Artificial Intelligence for Upper Gastrointestinal Endoscopy: A Roadmap from Technology Development to Clinical Practice
Abstract
:1. Introduction
2. Methods
3. Findings
3.1. UGIE Exam Quality Assessment
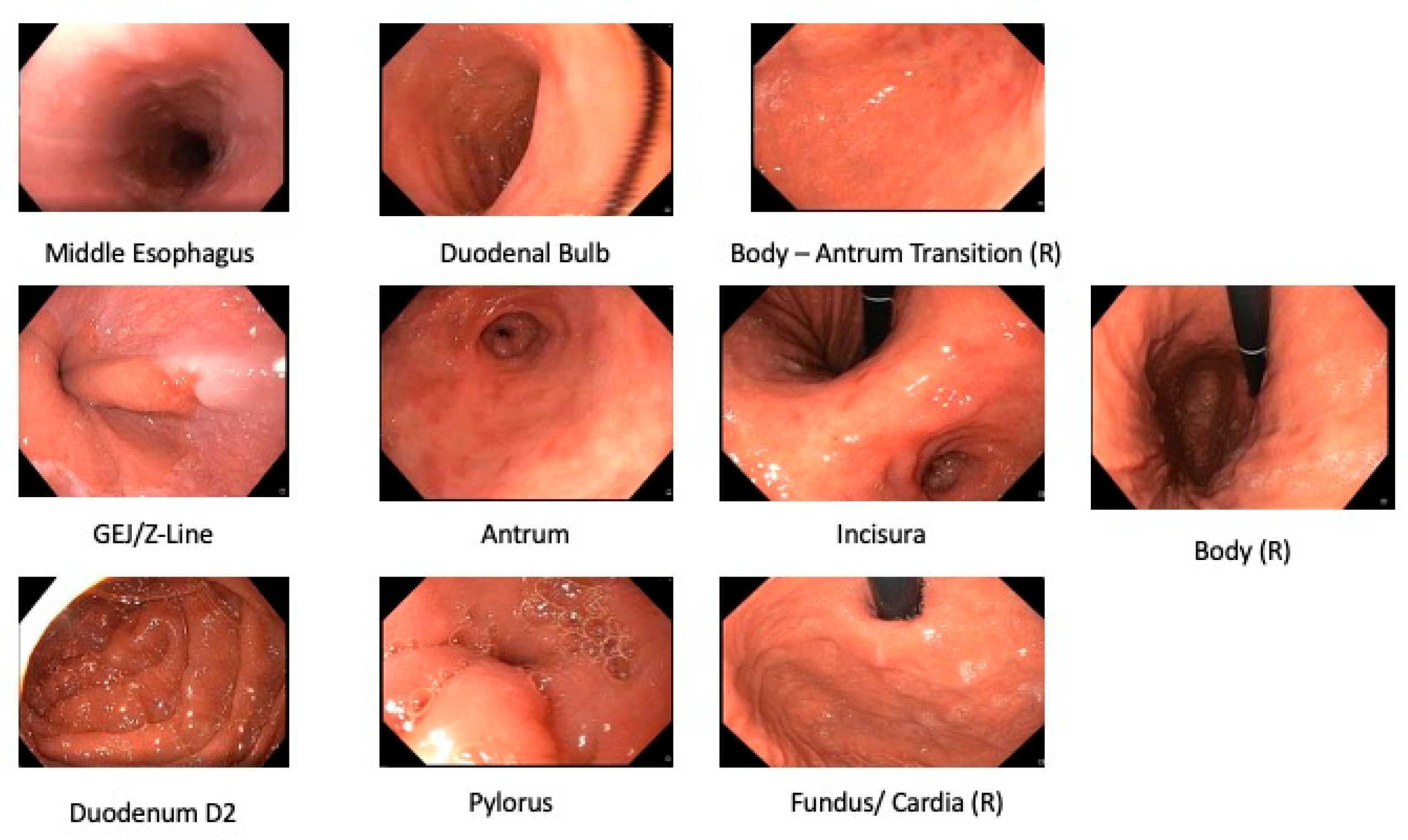
3.1.1. Anatomical Landmark Detection UGIE—Single Frame
| Authors | Data Annotation Protocol | Dataset | Classes | Algorithm | Pre-Processing | Validation | Average Performance | AI Impact (Clinical Setting) |
|---|---|---|---|---|---|---|---|---|
| Takiyama et al., 2018 [39] | Undefined | (Private Dataset) 44,416 UGIE images (optimal view WLI) | 4 sites + 3 gastric sites | GoogleLeNet | Black frame cropping | Holdout set | (4 anatomical classes) Accuracy: 99% Sensitivity: 94% (from 87% to 99%) 96% (from 96% to 97%) (3 gastric sub-classes) Sensitivity: 97% (from 96% to 97%) Specificity: 98% (from 98% to 99%) | N/A |
| Wu et al., 2019 [40] | 2 experts with >10 years of experience | (Private Dataset) 24,549 WLI images | 10 or 26 sites | VGG16-Resnet50 | CNN filters blurry frames | Holdout set | Accuracy: 90% (10 sites) 66% (26 sites) | (Single-center, retrospective trial) [40] **** Endoscopist accuracy: 90% (10 sites, experts) 63% (26 sites, experts) 87% (10 sites, seniors) 59% (26 sites, seniors) 83% (10 sites, novices) 46% (26 sites, novices) |
| Zhang Xu et al., 2019 [41] | 2 expert endoscopists (years of experience unknown) | (Private Dataset) 75,275 UGIE images (including non-informative and NBI frames) * | 10 sites + uninformative + NBI | Muli-Task Custom CNN + SSD | None | Holdout set | Average precision (mAP): 94% | N/A |
| He et al., 2019 ** [42] | 1 doctoral student 1 clinical gastroenterology research fellow | 3704 UGIE images (WLI+LCI frames) optimal views | 11 sites + N/A | Inception-v3 | Data-driven ROI cropping | 5-fold C.V. | Accuracy: 83% F1: 80% (from 53% to 94%) | N/A |
| Igarashi et al., 2020 [43] | 1 expert with >30 years of experience 1 endoscopists with >4 years of experience | (Private Dataset) 85,246 upper GI images | 10 sites from UGIE + 4 classes pertaining to specimens and other examinations | AlexNet | None | Holdout set | Accuracy: 97% | N/A |
| Sun et al., 2021 *** [44] | >1 endoscopist with >5 years of experience | (Private Dataset) 10,474 UGIE images including NBI | 11 sites + NBI | Custom CNN+RCF | ROI extraction + bilinear interpolation | 5-fold C.V. | Accuracy: 99% Precision: 93% F1 score: 92% | N/A |
| Chang et al., 2021 [27] | Unclear | (Private Dataset) 15,723 frames from asymptomatic patients | 8 classes | ResNeSt | None | Holdout set | Accuracy: 97% | N/A |
Algorithms
Datasets
Results
3.1.2. Anatomical Landmark Detection in UGIE—Multiple Frames
| Authors | Data Annotation Protocol | Dataset | Classes | Algorithm | Augmentation | Validation | Average Performance | AI Impact (Clinical Setting) |
|---|---|---|---|---|---|---|---|---|
| Wu et al., 2019 [45] | 2 seniors 1–5 years of experience 3 experts > 5 years of experience 2 doctoral students ** | (Private Dataset) 34,513 UGIE images 107 UGIE videos | 26 sites + N/A | VGG-16 + DRL | None | 10-fold C.V. | Accuracy: 90% (from 70% to 100%) Sensitivity: 88% (from 63% to 100%) Specificity 95% (from 75% to 100%) | (Single-center randomized controlled trial) [45] *** Blind spot rate (#Landmarks) Humans+Algorithm: 5.86 ± 6.89 Humans (control): 22.46 ± 14.38 (Mean performance improvement: 15.39 from 19.23 to 11.54) (Multicenter Randomized Controlled Trial) Wu et al., 2021 [46] Blind spot rate (#Landmarks): Humans+Algorithm: 5.38 ± 4.32 Humans (control): 9.82 ± 9.82 (Prospective, single-blind, 3-parallel-group, randomized, single-center trial) Chen et al., 2019 [47] *** Blind spot rate (#Landmarks): Humans+Algorithm: 3.42 ± 5.73 Humans (control): 22.46 ± 14.76 |
| Choi et al., 2020 [26] | 5 blinded seniors with >3 years of experience | (Private Dataset) 2599 images from 250 UGIEs | 8 sites | SENet + positional loss | Random scaling, cropping, rotation and horizontal flip | 10-fold C.V. | Accuracy: 98% (from 94% to 99%) Sensitivity: 97% (from 94% to 99%) Specificity: ~100% | N/A |
| Ding et al., 2021 [48] | UGIE images from clinical reports | (Private Dataset) 3605 UGIE anatomical site frames 2346 background class frames | 6 sites + background | EfficientNet-b3 + thresholded sliding window with exponential decay | Random shear, scaling and translation | Holdout set and weighted oversampling | Accuracy: 88% * Precision: 89% * (from 65% to 95%) Recall: 88% * (from 69% to 99%) | N/A |
| Yan-Dong Li et al., 2021 [47] | 3 blinded seniors with 5–10 years of experience 1 expert > 10 years of experience | 170,297 UGIE (Private Dataset) images 5779 UGIE videos | 31 sites | Inception-v3 + LSTM | Random HSV jittering and corner cropping | Holdout set | Accuracy: 95% (from 88% to 100%) Sensitivity: 96% (from 81% to 100%) Specificity: 99% (from 99% to 100%) | N/A |
Algorithms
Datasets
Results
3.2. Lesion Detection in UGIE
| Authors | Classes | Algorithm | Dataset | Image Modality | Results | AI Impact (Clinical Setting) |
|---|---|---|---|---|---|---|
| Zhang et al., 2020 [55] | Non atrophic gastritis Atrophic gastritis | DenseNet121 | (Private Dataset) 3042 atrophic gastritis images 2428 normal images | WLI | Accuracy:94% Sensitivity: 95% Specificity: 94% | N/A |
| Hirasawa et al., 2018 [53] | EGC | SSD | (Private Dataset) 13,584 EGC images | WLI NBI Indigo carmine | Sensitivity: 92% PPV: 31% | N/A |
| Yoon et al., 2019 [56] | non-EGC EGC | VGG-16 | (Private Dataset) 1705 EGC images 9834 non-EGC images | WLI | Specificity: 98% sensitivity: 91% | N/A |
| Yan et al., 2020 [57] | Intestinal Metaplasia (IM) | Xception NASNet EfficientNetB4 | (Private Dataset) 1048 IM images 832 non-IM | NBI Magnifying NBI | EfficientNetB4: Sensitivity: 93% Specificity: 85% Accuracy: 88% | N/A |
| Wu et al., 2021 [58] | neoplastic lesions non neoplastic lesions | YOLO v3 and ResNet50 | (Private Dataset) For YOLO v3: 15,341 lesion images 9363 non-lesion images For ResNet50: 4442 gastric neoplastic images 3859 non-neoplastic images | WLI | Accuracy: 89%; Sensitivity 92%; Specificity 88% | (Multi-center prospective controlled trial) Experts (n = 8): Sensitivity: 81% Specificity: 75% Accuracy: 78% Seniors (n = 19): Sensitivity: 84% Specificity: 77% Accuracy: 80% Juniors (n = 19): Sensitivity: 84% Specificity: 66% Accuracy: 73% ENDOANGEL: Sensitivity: 88% Specificity: 93% Accuracy 91% |
3.2.1. UGIE Lesion Detection in the Stomach
Algorithms
Datasets
Results
3.3. Lesion Characterization in UGIE
| Authors | Classes | Algorithm | Dataset | Image Modality | Results | AI Impact (Clinical Setting) |
|---|---|---|---|---|---|---|
| Wu et al., 2021 [58] | Invasion depth: mucosal and submucosal Differentiation Status: Differentiated and undifferentiated | ResNet-50 | (Private Dataset) * 3407 images of gastric cancer 1131 differentiated type images 1086 undifferentiated type images | WLI Magnifying NBI | EGC invasion WLI: Accuracy: 88%; Sensitivity: 91%; Specificity: 85% EGC differentiation M-NBI: Accuracy of 86%; Sensitivity: 79%; Specificity: 89% | (Multi-center prospective controlled trial) EGC invasion Experts(n = 8): Sensitivity: 57%% Specificity: 76% Accuracy: 69% EGC invasion Seniors (n = 19): Sensitivity: 60% Specificity: 66% Accuracy: 64% EGC invasion Juniors (n = 19): Sensitivity: 61% Specificity: 61% Accuracy: 61% EGC invasion AI predictions: Sensitivity: 70% Specificity: 83% Accuracy 79% EGC differentiation Experts(n = 8): Sensitivity: 47% Specificity: 83% Accuracy 72% EGC differentiation Seniors (n = 19): Sensitivity: 53% Specificity: 74% Accuracy: 67% EGC differentiation Juniors (n = 19): Sensitivity: 56% Specificity: 60% Accuracy: 59% EGC differentiation AI predictions: Sensitivity: 50% Specificity: 80% Accuracy 71% |
| Yoon et al., 2019 [56] | T1a T1b Non-EGC | VGG-16 | (Private Dataset) 1097 T1a-EGC 1005 T1b-EGC 9834 non-EGC | WLI | Specificity: 75% Sensitivity: 82% | N/A |
| Nagao et al., 2020 [63] | M-SM1 SM2 or deeper | ResNet-50 | (Private Dataset) 10,589 M-SM1 images 6968 SM2 or deeper images | WLI NBI Indigo | WLI Accuracy: 95% NBI Accuracy: 94% Indigo Accuracy: 96% | N/A |
| Zhu et al., 2019 [64] | P0 (M or SM1) P1 (deeper than SM1) | ResNet-50 | (Private Dataset) 545 P0 images 245 P1 images | WLI | Accuracy: 89% Sensitivity: 76% Specificity: 96% | CNN Accuracy: 89% Sensitivity:76% Specificity: 96% Experts(n = 8): Accuracy: 77% Sensitivity: 91% Specificity: 71% Junior (n = 9): Accuracy: 66% Sensitivity: 85% Specificity: 57% |
| Xu et al., 2021 [65] | GA IM | VGG-16 | (Private Dataset) 2149 GA images 3049 IM images | Magnifying NBI Magnifying BLI | GA Accuracy: 90% Sensitivity: 90% Specificity: 91% IM Accuracy: 91% Sensitivity: 89% Specificity: 93% | (Multi-center Prospective blinded trial) GA classification CAD System: Accuracy: 87% Sensitivity: 87% Specificity: 86% Experts(n = 4): Accuracy: 85% Sensitivity: 91% Specificity: 72% Non experts (n = 5): Accuracy: 75% Sensitivity: 83% Specificity: 59% IM classification: CAD System: Accuracy: 89% Sensitivity: 90% Specificity: 86% Experts(n = 4): Accuracy: 82% Sensitivity: 83% Specificity: 81% Non experts (n = 5): Accuracy: 74% Sensitivity: 74% Specificity: 73% |
3.3.1. UGIE Lesion Characterization in the Stomach
Algorithms
Datasets
Results
4. Discussion
4.1. UGIE Exam Quality Assessment
- Beyond CNNs: The adoption of novel deep learning architectures for visual understanding, as the recently proposed visual transformers [67], could provide a significant improvement at the backbone of the multi-frame classification approaches. This is due to their improved capacity in automatically selecting relevant features from the imaging data as well as better capacity in transferring efficiently information from a larger receptive field into deep layers.
- Hybrid models: The sequential nature of UGIE examinations and, in particular, the strong priors associated with the expected order in visiting the different anatomical landmarks open up the possibility of pairing highly discriminative deep learning models with robust time sequence statistical models able to model more explicitly time evolution [68]. These techniques are then expected to provide models that are more robust to input perturbation than purely data-driven methods, while at the same time requiring a lower amount of training data to generalize well.
- Enhanced explainability: Efforts to provide explainable and interpretable models for anatomical landmarks detection have focused mainly on the use of CAM-based approaches able to highlight which part of the considered frame is mostly responsible for the output of the CNN classifier. Increased explainability should be pursued, by starting with the inclusion of the temporal dimension in substantiating why a given landmark is detected by the algorithm at that particular time of the UGIE procedure. The produced explanations could be also enriched with example-based approaches able to showcase, in real-time, to the operator to most similar and significant landmarks encountered in other exams [69].
- 3D Photodocumentation: Assuming screening protocols are followed, machine learning algorithms can project the frames of an UGIE video onto a three-dimensional reconstruction of the UGI tract. These algorithms can potentially be applied to any photodocumentation protocol since they can in principle construct a complete representation of the UGI tract. They also allow one to query a bounded region of the produced 3D surface and retrieve the original 2-D frames used in the projective step. Not only is this functionality relevant from a usability perspective, but it can also convey a more accurate realization of the mental model required by endoscopists when interpreting a UGIE report. Blind spots become overwhelmingly evident in a 3D model, potentiating feedback loops for the physicians. The work of Widya et al. [70,71], which is based on classical methods for structure invariant feature transforms (SIFT) and structure-from-motion, has been seminal in this direction, leading towards reconstructing a virtual 3D model of the stomach from unlabeled UGIE data. More recently, Yuwei Xu et al. [72] introduced a CNN that is able to find more invariant anchor points that enable 3D reconstructions of higher resolution and under more challenging conditions. Moreover, advancements in the field of light field imaging and neural radiance fields are likely able to mitigate incomplete reconstructions through 3D view synthesis [73,74,75,76].
4.2. Lesion Detection in UGIE
- Beyond CNNs: Similar to what has been discussed for anatomical landmark detection, novel architectures for computer vision tasks can be considered in the design of lesion detection pipelines, also including visual transformers. As an example, [77] uses self-attention mechanisms of transformers which enables the models to selectively focus on certain parts of their input and thus reason more effectively. This type of architecture that relies on attention mechanisms also helps in improving the interpretability of the model.
- Generative models for data augmentation: To overcome the lack of publicly available data and the representativeness of multiple lesions classes, generative models can synthesize images of lesions. Generative models can also be used for explaining the attributes that are highly correlated with the model’s decision; for example, [77] trained and highlighted the most relevant attributes for the predictions. StyleGAN [78] models can learn the classifier-specific style space as explanatory attributes.
- Unsupervised and semi-supervised approaches: The performance of the deep learning solutions heavily depends on the quality of the data used for training. Possible new directions to guarantee better generalization of lesion detection models could lie in the usage of unsupervised and semi-supervised learning techniques that could leverage the huge amount of non-labeled UGIE video currently available. These can be achieved by considering first unsupervised feature extraction approaches based on the use of autoencoders or generative adversarial networks.
- Use spatial information: A possible approach to provide more reliable detection performance is envisioned by embedding prior spatial information into the lesion detection algorithm. For example, CAD systems that provide automatic photodocumentation assistance via anatomical landmark detection could use such detailed information regarding the UGI navigation process, thus providing context information to the lesion detection algorithms.
4.3. Lesion Characterization in UGIE
- Explainable CNNs: Model explainability is one of the main focal points for future research, due to the clear necessity of interpretable and transparent models when applied to sensitive applications domains, such as healthcare. Visual transformers can naturally enable model interpretability by leveraging self-attention layers.
- Generative models: The lack of available data and the representation of each class of interest is also a common problem pointed out in several sections of this review. Generative models can overcome this issue by creating synthetic images that enhance the representativeness of classes within the dataset. Beyond that, styles can be transferred from, for example, different image modalities, thus creating synthetic NBI images from WLI or synthetic indigo carmine dye contrast images from WLI, and so on. To this end, conditional generative adversarial approaches, such as the StyleGAN [78] can be used.
- Beyond 2D Characterization: Segmentation models can be used to precisely characterize the extension of the region of mucosa affected by a lesion. A possible evolution to this approach would be that of providing a description of the 3D volumetric characteristics of the abnormal tissue, which can give more information about the invasion depth throughout the lesion volume, thus providing a further step towards the actual implementation of what could be interpreted as a sort of 3D visual biopsy.
5. Conclusions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Vos, T.; Abajobir, A.A.; Abate, K.H.; Abbafati, C.; Abbas, K.M.; Abd-Allah, F.; Abdulkader, R.S.; Abdulle, A.M.; Abebo, T.A.; Abera, S.F.; et al. Global, Regional, and National Incidence, Prevalence, and Years Lived with Disability for 328 Diseases and Injuries for 195 Countries, 1990–2016: A Systematic Analysis for the Global Burden of Disease Study 2016. Lancet 2017, 390, 1211–1259. [Google Scholar] [CrossRef] [Green Version]
- Cancer Tomorrow. Available online: https://gco.iarc.fr/tomorrow/home (accessed on 31 March 2022).
- Săftoiu, A.; Hassan, C.; Areia, M.; Bhutani, M.S.; Bisschops, R.; Bories, E.; Cazacu, I.M.; Dekker, E.; Deprez, P.H.; Pereira, S.P.; et al. Role of Gastrointestinal Endoscopy in the Screening of Digestive Tract Cancers in Europe: European Society of Gastrointestinal Endoscopy (ESGE) Position Statement. Endoscopy 2020, 52, 293–304. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pimenta-Melo, A.R.; Monteiro-Soares, M.; Libânio, D.; Dinis-Ribeiro, M. Missing Rate for Gastric Cancer during Upper Gastrointestinal Endoscopy: A Systematic Review and Meta-Analysis. Eur. J. Gastroenterol. Hepatol. 2016, 28, 1041–1049. [Google Scholar] [CrossRef] [PubMed]
- Rutter, M.; Senore, C.; Bisschops, R.; Domagk, D.; Valori, R.; Kaminski, M.; Spada, C.; Bretthauer, M.; Bennett, C.; Bellisario, C.; et al. The European Society of Gastrointestinal Endoscopy Quality Improvement Initiative: Developing Performance Measures. Endoscopy 2015, 48, 81–89. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Frazzoni, L.; Arribas, J.; Antonelli, G.; Libanio, D.; Ebigbo, A.; van der Sommen, F.; de Groof, A.J.; Fukuda, H.; Ohmori, M.; Ishihara, R.; et al. Endoscopists’ Diagnostic Accuracy in Detecting Upper Gastrointestinal Neoplasia in the Framework of Artificial Intelligence Studies. Endoscopy 2022, 54, 403–411. [Google Scholar] [CrossRef]
- Arribas, J.; Antonelli, G.; Frazzoni, L.; Fuccio, L.; Ebigbo, A.; van der Sommen, F.; Ghatwary, N.; Palm, C.; Coimbra, M.; Renna, F.; et al. Standalone Performance of Artificial Intelligence for Upper GI Neoplasia: A Meta-Analysis. Gut 2021, 70, 1458–1468. [Google Scholar] [CrossRef]
- Lui, T.K.; Tsui, V.W.; Leung, W.K. Accuracy of Artificial Intelligence–Assisted Detection of Upper GI Lesions: A Systematic Review and Meta-Analysis. Gastrointest. Endosc. 2020, 92, 821–830. [Google Scholar] [CrossRef]
- Nagendran, M.; Chen, Y.; Lovejoy, C.A.; Gordon, A.C.; Komorowski, M.; Harvey, H.; Topol, E.J.; Ioannidis, J.P.; Collins, G.S.; Maruthappu, M. Artificial Intelligence versus Clinicians: Systematic Review of Design, Reporting Standards, and Claims of Deep Learning Studies. BMJ 2020, 368, m689. [Google Scholar] [CrossRef] [Green Version]
- Sharma, P.; Hassan, C. Artificial Intelligence and Deep Learning for Upper Gastrointestinal Neoplasia. Gastroenterology 2021, 162, 1056–1066. [Google Scholar] [CrossRef]
- Visaggi, P.; de Bortoli, N.; Barberio, B.; Savarino, V.; Oleas, R.; Rosi, E.M.; Marchi, S.; Ribolsi, M.; Savarino, E. Artificial Intelligence in the Diagnosis of Upper Gastrointestinal Diseases. J. Clin. Gastroenterol. 2022, 56, 23–35. [Google Scholar] [CrossRef]
- Pecere, S.; Milluzzo, S.M.; Esposito, G.; Dilaghi, E.; Telese, A.; Eusebi, L.H. Applications of Artificial Intelligence for the Diagnosis of Gastrointestinal Diseases. Diagnostics 2021, 11, 1575. [Google Scholar] [CrossRef] [PubMed]
- Anta, J.A.; Dinis-Ribeiro, M. Early Gastric Cancer and Artificial Intelligence: Is It Time for Population Screening? Best Pract. Res. Clin. Gastroenterol. 2021, 52, 101710. [Google Scholar] [CrossRef] [PubMed]
- Cogan, T.; Cogan, M.; Tamil, L. MAPGI: Accurate Identification of Anatomical Landmarks and Diseased Tissue in Gastrointestinal Tract Using Deep Learning. Comput. Biol. Med. 2019, 111, 103351. [Google Scholar] [CrossRef] [PubMed]
- Park, J.-W.; Kim, Y.; Kim, W.-J.; Nam, S.-J. Automatic Anatomical Classification Model of Esophagogastroduodenoscopy Images Using Deep Convolutional Neural Networks for Guiding Endoscopic Photodocumentation. J. Korea Soc. Comput. Inf. 2021, 26, 19–28. [Google Scholar] [CrossRef]
- Hashimoto, R.; Requa, J.; Dao, T.; Ninh, A.; Tran, E.; Mai, D.; Lugo, M.; El-Hage Chehade, N.; Chang, K.J.; Karnes, W.E.; et al. Artificial Intelligence Using Convolutional Neural Networks for Real-Time Detection of Early Esophageal Neoplasia in Barrett’s Esophagus (with Video). Gastrointest. Endosc. 2020, 91, 1264–1271.e1. [Google Scholar] [CrossRef]
- Liu, G.; Hua, J.; Wu, Z.; Meng, T.; Sun, M.; Huang, P.; He, X.; Sun, W.; Li, X.; Chen, Y. Automatic Classification of Esophageal Lesions in Endoscopic Images Using a Convolutional Neural Network. Ann. Transl. Med. 2020, 8, 486. [Google Scholar] [CrossRef]
- Shiroma, S.; Yoshio, T.; Kato, Y.; Horie, Y.; Namikawa, K.; Tokai, Y.; Yoshimizu, S.; Yoshizawa, N.; Horiuchi, Y.; Ishiyama, A.; et al. Ability of Artificial Intelligence to Detect T1 Esophageal Squamous Cell Carcinoma from Endoscopic Videos and the Effects of Real-Time Assistance. Sci. Rep. 2021, 11, 7759. [Google Scholar] [CrossRef]
- Wu, Z.; Ge, R.; Wen, M.; Liu, G.; Chen, Y.; Zhang, P.; He, X.; Hua, J.; Luo, L.; Li, S. ELNet:Automatic Classification and Segmentation for Esophageal Lesions Using Convolutional Neural Network. Med. Image Anal. 2021, 67, 101838. [Google Scholar] [CrossRef]
- Ghatwary, N.; Zolgharni, M.; Janan, F.; Ye, X. Learning Spatiotemporal Features for Esophageal Abnormality Detection From Endoscopic Videos. IEEE J. Biomed. Health Inform. 2021, 25, 131–142. [Google Scholar] [CrossRef]
- Guo, L.; Xiao, X.; Wu, C.; Zeng, X.; Zhang, Y.; Du, J.; Bai, S.; Xie, J.; Zhang, Z.; Li, Y.; et al. Real-Time Automated Diagnosis of Precancerous Lesions and Early Esophageal Squamous Cell Carcinoma Using a Deep Learning Model (with Videos). Gastrointest. Endosc. 2020, 91, 41–51. [Google Scholar] [CrossRef]
- de Groof, A.J.; Struyvenberg, M.R.; van der Putten, J.; van der Sommen, F.; Fockens, K.N.; Curvers, W.L.; Zinger, S.; Pouw, R.E.; Coron, E.; Baldaque-Silva, F.; et al. Deep-Learning System Detects Neoplasia in Patients With Barrett’s Esophagus With Higher Accuracy Than Endoscopists in a Multistep Training and Validation Study With Benchmarking. Gastroenterology 2020, 158, 915–929.e4. [Google Scholar] [CrossRef] [PubMed]
- Tokai, Y.; Yoshio, T.; Aoyama, K.; Horie, Y.; Yoshimizu, S.; Horiuchi, Y.; Ishiyama, A.; Tsuchida, T.; Hirasawa, T.; Sakakibara, Y.; et al. Application of Artificial Intelligence Using Convolutional Neural Networks in Determining the Invasion Depth of Esophageal Squamous Cell Carcinoma. Esophagus 2020, 17, 250–256. [Google Scholar] [CrossRef] [PubMed]
- Yao, K. The Endoscopic Diagnosis of Early Gastric Cancer. Ann. Gastroenterol. 2013, 26, 11–22. [Google Scholar] [PubMed]
- Yao, K.; Uedo, N.; Muto, M.; Ishikawa, H. Development of an E-Learning System for Teaching Endoscopists How to Diagnose Early Gastric Cancer: Basic Principles for Improving Early Detection. Gastric Cancer 2017, 20, 28–38. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Choi, S.J.; Khan, M.A.; Choi, H.S.; Choo, J.; Lee, J.M.; Kwon, S.; Keum, B.; Chun, H.J. Development of Artificial Intelligence System for Quality Control of Photo Documentation in Esophagogastroduodenoscopy. Surg. Endosc. 2022, 36, 57–65. [Google Scholar] [CrossRef] [PubMed]
- Chang, Y.-Y.; Li, P.-C.; Chang, R.-F.; Yao, C.-D.; Chen, Y.-Y.; Chang, W.-Y.; Yen, H.-H. Deep Learning-Based Endoscopic Anatomy Classification: An Accelerated Approach for Data Preparation and Model Validation. Surg. Endosc. 2021, 36, 3811–3821. [Google Scholar] [CrossRef]
- Bisschops, R.; Areia, M.; Coron, E.; Dobru, D.; Kaskas, B.; Kuvaev, R.; Pech, O.; Ragunath, K.; Weusten, B.; Familiari, P.; et al. Performance Measures for Upper Gastrointestinal Endoscopy: A European Society of Gastrointestinal Endoscopy (ESGE) Quality Improvement Initiative. Endoscopy 2016, 48, 843–864. [Google Scholar] [CrossRef] [Green Version]
- Bisschops, R.; Rutter, M.D.; Areia, M.; Spada, C.; Domagk, D.; Kaminski, M.F.; Veitch, A.; Khannoussi, W.; Gralnek, I.M.; Hassan, C.; et al. Overcoming the Barriers to Dissemination and Implementation of Quality Measures for Gastrointestinal Endoscopy: European Society of Gastrointestinal Endoscopy (ESGE) and United European Gastroenterology (UEG) Position Statement. Endoscopy 2021, 53, 196–202. [Google Scholar] [CrossRef]
- Cohen, J.; Pike, I.M. Defining and Measuring Quality in Endoscopy. Gastrointest. Endosc. 2015, 81, 1–2. [Google Scholar] [CrossRef]
- Beg, S.; Ragunath, K.; Wyman, A.; Banks, M.; Trudgill, N.; Pritchard, M.D.; Riley, S.; Anderson, J.; Griffiths, H.; Bhandari, P.; et al. Quality Standards in Upper Gastrointestinal Endoscopy: A Position Statement of the British Society of Gastroenterology (BSG) and Association of Upper Gastrointestinal Surgeons of Great Britain and Ireland (AUGIS). Gut 2017, 66, 1886–1899. [Google Scholar] [CrossRef] [Green Version]
- Rey, J.-F.; Lambert, R. ESGE Recommendations for Quality Control in Gastrointestinal Endoscopy: Guidelines for Image Documentation in Upper and Lower GI Endoscopy. Endoscopy 2001, 33, 901–903. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Teh, J.L.; Hartman, M.; Lau, L.; Tan, J.-R.; Wong, A.; Ng, J.-J.; Saxena, N.; Shabbir, A.; So, J.B. Mo1579 Duration of Endoscopic Examination Significantly Impacts Detection Rates of Neoplastic Lesions During Diagnostic Upper Endoscopy. Gastrointest. Endosc. 2011, 73, AB393. [Google Scholar] [CrossRef]
- Gastroscopy, T. committee for standardizing screening Gastric Cancer Screening Techniques. JSGCS Cho Handb. 2010, 1, 1–24. [Google Scholar]
- Lee, J.I.; Kim, J.S.; Kim, B.-W.; Huh, C.W. Taking More Gastroscopy Images Increases the Detection Rate of Clinically Significant Gastric Lesions: Validation of a Systematic Screening Protocol for the Stomach. Korean J. Helicobacter Up. Gastrointest. Res. 2020, 20, 225–232. [Google Scholar] [CrossRef]
- Emura, F.; Sharma, P.; Arantes, V.; Cerisoli, C.; Parra-Blanco, A.; Sumiyama, K.; Araya, R.; Sobrino, S.; Chiu, P.; Matsuda, K.; et al. Principles and Practice to Facilitate Complete Photodocumentation of the Upper Gastrointestinal Tract: World Endoscopy Organization Position Statement. Dig. Endosc. 2020, 32, 168–179. [Google Scholar] [CrossRef]
- Riegler, M.; Pogorelov, K.; Halvorsen, P.; Griwodz, C.; Lange, T.; Randel, K.; Eskeland, S.; Dang Nguyen, D.T.; Lux, M.; Spampinato, C. Multimedia for Medicine: The Medico Task at Mediaeval 2017. In Proceedings of the MediaEval’17, Dublin, Ireland, 13–15 September 2017. [Google Scholar]
- Pogorelov, K.; Randel, K.R.; Griwodz, C.; Eskeland, S.L.; de Lange, T.; Johansen, D.; Spampinato, C.; Dang-Nguyen, D.-T.; Lux, M.; Schmidt, P.T.; et al. KVASIR: A Multi-Class Image Dataset for Computer Aided Gastrointestinal Disease Detection. In Proceedings of the 8th ACM on Multimedia Systems Conference, Taipei, Taiwan, 20 June 2017; pp. 164–169. [Google Scholar]
- Takiyama, H.; Ozawa, T.; Ishihara, S.; Fujishiro, M.; Shichijo, S.; Nomura, S.; Miura, M.; Tada, T. Automatic Anatomical Classification of Esophagogastroduodenoscopy Images Using Deep Convolutional Neural Networks. Sci. Rep. 2018, 8, 7497. [Google Scholar] [CrossRef]
- Wu, L.; Zhou, W.; Wan, X.; Zhang, J.; Shen, L.; Hu, S.; Ding, Q.; Mu, G.; Yin, A.; Huang, X.; et al. A Deep Neural Network Improves Endoscopic Detection of Early Gastric Cancer without Blind Spots. Endoscopy 2019, 51, 522–531. [Google Scholar] [CrossRef] [Green Version]
- Xu, Z.; Tao, Y.; Wenfang, Z.; Ne, L.; Zhengxing, H.; Jiquan, L.; Weiling, H.; Huilong, D.; Jianmin, S. Upper Gastrointestinal Anatomy Detection with Multi-task Convolutional Neural Networks. Healthc. Technol. Lett. 2019, 6, 176–180. [Google Scholar] [CrossRef]
- He, Q.; Bano, S.; Ahmad, O.F.; Yang, B.; Chen, X.; Valdastri, P.; Lovat, L.B.; Stoyanov, D.; Zuo, S. Deep Learning-Based Anatomical Site Classification for Upper Gastrointestinal Endoscopy. Int. J. CARS 2020, 15, 1085–1094. [Google Scholar] [CrossRef]
- Igarashi, S.; Sasaki, Y.; Mikami, T.; Sakuraba, H.; Fukuda, S. Anatomical Classification of Upper Gastrointestinal Organs under Various Image Capture Conditions Using AlexNet. Comput. Biol. Med. 2020, 124, 103950. [Google Scholar] [CrossRef]
- Sun, M.; Ma, L.; Su, X.; Gao, X.; Liu, Z.; Ma, L. Channel Separation-Based Network for the Automatic Anatomical Site Recognition Using Endoscopic Images. Biomed. Signal Processing Control. 2022, 71, 103167. [Google Scholar] [CrossRef]
- Wu, L.; Zhang, J.; Zhou, W.; An, P.; Shen, L.; Liu, J.; Jiang, X.; Huang, X.; Mu, G.; Wan, X.; et al. Randomised Controlled Trial of WISENSE, a Real-Time Quality Improving System for Monitoring Blind Spots during Esophagogastroduodenoscopy. Gut 2019, 68, 2161–2169. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, L.; He, X.; Liu, M.; Xie, H.; An, P.; Zhang, J.; Zhang, H.; Ai, Y.; Tong, Q.; Guo, M.; et al. Evaluation of the Effects of an Artificial Intelligence System on Endoscopy Quality and Preliminary Testing of Its Performance in Detecting Early Gastric Cancer: A Randomized Controlled Trial. Endoscopy 2021, 53, 1199–1207. [Google Scholar] [CrossRef] [PubMed]
- Chen, D.; Wu, L.; Li, Y.; Zhang, J.; Liu, J.; Huang, L.; Jiang, X.; Huang, X.; Mu, G.; Hu, S.; et al. Comparing Blind Spots of Unsedated Ultrafine, Sedated, and Unsedated Conventional Gastroscopy with and without Artificial Intelligence: A Prospective, Single-Blind, 3-Parallel-Group, Randomized, Single-Center Trial. Gastrointest. Endosc. 2020, 91, 332–339.e3. [Google Scholar] [CrossRef] [PubMed]
- Ding, A.; Li, Y.; Chen, Q.; Cao, Y.; Liu, B.; Chen, S.; Liu, X. Gastric Location Classification During Esophagogastroduodenoscopy Using Deep Neural Networks. In Proceedings of the 2021 IEEE 21st International Conference on Bioinformatics and Bioengineering (BIBE), Kragujevac, Serbia, 25 October 2021; pp. 1–8. [Google Scholar]
- Li, Y.-D.; Zhu, S.-W.; Yu, J.-P.; Ruan, R.-W.; Cui, Z.; Li, Y.-T.; Lv, M.-C.; Wang, H.-G.; Chen, M.; Jin, C.-H.; et al. Intelligent Detection Endoscopic Assistant: An Artificial Intelligence-Based System for Monitoring Blind Spots during Esophagogastroduodenoscopy in Real-Time. Dig. Liver Dis. 2021, 53, 216–223. [Google Scholar] [CrossRef]
- Tan, M.; Le, Q.V. EfficientNet: Rethinking Model Scaling for Convolutional Neural Networks. arXiv 2020, arXiv:1905.11946. [Google Scholar]
- Choi, K.S.; Suh, M. Screening for Gastric Cancer: The Usefulness of Endoscopy. Clin. Endosc. 2014, 47, 490. [Google Scholar] [CrossRef] [Green Version]
- Ishioka, M.; Hirasawa, T.; Tada, T. Detecting gastric cancer from video images using convolutional neural networks. Dig. Endosc. 2019, 31, e34–e35. [Google Scholar] [CrossRef] [Green Version]
- Hirasawa, T.; Aoyama, K.; Tanimoto, T.; Ishihara, S.; Shichijo, S.; Ozawa, T.; Ohnishi, T.; Fujishiro, M.; Matsuo, K.; Fujisaki, J.; et al. Application of Artificial Intelligence Using a Convolutional Neural Network for Detecting Gastric Cancer in Endoscopic Images. Gastric Cancer 2018, 21, 653–660. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.-D.; Li, H.-Z.; Chen, S.-S.; Jin, C.-H.; Chen, M.; Cheng, M.; Ma, M.-J.; Zhang, X.-P.; Wang, X.; Zhou, J.-B.; et al. Correlation of the Detection Rate of Upper GI Cancer with Artificial Intelligence Score: Results from a Multicenter Trial (with Video). Gastrointest. Endosc. 2021, 95, 1138–1146.E2. [Google Scholar] [CrossRef]
- Zhang, Y.; Li, F.; Yuan, F.; Zhang, K.; Huo, L.; Dong, Z.; Lang, Y.; Zhang, Y.; Wang, M.; Gao, Z.; et al. Diagnosing Chronic Atrophic Gastritis by Gastroscopy Using Artificial Intelligence. Dig. Liver Dis. 2020, 52, 566–572. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yoon, H.J.; Kim, S.; Kim, J.-H.; Keum, J.-S.; Oh, S.-I.; Jo, J.; Chun, J.; Youn, Y.H.; Park, H.; Kwon, I.G.; et al. A Lesion-Based Convolutional Neural Network Improves Endoscopic Detection and Depth Prediction of Early Gastric Cancer. JCM 2019, 8, 1310. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yan, T.; Wong, P.K.; Choi, I.C.; Vong, C.M.; Yu, H.H. Intelligent Diagnosis of Gastric Intestinal Metaplasia Based on Convolutional Neural Network and Limited Number of Endoscopic Images. Comput. Biol. Med. 2020, 126, 104026. [Google Scholar] [CrossRef] [PubMed]
- Wu, L.; Wang, J.; He, X.; Zhu, Y.; Jiang, X.; Chen, Y.; Wang, Y.; Huang, L.; Shang, R.; Dong, Z.; et al. Deep Learning System Compared with Expert Endoscopists in Predicting Early Gastric Cancer and Its Invasion Depth and Differentiation Status (with Videos). Gastrointest. Endosc. 2022, 95, 92–104.e3. [Google Scholar] [CrossRef]
- Van Cutsem, E.; Sagaert, X.; Topal, B.; Haustermans, K.; Prenen, H. Gastric Cancer. Lancet 2016, 388, 2654–2664. [Google Scholar] [CrossRef]
- Correa, P.; Piazuelo, M.B. The Gastric Precancerous Cascade: The Gastric Precancerous Cascade. J. Dig. Dis. 2012, 13, 2–9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chung, C.-S.; Wang, H.-P. Screening for Precancerous Lesions of Upper Gastrointestinal Tract: From the Endoscopists’ Viewpoint. Gastroenterol. Res. Pract. 2013, 2013, 1–18. [Google Scholar] [CrossRef] [Green Version]
- de Vries, A.C.; van Grieken, N.C.T.; Looman, C.W.N.; Casparie, M.K.; de Vries, E.; Meijer, G.A.; Kuipers, E.J. Gastric Cancer Risk in Patients With Premalignant Gastric Lesions: A Nationwide Cohort Study in the Netherlands. Gastroenterology 2008, 134, 945–952. [Google Scholar] [CrossRef]
- Nagao, S.; Tsuji, Y.; Sakaguchi, Y.; Takahashi, Y.; Minatsuki, C.; Niimi, K.; Yamashita, H.; Yamamichi, N.; Seto, Y.; Tada, T.; et al. Highly Accurate Artificial Intelligence Systems to Predict the Invasion Depth of Gastric Cancer: Efficacy of Conventional White-Light Imaging, Nonmagnifying Narrow-Band Imaging, and Indigo-Carmine Dye Contrast Imaging. Gastrointest. Endosc. 2020, 92, 866–873.e1. [Google Scholar] [CrossRef]
- Zhu, Y.; Wang, Q.-C.; Xu, M.-D.; Zhang, Z.; Cheng, J.; Zhong, Y.-S.; Zhang, Y.-Q.; Chen, W.-F.; Yao, L.-Q.; Zhou, P.-H.; et al. Application of convolutional neural network in the diagnosis of the invasion depth of gastric cancer based on conventional endoscopy. Gastrointest. Endosc. 2019, 89, 806–815.e1. [Google Scholar] [CrossRef]
- Xu, M.; Zhou, W.; Wu, L.; Zhang, J.; Wang, J.; Mu, G.; Huang, X.; Li, Y.; Yuan, J.; Zeng, Z.; et al. Artificial Intelligence in the Diagnosis of Gastric Precancerous Conditions by Image-Enhanced Endoscopy: A Multicenter, Diagnostic Study (with Video). Gastrointest. Endosc. 2021, 94, 540–548.e4. [Google Scholar] [CrossRef] [PubMed]
- Deng, J.; Dong, W.; Socher, R.; Li, L.J.; Li, K.; Fei-Fei, L. ImageNet: A Large-Scale Hierarchical Image Database. In Proceedings of the 2009 IEEE Conference on Computer Vision and Pattern Recognition, Miami, FL, USA, 20–25 June 2009; pp. 248–255. [Google Scholar]
- Khan, S.; Naseer, M.; Hayat, M.; Zamir, S.W.; Khan, F.S.; Shah, M. Transformers in Vision: A Survey. ACM Comput. Surv. (CSUR) 2021. [Google Scholar] [CrossRef]
- Shlezinger, N.; Whang, J.; Eldar, Y.C.; Dimakis, A.G. Model-Based Deep Learning. arXiv 2020, arXiv:2012.08405. [Google Scholar]
- Tjoa, E.; Guan, C. A Survey on Explainable Artificial Intelligence (XAI): Towards Medical XAI. IEEE Trans. Neural Netw. Learn. Syst. 2021, 32, 4793–4813. [Google Scholar] [CrossRef] [PubMed]
- Widya, A.R.; Monno, Y.; Okutomi, M.; Suzuki, S.; Gotoda, T.; Miki, K. Whole Stomach 3D Reconstruction and Frame Localization from Monocular Endoscope Video. IEEE J. Transl. Eng. Health Med. 2019, 7, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Widya, A.R.; Monno, Y.; Okutomi, M.; Suzuki, S.; Gotoda, T.; Miki, K. Stomach 3D Reconstruction Based on Virtual Chromoendoscopic Image Generation. In Proceedings of the 2020 42nd Annual International Conference of the IEEE Engineering in Medicine & Biology Society (EMBC), Montreal, QC, Canada, 20–24 July 2020; pp. 1848–1852. [Google Scholar]
- Xu, Y.; Feng, L.; Xia, Z.; Xiong, J. Camera Pose Estimation Based on Feature Extraction and Description for Robotic Gastrointestinal Endoscopy. In Proceedings of the International Conference on Intelligent Robotics and Applications, Yantai, China, 22–25 October 2021; Springer: Berlin/Heidelberg, Germany, 2021; pp. 113–122. [Google Scholar]
- Kalantari, N.K.; Wang, T.-C.; Ramamoorthi, R. Learning-Based View Synthesis for Light Field Cameras. ACM Trans. Graph. (TOG) 2016, 35, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Yu, A.; Ye, V.; Tancik, M.; Kanazawa, A. Pixelnerf: Neural Radiance Fields from One or Few Images. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Nashville, TN, USA, 20–25 June 2021; pp. 4578–4587. [Google Scholar]
- Wang, Z.; Wu, S.; Xie, W.; Chen, M.; Prisacariu, V.A. NeRF–: Neural Radiance Fields without Known Camera Parameters. arXiv 2021, arXiv:2102.07064. [Google Scholar]
- Mildenhall, B.; Srinivasan, P.P.; Tancik, M.; Barron, J.T.; Ramamoorthi, R.; Ng, R. Nerf: Representing Scenes as Neural Radiance Fields for View Synthesis. In Proceedings of the European Conference on Computer Vision, Glasgow, UK, 23–28 August 2020; Springer: Berlin/Heidelberg, Germany, 2020; pp. 405–421. [Google Scholar]
- Lang, O.; Gandelsman, Y.; Yarom, M.; Wald, Y.; Elidan, G.; Hassidim, A.; Freeman, W.T.; Isola, P.; Globerson, A.; Irani, M.; et al. Explaining in Style: Training a GAN to Explain a Classifier in StyleSpace. arXiv 2021, arXiv:2104.13369. [Google Scholar]
- Karras, T.; Laine, S.; Aittala, M.; Hellsten, J.; Lehtinen, J.; Aila, T. Analyzing and Improving the Image Quality of StyleGAN. arXiv 2020, arXiv:1912.04958. [Google Scholar]




Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Renna, F.; Martins, M.; Neto, A.; Cunha, A.; Libânio, D.; Dinis-Ribeiro, M.; Coimbra, M. Artificial Intelligence for Upper Gastrointestinal Endoscopy: A Roadmap from Technology Development to Clinical Practice. Diagnostics 2022, 12, 1278. https://doi.org/10.3390/diagnostics12051278
Renna F, Martins M, Neto A, Cunha A, Libânio D, Dinis-Ribeiro M, Coimbra M. Artificial Intelligence for Upper Gastrointestinal Endoscopy: A Roadmap from Technology Development to Clinical Practice. Diagnostics. 2022; 12(5):1278. https://doi.org/10.3390/diagnostics12051278
Chicago/Turabian StyleRenna, Francesco, Miguel Martins, Alexandre Neto, António Cunha, Diogo Libânio, Mário Dinis-Ribeiro, and Miguel Coimbra. 2022. "Artificial Intelligence for Upper Gastrointestinal Endoscopy: A Roadmap from Technology Development to Clinical Practice" Diagnostics 12, no. 5: 1278. https://doi.org/10.3390/diagnostics12051278
APA StyleRenna, F., Martins, M., Neto, A., Cunha, A., Libânio, D., Dinis-Ribeiro, M., & Coimbra, M. (2022). Artificial Intelligence for Upper Gastrointestinal Endoscopy: A Roadmap from Technology Development to Clinical Practice. Diagnostics, 12(5), 1278. https://doi.org/10.3390/diagnostics12051278








