Rauschecker, A.M.; Rudie, J.D.; Xie, L.; Wang, J.; Duong, M.T.; Botzolakis, E.J.; Kovalovich, A.M.; Egan, J.; Cook, T.C.; Nick Bryan, R.; et al. Artificial Intelligence System Approaching Neuroradiologist-Level Diagnosis Accuracy at Brain MRI. Radiology 2020, 295, 626. [Google Scholar] [CrossRef]
- Vidoni, E.D. The Whole Brain Atlas. J. Neurol. Phys. Ther. 2012, 36, 108. [Google Scholar] [CrossRef]
- Clark, K.; Vendt, B.; Smith, K.; Freymann, J.; Kirby, J.; Koppel, P.; Moore, S.; Phillips, S.; Maffitt, D.; Pringle, M.; et al. The Cancer Imaging Archive (TCIA): Maintaining and Operating a Public Information Repository. J. Digit. Imaging 2013, 26, 1045–1057. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Menze, B.H.; Jakab, A.; Bauer, S.; Kalpathy-Cramer, J.; Farahani, K.; Kirby, J.; Burren, Y.; Porz, N.; Slotboom, J.; Wiest, R.; et al. The Multimodal Brain Tumor Image Segmentation Benchmark (BRATS). IEEE Trans. Med. Imaging 2015, 34, 1993–2024. [Google Scholar] [CrossRef] [PubMed]
- Maier, O.; Menze, B.H.; von der Gablentz, J.; Häni, L.; Heinrich, M.P.; Liebrand, M.; Winzeck, S.; Basit, A.; Bentley, P.; Chen, L.; et al. ISLES 2015—A Public Evaluation Benchmark for Ischemic Stroke Lesion Segmentation from Multispectral MRI. Med. Image Anal. 2017, 35, 250. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ahmadi, M.; Sharifi, A.; Jafarian Fard, M.; Soleimani, N. Detection of Brain Lesion Location in MRI Images Using Convolutional Neural Network and Robust PCA. Int. J. Neurosci. 2021, 1–12. [Google Scholar] [CrossRef]
- Baur, C.; Wiestler, B.; Muehlau, M.; Zimmer, C.; Navab, N.; Albarqouni, S. Modeling Healthy Anatomy with Artificial Intelligence for Unsupervised Anomaly Detection in Brain MRI. Radiol. Artif. Intell. 2021, 3, e190169. [Google Scholar] [CrossRef] [PubMed]
- Duong, M.T.; Rudie, J.D.; Wang, J.; Xie, L.; Mohan, S.; Gee, J.C.; Rauschecker, A.M. Convolutional Neural Network for Automated FLAIR Lesion Segmentation on Clinical Brain MR Imaging. Am. J. Neuroradiol. 2019, 40, 1282–1290. [Google Scholar] [CrossRef] [Green Version]
- Fayaz, M.; Torokeldiev, N.; Turdumamatov, S.; Qureshi, M.S.; Qureshi, M.B.; Gwak, J. An Efficient Methodology for Brain Mri Classification Based on Dwt and Convolutional Neural Network. Sensors 2021, 21, 7480. [Google Scholar] [CrossRef]
- Felipe Fattori Alves, A.; Ricardo de Arruda Miranda, J.; Reis, F.; Augusto Santana de Souza, S.; Luchesi Rodrigues Alves, L.; de Moura Feitoza, L.; Thiago de Souza de Castro, J.; Rodrigues de Pina, D. Inflammatory Lesions and Brain Tumors: Is It Possible to Differentiate Them Based on Texture Features in Magnetic Resonance Imaging? J. Venom. Anim. Toxins Incl. Trop. Dis. 2020, 26, 20200011. [Google Scholar] [CrossRef]
- Gauriau, R.; Bizzo, B.C.; Kitamura, F.C.; Junior, O.L.; Ferraciolli, S.F.; Macruz, F.B.C.; Sanchez, T.A.; Garcia, M.R.T.; Vedolin, L.M.; Domingues, R.C.; et al. A Deep Learning–Based Model for Detecting Abnormalities on Brain MR Images for Triaging: Preliminary Results from a Multisite. Radiol. Artif. Intell. 2021, 3, e200184. [Google Scholar] [CrossRef]
- Gilanie, G.; Bajwa, U.I.; Waraich, M.M.; Habib, Z.; Ullah, H.; Nasir, M. Classification of Normal and Abnormal Brain MRI Slices Using Gabor Texture and Support Vector Machines. Signal Image Video Processing 2018, 12, 479–487. [Google Scholar] [CrossRef]
- Han, C.; Rundo, L.; Murao, K.; Noguchi, T.; Shimahara, Y.; Milacski, Z.Á.; Koshino, S.; Sala, E.; Nakayama, H.; Satoh, S. MADGAN: Unsupervised Medical Anomaly Detection GAN Using Multiple Adjacent Brain MRI Slice Reconstruction. BMC Bioinform. 2021, 22, 31. [Google Scholar] [CrossRef] [PubMed]
- Hu, X.; Luo, W.; Hu, J.; Guo, S.; Huang, W.; Scott, M.R.; Wiest, R.; Dahlweid, M.; Reyes, M. Brain SegNet: 3D Local Refinement Network for Brain Lesion Segmentation. BMC Med. Imaging 2020, 20, 17. [Google Scholar] [CrossRef] [PubMed]
- Kamnitsas, K.; Ledig, C.; Newcombe, V.F.J.; Simpson, J.P.; Kane, A.D.; Menon, D.K.; Rueckert, D.; Glocker, B. Efficient Multi-Scale 3D CNN with Fully Connected CRF for Accurate Brain Lesion Segmentation. Med. Image Anal. 2017, 36, 61–78. [Google Scholar] [CrossRef] [PubMed]
- Kim, B.; Kwon, K.; Oh, C.; Park, H. Unsupervised Anomaly Detection in MR Images Using Multi-Contrast Information. Med. Phys. 2021, 48, 7346–7359. [Google Scholar] [CrossRef]
- Lu, P.J.; Yoo, Y.; Rahmanzadeh, R.; Galbusera, R.; Weigel, M.; Ceccaldi, P.; Nguyen, T.D.; Spincemaille, P.; Wang, Y.; Daducci, A.; et al. GAMER MRI: Gated-Attention Mechanism Ranking of Multi-Contrast MRI in Brain Pathology. NeuroImage Clin. 2021, 29, 102522. [Google Scholar] [CrossRef] [PubMed]
- Lu, S.; Lu, Z.; Zhang, Y.D. Pathological Brain Detection Based on AlexNet and Transfer Learning. J. Comput. Sci. 2019, 30, 41–47. [Google Scholar] [CrossRef]
- Nael, K.; Gibson, E.; Yang, C.; Ceccaldi, P.; Yoo, Y.; Das, J.; Doshi, A.; Georgescu, B.; Janardhanan, N.; Odry, B.; et al. Automated Detection of Critical Findings in Multi-Parametric Brain MRI Using a System of 3D Neural Networks. Sci. Rep. 2021, 11, 6876. [Google Scholar] [CrossRef]
- Nayak, D.R.; Dash, R.; Majhi, B. Automated Diagnosis of Multi-Class Brain Abnormalities Using MRI Images: A Deep Convolutional Neural Network Based Method. Pattern Recognit. Lett. 2020, 138, 385–391. [Google Scholar] [CrossRef]
- Nayak, D.R.; Das, D.; Dash, R.; Majhi, S.; Majhi, B. Deep Extreme Learning Machine with Leaky Rectified Linear Unit for Multiclass Classification of Pathological Brain Images. Multimed. Tools Appl. 2020, 79, 15381–15396. [Google Scholar] [CrossRef]
- Pereira, S.; Pinto, A.; Amorim, J.; Ribeiro, A.; Alves, V.; Silva, C.A. Adaptive Feature Recombination and Recalibration for Semantic Segmentation with Fully Convolutional Networks. IEEE Trans. Med. Imaging 2019, 38, 2914–2925. [Google Scholar] [CrossRef] [PubMed]
- Wood, D.A.; Kafiabadi, S.; Al Busaidi, A.; Guilhem, E.; Montvila, A.; Lynch, J.; Townend, M.; Agarwal, S.; Mazumder, A.; Barker, G.J.; et al. Deep Learning Models for Triaging Hospital Head MRI Examinations. Med. Image Anal. 2022, 78, 102391. [Google Scholar] [CrossRef] [PubMed]
- Wood, D.A.; Kafiabadi, S.; al Busaidi, A.; Guilhem, E.L.; Lynch, J.; Townend, M.K.; Montvila, A.; Kiik, M.; Siddiqui, J.; Gadapa, N.; et al. Deep Learning to Automate the Labelling of Head MRI Datasets for Computer Vision Applications. Eur. Radiol. 2022, 32, 725–736. [Google Scholar] [CrossRef]
- Daugaard Jørgensen, M.; Antulov, R.; Hess, S.; Lysdahlgaard, S. Convolutional Neural Network Performance Compared to Radiologists in Detecting Intracranial Hemorrhage from Brain Computed Tomography: A Systematic Review and Meta-Analysis. Eur. J. Radiol. 2022, 146, 110073. [Google Scholar] [CrossRef] [PubMed]
- Rajpurkar, P.; Chen, E.; Banerjee, O.; Topol, E.J. AI in Health and Medicine. Nat. Med. 2022, 28, 31–38. [Google Scholar] [CrossRef]
- Zhang, Y.; Liu, S.; Li, C.; Wang, J. Application of Deep Learning Method on Ischemic Stroke Lesion Segmentation. J. Shanghai Jiaotong Univ. (Sci.) 2018, 2022, 99–111. [Google Scholar] [CrossRef]
- van Kempen, E.J.; Post, M.; Mannil, M.; Witkam, R.L.; ter Laan, M.; Patel, A.; Meijer, F.J.A.; Henssen, D. Performance of Machine Learning Algorithms for Glioma Segmentation of Brain MRI: A Systematic Literature Review and Meta-Analysis. Eur. Radiol. 2021, 31, 9638–9653. [Google Scholar] [CrossRef]
- Hickman, S.E.; Woitek, R.; Le, E.P.V.; Im, Y.R.; Luxhøj, C.M.; Aviles-Rivero, A.I.; Baxter, G.C.; MacKay, J.W.; Gilbert, F.J. Machine Learning for Workflow Applications in Screening Mammography: Systematic Review and Meta-Analysis. Radiology 2022, 302, 88–104. [Google Scholar] [CrossRef]
- Kelly, B.S.; Judge, C.; Bollard, S.M.; Clifford, S.M.; Healy, G.M.; Aziz, A.; Mathur, P.; Islam, S.; Yeom, K.W.; Lawlor, A.; et al. Radiology Artificial Intelligence: A Systematic Review and Evaluation of Methods (RAISE). Eur. Radiol. 2022, 1–10. [Google Scholar] [CrossRef]
- Yu, A.C.; Mohajer, B.; Eng, J. External Validation of Deep Learning Algorithms for Radiologic Diagnosis: A Systematic Review. Radiol. Artif. Intell. 2022, 4, e210064. [Google Scholar] [CrossRef]
- Choi, K.S.; Sunwoo, L. Artificial Intelligence in Neuroimaging: Clinical Applications. Investig. Magn. Reson. Imaging 2022, 26, 1–9. [Google Scholar] [CrossRef]
- Kotter, E.; Ranschaert, E. Challenges and Solutions for Introducing Artificial Intelligence (AI) in Daily Clinical Workflow. Eur. Radiol. 2020, 31, 5–7. [Google Scholar] [CrossRef] [PubMed]
- Olthof, A.W.; van Ooijen, P.M.A.; Rezazade Mehrizi, M.H. Promises of Artificial Intelligence in Neuroradiology: A Systematic Technographic Review. Neuroradiology 2020, 62, 1265–1278. [Google Scholar] [CrossRef] [PubMed]
- Ghaffari, M.; Sowmya, A.; Oliver, R. Automated Brain Tumor Segmentation Using Multimodal Brain Scans: A Survey Based on Models Submitted to the BraTS 2012–2018 Challenges. IEEE Rev. Biomed. Eng. 2020, 13, 156–168. [Google Scholar] [CrossRef]
- Heiss, W.D.; Kidwell, C.S. Imaging for Prediction of Functional Outcome and Assessment of Recovery in Ischemic Stroke. Stroke 2014, 45, 1195–1201. [Google Scholar] [CrossRef] [Green Version]
- Wallis, D.; Buvat, I. Clever Hans Effect Found in a Widely Used Brain Tumour MRI Dataset. Med. Image Anal. 2022, 77, 102368. [Google Scholar] [CrossRef]
- Mongan, J.; Moy, L.; Kahn, C.E. Checklist for Artificial Intelligence in Medical Imaging (CLAIM): A Guide for Authors and Reviewers. Radiol. Artif. Intell. 2020, 2, e200029. [Google Scholar] [CrossRef] [Green Version]
- Bianchetti, G.; Taralli, S.; Vaccaro, M.; Indovina, L.; Mattoli, M.V.; Capotosti, A.; Scolozzi, V.; Calcagni, M.L.; Giordano, A.; de Spirito, M.; et al. Automated Detection and Classification of Tumor Histotypes on Dynamic PET Imaging Data through Machine-Learning Driven Voxel Classification. Comput. Biol. Med. 2022, 145, 105423. [Google Scholar] [CrossRef]
| Inclusion Criteria: | Exclusion Criteria: |
|---|---|
| Studies focusing on abnormal brain diseases that included either brain infarct, hemorrhage, or tumor on brain MRI | Studies focusing on tasks not relevant for identification of brain diseases |
| Studies developing algorithms tested on a dataset that was separate from the training dataset | Studies focusing on identification of a single brain disease only |
| Peer-reviewed studies in English | Studies focusing on development of ML for specialized MR sequences (e.g., MR elastography, functional MRI) or other imaging modalities (e.g., SPECT, PET, CT, US) |
| Studies with primarily non-adult populations | |
| Editorials, case series, letters, conference proceedings, reviews, and inaccessible papers |
| Author | Data Source | No. Patients /Images | Training Data | Validation Data | Testing Data | Disease Distribution in Data | MR Sequences Utilized | MR Field Strength |
|---|---|---|---|---|---|---|---|---|
| Ahmadi et al., 2021 [28] | Private + Harvard Medical School Whole Brain Atlas | 1200 images | 1120 | N/A | 80 | 12.5% normals 87.5% abnormal incl. glioma, Huntington’s disease, Meningioma, and Alzheimer | 2D single slice of: Ax T2 | 1.5 T |
| Baur et al., 2021 [29] | Private WMH TCIA | 259 patients | 100 | 18 | 141 | 42% normal used for unsupervised training 19% multiple sclerosis 15% glioma & glioblastoma 4% microangiopathy 20% WMH | Ax T2-FLAIR | 1.5 T 3 T |
| Duong et al., 2019 [30] | Private | 387 patients | 295 | N/A | 92 | Normal and 19 different abnormalities incl. MS, high grade glioma, and vascular (acute or subacute ischemia) | Ax T2-FLAIR | 1.5 T 3 T |
| Fayaz et al., 2021 [31] | Harvard Medical School Whole Brain Atlas | 4100 images | 2870 | N/A | 1230 | 50% normal 50% abnormal incl. glioma, meningioma, and Alzheimer | 2D single slice of: Ax T2 | 1.5 T |
| Felipe Fattori Alves et al., 2020 [32] | Private | 67 patients | 50 | N/A | 17 | 45% inflammatory lesion (incl. MS, vasculitis, toxoplasmosis, pyogenic and septic-embolic brain abscess, etc.) 55% brain tumors (incl. Glioblastoma, anaplastic astrocytoma, anaplastic ependymoma) | Ax T1 & T1 + C Ax T2 Ax T2-FLAIR Ax DWI | 1.5 T 3 T |
| Gauriau et al., 2021 [33] | Private | 10,770 patients | 7795 | 473 | 2502 | Normal and 8+ different abnormalities including infarct, hemorrhage, neoplasm, demyelination, and infections | Ax T2-FLAIR | 1.5 T 3 T |
| Gilanie et al., 2018 [34] | Harvard Medical School Whole Brain Atlas | 4589 images | 3029 | N/A | 1560 | 11% normal 89% abnormal incl. cerebrovascular, neoplasm, neurodegenerative, and inflammatory disease | 2D single slices of: Ax T1 & T1 + C Ax T2 & Ax PD Ax T2-FLAIR | 1.5 T |
| Han et al., 2020 [35] | OASIS-3 Private | 1162 patients | 543 | N/A | 619 | 47% normals used for unsupervised training 19% normals used for testing 21% dementia of varying degree 7% brain metastasis 6% various disease incl. small infarct, hemorrhage, and white matter lesions | Ax T1 & Ax T1 + c | 1.5 T 3 T |
| Hu et al., 2020 [36] | BRATS 2019 ISLES 2017 | 459 patients | 317 | N/A | 142 | 84% glioma (HGG, LGG) 16% acute & subacute infarct | Ax T1 & T1 + C Ax T2 Ax T2-FLAIR Ax DWI, ADC, perfusion | 1.5 T 3 T |
| Kamnitsas et al., 2017 [37] | Private BRATS 2015 ISLES 2015 | 509 patients | 348 | N/A | 161 | 75% tumor (high grade glioma, low grade glioma) 13% acute & subacute infarct 12% traumatic brain injury | Ax or Sag T1 & T1 + C Ax T2 & Ax PD Ax T2-FLAIR Ax T2 * GRE Ax DWI & ADC | 1.5 T 3 T |
| Kim et al., 2021 [38] | BRATS 2019 ISLES 2015 | 259 patients | 239 | N/A | 26 | 36% normal 60% glioma 4% acute & subacute infarct | 2D slices of Ax T1 & T1 + C Ax T2 Ax T2-FLAIR Ax DWI, ADC, perfusion | 1.5 T 3 T |
| Lu et al., 2021 [39] | Private | 7134 patients | * 5002 | 1061 | 1071 | 13% acute/subacute stroke 87% non-stroke abnormalities incl. tumor, hemorrhage and normals | Axial T2-FLAIR Axial DWI + ADC | 1.5 T 3 T |
| Lu, Lu et Zhang., 2019 [40] | Harvard Medical School Whole Brain Atlas | 291 images | 204 | N/A | 87 | 39% normal 61% abnormal incl. neoplasm, neurodegenerative, and inflammatory disease | 2D single slice of: Ax T2 | 1.5 T |
| Nael et al., 2021 [41] | Private | 13,215 patients | 9845 | 1248 | 2122 | 17% normal 11% acute infarction 5% acute hemorrhage 4% intracranial mass effect 63% other abnormalities including white matter lesions | Ax or Sag T1 & T1 + C Ax T2 Ax T2-FLAIR Ax ADC & DWI Ax T2 * GRE | 1.5 T 3 T |
| Nayak et al., 2020 [42] | Harvard Medical School Whole Brain Atlas & | 275 images | 165 | N/A | 110 | 20% normal 20% stroke 20% neurodegenerative 20% infectious 20% neoplasm | 2D single slice of: Ax T2 | 1.5 T |
| Nayak et al., 2020 [43] | Havard Medical School | 200 images | 120 | N/A | 80 | 20% normal 20% stroke 20% neurodegenerative 20% infectious 20% neoplasm | 2D single slice of: Ax T2 | 1.5 T |
| Pereira et al., 2019 [44] | BRATS 2013 BRATS 2017 ISLES 2017 | 471 patients | 358 | 10 | 103 | 89% tumor (high grade glioma, low grade glioma) 11% acute & subacute infarct | Ax T1 & T1 + C Ax T2 Ax T2-FLAIR Ax DWI, ADC, perfusion | 1.5 T 3 T |
| Rauschecker et al., 2020 [23] | Private | 178 patients | 86 | N/A | 92 | 19 different abnormalities incl. MS, high grade glioma, and vascular (acute or subacute ischemia) | Ax T1 + C Ax T2 Ax T2-FLAIR Ax T2 * GRE Ax DWI & ADC | 1.5 T 3 T |
| Wood et al., 2022 [45] | Private | 71,206 patients | 53,409 | 9425 | 7372 | Normal and 90+ different abnormalities including vascular disease, neoplasms, demyelination, and atrophy | Ax T2-FLAIR Ax DWI | 1.5 T 3 T |
| Author | Aim of Algorithm | Type of Algorithm | Ground Truth | Testing Strategy | Performance Results | Workflow Applicability | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AUC | Acc (%) | Sens (%) | Spec (%) | F1 (%) | PPV (%) | NPV (%) | ||||||
| (a) | ||||||||||||
| Fayaz et al., 2021 [31] | Binary classification of normal and abnormal | CNN + DWT | Expert labels | Train-test split | 0.997 | N/A | 99.7 | N/A | N/A | N/A | N/A | (A) Reflecting clinical practice: NS (B) External validation: NS (C) Performance: S Note: High performance observed on single 2D MR slices |
| Felipe Fattori Alves et al., 2020 [32] | Binary classification of inflammatory lesions and brain tumors | RF SVM k-NN | Expert delineation | Train-test split | * 0.906 | * 82.7 | * 91.2 | N/A | * 87.5 | N/A | N/A | (A) Reflecting clinical practice: NS (B) External validation: NS (C) Performance: S |
| Gauriau et al., 2021 [33] | Binary classification of normal and abnormal | CNN | Radiological report | Train-test split incl. external test set | 0.800 [0.770; 0.820] | N/A | 77.0 [75; 80] | 65.0 [61; 69] | 78.0 [76; 80] | N/A | N/A | (A) Reflecting clinical practice: S (B) External validation: S (C) Performance: NS |
| Gilanie et al., 2018 [34] | Binary classification of normal and abnormal | Gabor filter SVM | Expert labels | Train-test split | 0.970 | 96.5 | 98.0 | 92.0 | N/A | N/A | N/A | (A) Reflecting clinical practice: NS (B) External validation: NS (C) Performance: S |
| Lu et al., 2021 [39] | Binary classification of stroke/non- stroke patients | CNN + Gating attention mechanism ranking of multi-contrast MRI | Expert labels | Train-test split | ** 0.881 | N/A | N/A | N/A | N/A | N/A | N/A | (A) Reflecting clinical practice: NS (B) External validation: NS (C) Performance: S |
| Lu, Lu et Zhang., 2019 [40] | Binary classification of normal and abnormal | CNN + transfer learning | Expert labels | Train-test split | N/A | 100.0 | 100.0 | 100.0 | N/A | N/A | N/A | (A) Reflecting clinical practice: NS (B) External validation: NS (C) Performance: S Note: High test performance result on small test set <100 2D MR slices |
| Wood et al., 2022 [45] | Binary classification of normal and abnormal | Ensemble CNN | NLP labelled radiological report | Train-test split incl. external test set | 0.948 [0.945; 0.951] | N/A | 91.9 [89.9; 93.9] | 84.2 [82.2; 86.2] | 92.3 [90.3; 94.3] | N/A | N/A | (A) Reflecting clinical practice: NS (B) External validation: S (C) Performance: S |
| (b) | ||||||||||||
| Han et al., 2020 [35] | Multiple binary classification of normal/clinical dementia (Dem), normal/brain metastasis (BM), and normal/various diseases (VD) incl. small infarct and hemorrhage. | Unsupervised GAN + 7 Self-attention (SA) modules | Expert label | Train-test split | Dem: 0.765 BM: 0.921 VD: 0.613 | N/A | N/A | N/A | N/A | N/A | N/A | (A) Reflecting clinical practice: NS (B) External validation: NS (C) Performance: NS |
| Nael et al., 2021 [41] | Multiple binary classification of normal/any abnormalities (abn), infarct (inf)/non-infarct, hemorrhage (hem)/non-hemorrhage, and mass effect (ME)/non-mass effect | CNN | Radiological report Expert image delineation | Train-test split incl. external test set | Abn: 0.880 Inf.: 0.970 Hem.: 0.830 ME: 0.870 | Abn: 80.0 Inf.: 95.0 Hem: 87.0 ME: 81.0 | Abn: 80.0 Inf.: 90.0 Hem.: 72.0 ME: 79.0 | Abn: 80.0 Inf.: 97.0 Hem.: 88.0 ME: 81.0 | N/A | Abn: 94.0 Inf.: 92.0 Hem.: 32.0 ME: 12.0 | Abn: 48.0 Inf.: 96.0 Hem.: 98.0 ME: 99.0 | (A) Reflecting clinical practice: NS (B) External validation: S (C) Performance: S |
| Nayak et al., 2020 [42] | Multiclass classification of normal, stroke, tumor, infectious, degenerative | CNN | Expert labels | Train-test split | N/A | *** 97.5 | N/A | N/A | N/A | N/A | N/A | (A) Reflecting clinical practice: NS (B) External validation: NS (C) Performance: S Note: High test performance result on small test set of 2D MR slices |
| Nayak et al., 2020 [43] | Multiclass classification of normal, stroke, tumor, infectious, degenerative | CNN + ELM | Expert labels | Train-test split | N/A | 93.8 | N/A | N/A | N/A | N/A | N/A | (A) Reflecting clinical practice: NS (B) External validation: NS (C) Performance: S Note: High test performance result on small test set <100 2D MR slices |
| Rauschecker et al., 2020 [23] | Multiclass classification of 19 brain diseases incl multiple sclerosis (MS), high grade glioma, and vascular infarct defined as correctly classified within top 3 differential diagnosis | CNN + Bayesian inference | Expert labels | Train-test split | 0.920 [0.880; 0.950] | 91.0 [84; 96] | N/A | N/A | N/A | N/A | N/A | (A) Reflecting clinical practice: NS (B) External validation: NS (C) Performance: S |
| Author | Aim of Algorithm | Type of Algorithm | Ground Truth | Testing Strategy | Performance Results | Workflow Applicability | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| DSC | Sens (%) | Spec (%) | PPV (%) | NPV (%) | ||||||
| Ahmadi et al., 2021 [28] | Multiclass segmentation incl. neoplasm and neurodegenerative disease | CNN | Synthetic labels via robust PCA | Train-test split | 0.912 | 99.9 | 99.8 | N/A | N/A | (A) Reflecting clinical practice: NS (B) External validation: NS (C) Performance: S |
| Baur et al., 2021 [29] | Multiclass segmentation of normal, MS, glioblastoma (GBM), glioma, microangiopathy (MA), and WMH | Unsupervised VAE | Radiological report Expert image delineation | Train-test split | MS: 0.650 GBM: 0.390 Glioma: 0.350 MA: 0.730 WMH: 0.450 | MS: 62.0 GBM: 56.0 Glioma: 29.0 MA: 36.0 WMH: 13.0 | N/A | MS: 67.0 GBM: 14.0 Glioma: 28.0 MA: 17.0 WMH: 49.0 | N/A | (A) Reflecting clinical practice: NS (B) External validation: NS (C) Performance: NS |
| Duong et al., 2019 [30] | Multiclass segmentation of 19+ different abnormalities incl. MS, high grade glioma, and infarcts | CNN | Expert image delineation | Train-test split | 0.789 [0.767; 0.811] | 76.7 [74.2; 79.2] | 99.9 [99; 99] | 76.9 [75.1; 78.7] | 99.0 [99; 99] | (A) Reflecting clinical practice: NS (B) External validation: NS (C) Performance: NS |
| Hu et al., 2020 [36] | Multiclass segmentation of infarct and glioma | CNN | Expert image delineation | Train-test split | Infarct: 0.300 [0.120; 0.520] Glioma: 0.860 | Infarct: 43.0 [16; 70] Glioma: 87.0 | Infarct: N/A Glioma: 87.0 | Infarct: 35.0 [8; 62] Glioma: N/A | N/A | (A) Reflecting clinical practice: NS (B) External validation: NS (C) Performance: NS |
| Kamnitsas et al., 2017 [37] | Multiclass segmentation of infarct, traumatic brain injury (TBI), and glioma | Ensemble CNN | Expert image delineation | Train-test split | Infarct: 0.590 [0.280; 0.900] TBI: 0.645 [0.480; 0.810] Glioma: 0.849 | Infarct: 60.0 [33; 87] TBI: 63.9 [47; 81] Glioma: 87.7 | N/A | Infarct: 68.0 [35; 100] TBI: 69.8 [52; 88] Glioma: 85.3 | N/A | (A) Reflecting clinical practice: NS (B) External validation: NS (C) Performance: NS |
| Kim et al., 2021 [38] | Multiclass segmentation of infarct and glioma | Unsupervised VAE | Expert image delineation | Train-test split | Infarct: 0.278 [0.273; 0.283] Glioma: 0.692 [0.686; 0.698] | Infarct: 42.9 [42.2; 43.6] Glioma: 67.5 [65.1; 69.9] | N/A | Infarct: 20.5 [19.8; 21.2] Glioma: 71.1 [67.2; 75.0] | N/A | (A) Reflecting clinical practice: NS (B) External validation: NS (C) Performance: NS |
| Pereira et al., 2019 [44] | Multiclass segmentation of infarct incl. penumbra and glioma | CNN | Expert image delineation | Train-test split | Infarct: 0.340 [0.140; 0.540] Penumbra: 0.820 [0.730; 0.910] Glioma: 0.866 | Infarct: 55.0 [25; 85] Glioma: 84.6 | N/A | Infarct: 36.0 [11; 61] Glioma: 89.8 | N/A | (A) Reflecting clinical practice: NS (B) External validation: NS (C) Performance: NS |
| Source | Risk of Bias | Concern for Applicability | |||||
|---|---|---|---|---|---|---|---|
| Patient Selection | Index Test | Reference Test | Flow and Timing | Patient Selection | Index Test | Reference Test | |
| Ahmadi et al., 2021 [28] |  |  |  |  |  |  |  |
| Baur et al., 2021 [29] |  |  |  |  |  |  |  |
| Duong et al., 2019 [30] |  |  |  |  |  |  |  |
| Fayaz et al., 2021 [31] |  |  |  |  |  |  |  |
| Felipe Fattori Alves et al., 2020 [32] |  |  |  |  |  |  |  |
| Gauriau et al., 2021 [33] |  |  |  |  |  |  |  |
| Gilanie et al., 2018 [34] |  |  |  |  |  |  |  |
| Han et al., 2020 [35] |  |  |  |  |  |  |  |
| Hu et al., 2020 [36] |  |  |  |  |  |  |  |
| Kamnitsas et al., 2017 [37] |  |  |  |  |  |  |  |
| Kim et al., 2021 [38] |  |  |  |  |  |  |  |
| Lu et al., 2021 [39] |  |  |  |  |  |  |  |
| Lu, Lu et Zhang., 2019 [40] |  |  |  |  |  |  |  |
| Nael et al., 2021 [41] |  |  |  |  |  |  |  |
| Nayak et al., 2020 [42] |  |  |  |  |  |  |  |
| Nayak et al., 2020 [43] |  |  |  |  |  |  |  |
| Pereira et al., 2019 [44] |  |  |  |  |  |  |  |
| Rauschecker et al., 2020 [23] |  |  |  |  |  |  |  |
| Wood et al., 2022 [45] |  |  |  |  |  |  |  |
 = low risk of bias and concern for applicability;
= low risk of bias and concern for applicability;  = high risk of bias and concern for applicability;
= high risk of bias and concern for applicability;  = unclear risk of bias and concern for applicability.
= unclear risk of bias and concern for applicability.Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sheng, K.; Offersen, C.M.; Middleton, J.; Carlsen, J.F.; Truelsen, T.C.; Pai, A.; Johansen, J.; Nielsen, M.B. Automated Identification of Multiple Findings on Brain MRI for Improving Scan Acquisition and Interpretation Workflows: A Systematic Review. Diagnostics 2022, 12, 1878. https://doi.org/10.3390/diagnostics12081878
Sheng K, Offersen CM, Middleton J, Carlsen JF, Truelsen TC, Pai A, Johansen J, Nielsen MB. Automated Identification of Multiple Findings on Brain MRI for Improving Scan Acquisition and Interpretation Workflows: A Systematic Review. Diagnostics. 2022; 12(8):1878. https://doi.org/10.3390/diagnostics12081878
Chicago/Turabian StyleSheng, Kaining, Cecilie Mørck Offersen, Jon Middleton, Jonathan Frederik Carlsen, Thomas Clement Truelsen, Akshay Pai, Jacob Johansen, and Michael Bachmann Nielsen. 2022. "Automated Identification of Multiple Findings on Brain MRI for Improving Scan Acquisition and Interpretation Workflows: A Systematic Review" Diagnostics 12, no. 8: 1878. https://doi.org/10.3390/diagnostics12081878
APA StyleSheng, K., Offersen, C. M., Middleton, J., Carlsen, J. F., Truelsen, T. C., Pai, A., Johansen, J., & Nielsen, M. B. (2022). Automated Identification of Multiple Findings on Brain MRI for Improving Scan Acquisition and Interpretation Workflows: A Systematic Review. Diagnostics, 12(8), 1878. https://doi.org/10.3390/diagnostics12081878









