Genotype Triad for HOTAIR rs10783618, LINC-ROR rs1942347, and MALAT1 rs3200401 as Molecular Markers in Systemic Lupus Erythematous
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Subjects
2.2. Selection of the Study Genetic Variants
2.3. Allelic Discrimination Analysis
2.4. Statistical Analysis
3. Results
3.1. Characteristics of the Study Population
3.2. LncRNAs Genotype and Allelic Frequencies
3.3. Association of lncRNA Variants with SLE Development
3.4. Association of lncRNA Variants with Clinic-Laboratory Variables
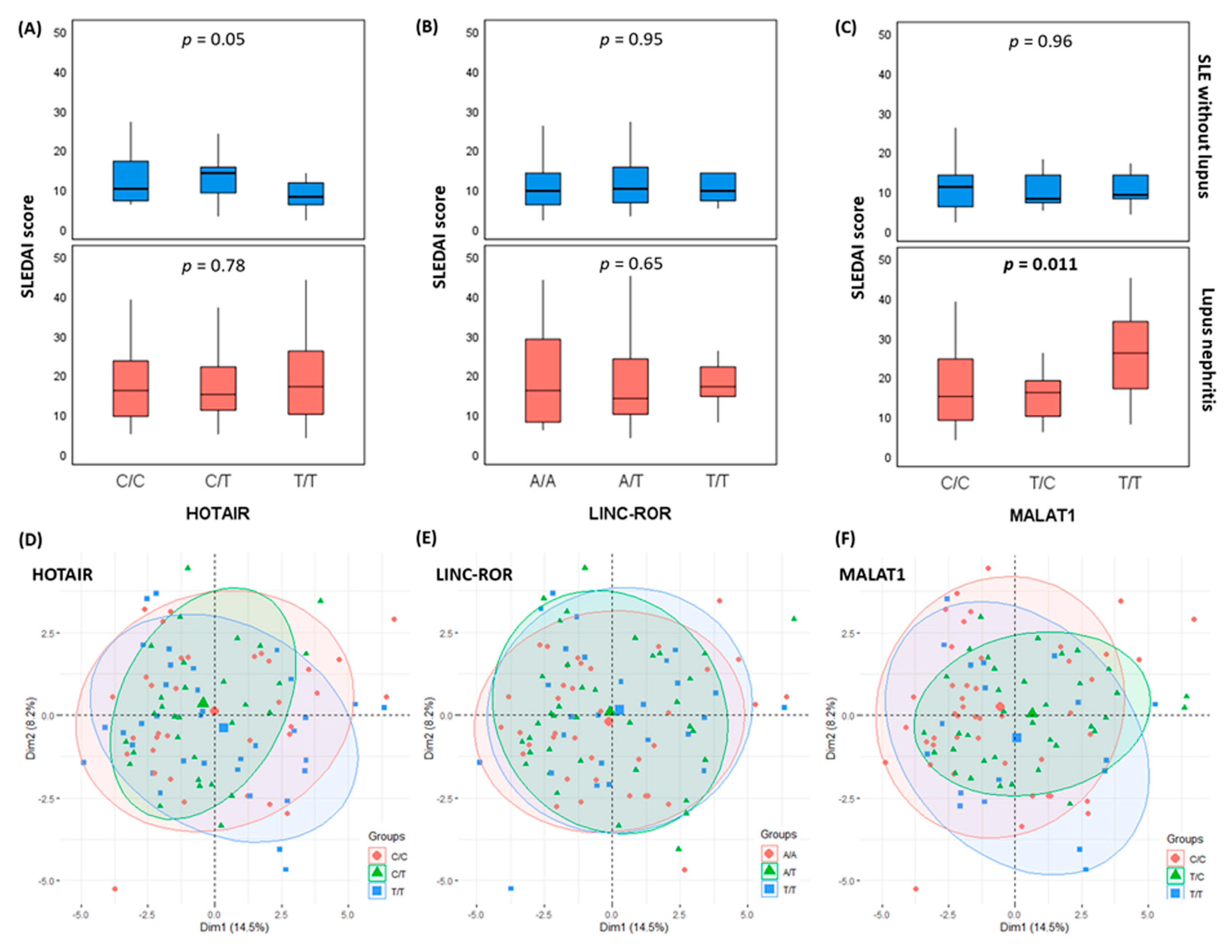
3.5. Impact of lncRNA Variants on the Disease Activity Index
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Leffers, H.C.B.; Lange, T.; Collins, C.; Ulff-Møller, C.J.; Jacobsen, S. The study of interactions between genome and exposome in the development of systemic lupus erythematosus. Autoimmun. Rev. 2019, 18, 382–392. [Google Scholar] [CrossRef] [PubMed]
- Yurkovich, M.; Vostretsova, K.; Chen, W.; Aviña-Zubieta, J.A. Overall and cause-specific mortality in patients with systemic lupus erythematosus: A meta-analysis of observational studies. Arthritis Care Res. 2014, 66, 608–616. [Google Scholar] [CrossRef] [PubMed]
- Faurschou, M.; Starklint, H.; Halberg, P.; Jacobsen, S. Prognostic factors in lupus nephritis: Diagnostic and therapeutic delay increases the risk of terminal renal failure. J. Rheumatol. 2006, 33, 1563–1569. [Google Scholar] [PubMed]
- Ramos, P.S.; Brown, E.E.; Kimberly, R.P.; Langefeld, C.D. Genetic factors predisposing to systemic lupus erythematosus and lupus nephritis. Semin. Nephrol. 2010, 30, 164–176. [Google Scholar] [CrossRef] [Green Version]
- Tiffin, N.; Adeyemo, A.; Okpechi, I. A diverse array of genetic factors contribute to the pathogenesis of systemic lupus erythematosus. Orphanet. J. Rare Dis. 2013, 8, 2. [Google Scholar] [CrossRef] [Green Version]
- Kamen, D.L. Environmental influences on systemic lupus erythematosus expression. Rheum. Dis. Clin. 2014, 40, 401–412. [Google Scholar] [CrossRef] [Green Version]
- Wu, H.; Zhao, M.; Yoshimura, A.; Chang, C.; Lu, Q. Critical Link Between Epigenetics and Transcription Factors in the Induction of Autoimmunity: A Comprehensive Review. Clin. Rev. Allergy Immunol. 2016, 50, 333–344. [Google Scholar] [CrossRef]
- Gao, Y.; Li, S.; Zhang, Z.; Yu, X.; Zheng, J. The Role of Long Non-coding RNAs in the Pathogenesis of RA, SLE, and SS. Front. Med. 2018, 5, 193. [Google Scholar] [CrossRef]
- Tsai, C.Y.; Shen, C.Y.; Liu, C.W.; Hsieh, S.C.; Liao, H.T.; Li, K.J.; Lu, C.S.; Lee, H.T.; Lin, C.S.; Wu, C.H.; et al. Aberrant Non-Coding RNA Expression in Patients with Systemic Lupus Erythematosus: Consequences for Immune Dysfunctions and Tissue Damage. Biomolecules 2020, 10, 1641. [Google Scholar] [CrossRef]
- Franks, A.L.; Slansky, J.E. Multiple associations between a broad spectrum of autoimmune diseases, chronic inflammatory diseases and cancer. Anticancer Res. 2012, 32, 1119–1136. [Google Scholar]
- Boussios, S.; Pentheroudakis, G.; Somarakis, G.; Markatseli, T.E.; Drosos, A.A.; Pavlidis, N. Cancer diagnosis in a cohort of patients with Sjogren’s syndrome and rheumatoid arthritis: A single-center experience and review of the literature. Anticancer Res. 2014, 34, 6669–6676. [Google Scholar] [PubMed]
- Yadlapati, S.; Efthimiou, P. Autoimmune/Inflammatory Arthritis Associated Lymphomas: Who Is at Risk? Biomed. Res. Int. 2016, 2016, 8631061. [Google Scholar] [CrossRef] [Green Version]
- Deng, Y.; Tsao, B.P. Advances in lupus genetics and epigenetics. Curr. Opin. Rheumatol. 2014, 26, 482–492. [Google Scholar] [CrossRef] [PubMed]
- Kwon, Y.C.; Chun, S.; Kim, K.; Mak, A. Update on the Genetics of Systemic Lupus Erythematosus: Genome-Wide Association Studies and Beyond. Cells 2019, 8, 1180. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fike, A.J.; Elcheva, I.; Rahman, Z.S.M. The Post-GWAS Era: How to Validate the Contribution of Gene Variants in Lupus. Curr. Rheumatol. Rep. 2019, 21, 3. [Google Scholar] [CrossRef] [PubMed]
- Clarke, J. Risk variant in long noncoding RNA linked to SLE. Nat. Rev. Rheumatol. 2020, 16, 126. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Atianand, M.K.; Caffrey, D.R.; Fitzgerald, K.A. Immunobiology of Long Noncoding RNAs. Annu. Rev. Immunol. 2017, 35, 177–198. [Google Scholar] [CrossRef]
- Lai, N.S.; Koo, M.; Yu, C.L.; Lu, M.C. Immunopathogenesis of systemic lupus erythematosus and rheumatoid arthritis: The role of aberrant expression of non-coding RNAs in T cells. Clin. Exp. Immunol. 2017, 187, 327–336. [Google Scholar] [CrossRef] [Green Version]
- Fan, Z.; Chen, X.; Liu, L.; Zhu, C.; Xu, J.; Yin, X.; Sheng, Y.; Zhu, Z.; Wen, L.; Zuo, X.; et al. Association of the Polymorphism rs13259960 in SLEAR With Predisposition to Systemic Lupus Erythematosus. Arthritis Rheumatol. 2020, 72, 985–996. [Google Scholar] [CrossRef]
- Gao, F.; Tan, Y.; Luo, H. MALAT1 is involved in type I IFNs-mediated systemic lupus erythematosus by up-regulating OAS2, OAS3, and OASL. Braz. J. Med. Biol. Res. 2020, 53, e9292. [Google Scholar] [CrossRef] [Green Version]
- Zhao, C.-N.; Mao, Y.-M.; Liu, L.-N.; Li, X.-M.; Wang, D.-G.; Pan, H.-F. Emerging role of lncRNAs in systemic lupus erythematosus. Biomed. Pharmacother. 2018, 106, 584–592. [Google Scholar] [CrossRef] [PubMed]
- Mao, Y.M.; He, Y.S.; Wu, G.C.; Hu, Y.Q.; Xiang, K.; Liao, T.; Yan, Y.L.; Yang, X.K.; Shuai, Z.W.; Wang, G.H.; et al. Association of MALAT-1 gene single nucleotide polymorphisms with genetic susceptibility to systemic lupus erythematosus. Lupus 2021, 30, 1923–1930. [Google Scholar] [CrossRef] [PubMed]
- Song, J.; Kim, D.; Han, J.; Kim, Y.; Lee, M.; Jin, E.J. PBMC and exosome-derived Hotair is a critical regulator and potent marker for rheumatoid arthritis. Clin. Exp. Med. 2015, 15, 121–126. [Google Scholar] [CrossRef] [PubMed]
- Bi, X.; Pan, Y.F.; Luo, X.Q.; Wang, M.L.; Chen, Y.X. P053 LncRNA HOTAIR promotes proliferation and invasion of fibroblast-like synoviocytes as microrna sponging in RA patients. Ann. Rheum. Dis. 2019, 78, A23. [Google Scholar]
- Safa, A.; Taheri, M.; Fallah, H.; Salmani, T.; Arsang-Jang, S.; Ghafouri-Fard, S.; Omrani, M.D. Downregulation of Cancer-Associated lncRNAs in Peripheral Blood of Multiple Sclerosis Patients. J. Mol. Neurosci. 2020, 70, 1533–1540. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Liang, N.; Wang, M.; Fei, Y.; Sun, J.; Li, Z.; Xu, Y.; Guo, C.; Cao, Z.; Li, S.; et al. Long noncoding RNA MALAT-1 is a novel inflammatory regulator in human systemic lupus erythematosus. Oncotarget 2017, 8, 77400–77406. [Google Scholar] [CrossRef] [Green Version]
- Ismail, N.M.; Toraih, E.A.; Mohammad, M.H.S.; Alshammari, E.M.; Fawzy, M.S. Association of microRNA-34a rs2666433 (A/G) Variant with Systemic Lupus Erythematosus in Female Patients: A Case-Control Study. J. Clin. Med. 2021, 10, 5095. [Google Scholar] [CrossRef]
- Bombardier, C.; Gladman, D.D.; Urowitz, M.B.; Caron, D.; Chang, C.H. Derivation of the SLEDAI. A disease activity index for lupus patients. The Committee on Prognosis Studies in SLE. Arthritis Rheum. 1992, 35, 630–640. [Google Scholar] [CrossRef]
- Aringer, M.; Costenbader, K.; Daikh, D.; Brinks, R.; Mosca, M.; Ramsey-Goldman, R.; Smolen, J.S.; Wofsy, D.; Boumpas, D.T.; Kamen, D.L.; et al. 2019 European League Against Rheumatism/American College of Rheumatology Classification Criteria for Systemic Lupus Erythematosus. Arthritis Rheumatol. 2019, 71, 1400–1412. [Google Scholar] [CrossRef] [Green Version]
- Fawzy, M.S.; Hussein, M.H.; Abdelaziz, E.Z.; Yamany, H.A.; Ismail, H.M.; Toraih, E.A. Association of MicroRNA-196a2 Variant with Response to Short-Acting β2-Agonist in COPD: An Egyptian Pilot Study. PLoS ONE 2016, 11, e0152834. [Google Scholar] [CrossRef]
- Solé, X.; Guinó, E.; Valls, J.; Iniesta, R.; Moreno, V. SNPStats: A web tool for the analysis of association studies. Bioinformatics 2006, 22, 1928–1929. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Harries, L.W. Long non-coding RNAs and human disease. Biochem. Soc. Trans. 2012, 40, 902–906. [Google Scholar] [CrossRef] [PubMed]
- Zhang, F.; Wu, L.; Qian, J.; Qu, B.; Xia, S.; La, T.; Wu, Y.; Ma, J.; Zeng, J.; Guo, Q.; et al. Identification of the long noncoding RNA NEAT1 as a novel inflammatory regulator acting through MAPK pathway in human lupus. J. Autoimmun. 2016, 75, 96–104. [Google Scholar] [CrossRef] [PubMed]
- Revythis, A.; Shah, S.; Kutka, M.; Moschetta, M.; Ozturk, M.A.; Pappas-Gogos, G.; Ioannidou, E.; Sheriff, M.; Rassy, E.; Boussios, S. Unraveling the Wide Spectrum of Melanoma Biomarkers. Diagnostics 2021, 11, 1341. [Google Scholar] [CrossRef]
- Cantile, M.; Scognamiglio, G.; Marra, L.; Aquino, G.; Botti, C.; Falcone, M.R.; Malzone, M.G.; Liguori, G.; Di Bonito, M.; Franco, R.; et al. HOTAIR role in melanoma progression and its identification in the blood of patients with advanced disease. J. Cell Physiol. 2017, 232, 3422–3432. [Google Scholar] [CrossRef]
- Xu, F.; Jin, L.; Jin, Y.; Nie, Z.; Zheng, H. Long noncoding RNAs in autoimmune diseases. J. Biomed. Mater. Res. A 2019, 107, 468–475. [Google Scholar] [CrossRef]
- Tang, L.; Zhang, W.; Su, B.; Yu, B. Long noncoding RNA HOTAIR is associated with motility, invasion, and metastatic potential of metastatic melanoma. Biomed. Res. Int. 2013, 2013, 251098. [Google Scholar] [CrossRef] [Green Version]
- Tian, Y.; Zhang, X.; Hao, Y.; Fang, Z.; He, Y. Potential roles of abnormally expressed long noncoding RNA UCA1 and Malat-1 in metastasis of melanoma. Melanoma. Res. 2014, 24, 335–341. [Google Scholar] [CrossRef]
- Lazăr, A.D.; Dinescu, S.; Costache, M. The Non-Coding Landscape of Cutaneous Malignant Melanoma: A Possible Route to Efficient Targeted Therapy. Cancers 2020, 12, 3378. [Google Scholar] [CrossRef]
- Taheri, M.; Eghtedarian, R.; Dinger, M.E.; Ghafouri-Fard, S. Exploring the Role of Non-Coding RNAs in the Pathophysiology of Systemic Lupus Erythematosus. Biomolecules 2020, 10, 937. [Google Scholar] [CrossRef]
- Xue, Z.; Cui, C.; Liao, Z.; Xia, S.; Zhang, P.; Qin, J.; Guo, Q.; Chen, S.; Fu, Q.; Yin, Z.; et al. Identification of LncRNA Linc00513 Containing Lupus-Associated Genetic Variants as a Novel Regulator of Interferon Signaling Pathway. Front. Immunol. 2018, 9, 2967. [Google Scholar] [CrossRef]
- Mohammadpour-Gharehbagh, A.; Jahantigh, D.; Saravani, M.; Harati-Sadegh, M.; Maruie-Milan, R.; Teimoori, B.; Salimi, S. Impact of HOTAIR variants on preeclampsia susceptibility based on blood and placenta and in silico analysis. IUBMB Life 2019, 71, 1367–1381. [Google Scholar] [CrossRef] [PubMed]
- Guo, W.; Dong, Z.; Bai, Y.; Guo, Y.; Shen, S.; Kuang, G.; Xu, J. Associations between polymorphisms of HOTAIR and risk of gastric cardia adenocarcinoma in a population of north China. Tumour. Biol. 2015, 36, 2845–2854. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Liu, Y.; Huang, G.; Cui, P.; Zhang, W.; Zhang, Y. Long non-coding RNAs in gastric cancer: Versatile mechanisms and potential for clinical translation. Am. J. Cancer Res. 2015, 5, 907–927. [Google Scholar]
- Marchese, F.P.; Huarte, M. Long non-coding RNAs and chromatin modifiers: Their place in the epigenetic code. Epigenetics 2014, 9, 21–26. [Google Scholar] [CrossRef]
- Hu, J.; Wang, Z.; Shan, Y.; Pan, Y.; Ma, J.; Jia, L. Long non-coding RNA HOTAIR promotes osteoarthritis progression via miR-17-5p/FUT2/β-catenin axis. Cell Death Dis. 2018, 9, 711. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, Y.; Xing, D.; Wang, Y.; Jia, H.; Li, B.; Li, J.J. A long non-coding RNA, HOTAIR, promotes cartilage degradation in osteoarthritis by inhibiting WIF-1 expression and activating Wnt pathway. BMC Mol. Cell Biol. 2020, 21, 53. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Sun, Y.; Liu, N.; Liu, H. LncRNA HOTAIR modulates chondrocyte apoptosis and inflammation in osteoarthritis via regulating miR-1277-5p/SGTB axis. Wound Repair Regen 2021, 29, 495–504. [Google Scholar] [CrossRef] [PubMed]
- Wasson, C.W.; Ross, R.L.; Wells, R.; Corinaldesi, C.; Georgiou, I.C.; Riobo-Del Galdo, N.A.; Del Galdo, F. Long non-coding RNA HOTAIR induces GLI2 expression through Notch signalling in systemic sclerosis dermal fibroblasts. Arthritis Res. Ther. 2020, 22, 286. [Google Scholar] [CrossRef]
- Zhou, H.; Gao, L.; Yu, Z.H.; Hong, S.J.; Zhang, Z.W.; Qiu, Z.Z. LncRNA HOTAIR promotes renal interstitial fibrosis by regulating Notch1 pathway via the modulation of miR-124. Nephrology 2019, 24, 472–480. [Google Scholar] [CrossRef]
- Wang, J.; Luo, X.; Cai, S.; Sun, J.; Wang, S.; Wei, X. Blocking HOTAIR protects human chondrocytes against IL-1β-induced cell apoptosis, ECM degradation, inflammatory response and oxidative stress via regulating miR-222-3p/ADAM10 axis. Int. Immunopharmacol. 2021, 98, 107903. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.F.; Xing, G.L.; Chen, Z.; Tu, S.H. Long non-coding RNA HOTAIR knockdown alleviates gouty arthritis through miR-20b upregulation and NLRP3 downregulation. Cell Cycle 2021, 20, 332–344. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Zhang, H.; Zhang, Z.; Li, S.; Jiang, W.; Li, X.; Lv, J. LncRNA MALAT1 cessation antagonizes hypoxia/reoxygenation injury in hepatocytes by inhibiting apoptosis and inflammation via the HMGB1-TLR4 axis. Mol. Immunol. 2019, 112, 22–29. [Google Scholar] [CrossRef]
- Gupta, S.C.; Awasthee, N.; Rai, V.; Chava, S.; Gunda, V.; Challagundla, K.B. Long non-coding RNAs and nuclear factor-κB crosstalk in cancer and other human diseases. Biochim. Biophys. Acta Rev. Cancer 2020, 1873, 188316. [Google Scholar] [CrossRef]
- Liu, C.; Ren, S.; Zhao, S.; Wang, Y. LncRNA MALAT1/MiR-145 Adjusts IL-1β-Induced Chondrocytes Viability and Cartilage Matrix Degradation by Regulating ADAMTS5 in Human Osteoarthritis. Yonsei Med. J. 2019, 60, 1081–1092. [Google Scholar] [CrossRef] [PubMed]
- Ou, M.; Zhao, H.; Ji, G.; Zhao, X.; Zhang, Q. Long noncoding RNA MALAT1 contributes to pregnancy-induced hypertension development by enhancing oxidative stress and inflammation through the regulation of the miR-150-5p/ET-1 axis. FASEB J. 2020, 34, 6070–6085. [Google Scholar] [CrossRef] [PubMed]
- Jiang, H.; Zhu, M.; Wang, H.; Liu, H. Suppression of lncRNA MALAT1 reduces pro-inflammatory cytokines production by regulating miR-150-5p/ZBTB4 axis through JAK/STAT signal pathway in systemic juvenile idiopathic arthritis. Cytokine 2021, 138, 155397. [Google Scholar] [CrossRef]
- Liu, W.; Zhang, Q.; Zhang, J.; Pan, W.; Zhao, J.; Xu, Y. Long non-coding RNA MALAT1 contributes to cell apoptosis by sponging miR-124 in Parkinson disease. Cell Biosci. 2017, 7, 19. [Google Scholar] [CrossRef] [Green Version]
- McCown, P.J.; Wang, M.C.; Jaeger, L.; Brown, J.A. Secondary Structural Model of Human MALAT1 Reveals Multiple Structure-Function Relationships. Int. J. Mol. Sci. 2019, 20, 5610. [Google Scholar] [CrossRef] [Green Version]

| Characteristics | Total (N = 163) | SLE without Nephritis (N = 65) | Lupus Nephritis (N = 98) | p-Value | |
|---|---|---|---|---|---|
| Demographics | |||||
| Age, years | Mean ± SD | 35.81 ± 9.60 | 34.86 ± 9.59 | 36.45 ± 9.61 | 0.38 |
| Sex | Male | 16 (9.8) | 11 (16.9) | 5 (5.1) | 0.016 |
| Female | 147 (90.2) | 54 (83.1) | 93 (94.9) | ||
| Family history | Negative | 105 (64.4) | 40 (61.5) | 65 (66.3) | 0.61 |
| Positive | 58 (35.6) | 25 (38.5) | 33 (33.7) | ||
| Clinical manifestations | |||||
| Organ involvement | Malar rash | 109 (66.9) | 45 (69.2) | 64 (65.3) | 0.61 |
| Discoid rash | 77 (47.2) | 26 (40) | 51 (52) | 0.15 | |
| Photosensitivity | 61 (37.4) | 23 (35.4) | 38 (38.8) | 0.74 | |
| Hair loss | 129 (79.1) | 52 (80) | 77 (78.6) | 0.82 | |
| Oral ulcer | 46 (28.2) | 19 (29.2) | 27 (27.6) | 0.86 | |
| Arthritis | 81 (49.7) | 31 (47.7) | 50 (51) | 0.75 | |
| Ecchymosis | 19 (11.7) | 8 (12.3) | 11 (11.2) | 0.83 | |
| Fever | 30 (18.4) | 12 (18.5) | 18 (18.4) | 0.98 | |
| Infection | 26 (16) | 9 (13.8) | 17 (17.3) | 0.55 | |
| Dyspnea | 68 (41.7) | 30 (46.2) | 38 (38.8) | 0.42 | |
| Chest pain | 35 (21.5) | 11 (16.9) | 24 (24.5) | 0.33 | |
| Cough | 35 (21.5) | 14 (21.5) | 21 (21.4) | 0.98 | |
| CNS | 41 (25.2) | 18 (27.7) | 23 (23.5) | 0.54 | |
| Peripheral neuropathy | 70 (42.9) | 28 (43.1) | 42 (42.9) | 0.98 | |
| Hematuria | 56 (34.4) | 20 (30.8) | 36 (36.7) | 0.43 | |
| Weight loss | 76 (46.6) | 36 (55.4) | 40 (40.8) | 0.07 | |
| Severity | |||||
| SLEDAI score | Mean ± SD | 15.97 ± 9.82 | 12.03 ± 7.75 | 18.63 ± 10.21 | <0.001 |
| Grade 1 | 12 (7.4) | 9 (13.8) | 3 (3.1) | <0.001 | |
| Grade 2 | 50 (30.7) | 26 (40) | 24 (24.5) | ||
| Grade 3 | 56 (34.4) | 22 (33.8) | 34 (34.7) | ||
| Grade 4 | 45 (27.6) | 8 (12.3) | 37 (37.8) | ||
| Markers for severity | Elevated inflammatory markers | 67 (41.1) | 30 (46.2) | 37 (37.8) | 0.33 |
| Thrombocytopenia | 5 (3.1) | 1 (1.5) | 4 (4.1) | 0.64 | |
| Hypocomplementemia | 47 (28.8) | 16 (24.6) | 31 (31.6) | 0.38 | |
| High serum creatinine | 72 (44.2) | 16 (24.6) | 56 (57.1) | <0.001 | |
| Proteinuria | 92 (56.4) | 15 (23.1) | 77 (78.6) | <0.001 | |
| Cast in the urine | 29 (17.8) | 4 (6.2) | 25 (25.5) | 0.001 | |
| Laboratory data | |||||
| Autoantibodies | Positive dsDNA | 147 (90.2) | 51 (78.5) | 96 (98) | <0.001 |
| Positive ANA titer | 162 (99.4) | 64 (98.5) | 98 (100) | 0.21 | |
| Biochemical tests | Hemoglobin (g/dL) | 11.66 ± 2.89 | 11.77 ± 1.50 | 11.58 ± 3.54 | 0.69 |
| RBC (×106 per mm3) | 4.09 ± 0.74 | 4.16 ± 0.72 | 4.05 ± 0.75 | 0.42 | |
| HCT (%) | 38.18 ± 6.05 | 38.53 ± 6.38 | 37.95 ± 5.85 | 0.42 | |
| MCV (fl) | 81.42 ± 6.36 | 81.87 ± 6.72 | 81.11 ± 6.12 | 0.42 | |
| Platelet count (×103/mm3) | 264.51 ± 77.59 | 256.48 ± 80.57 | 269.92 ± 75.46 | 0.37 | |
| WBC (×103/uL) | 6.58 ± 2.22 | 6.64 ± 2.21 | 6.55 ± 2.24 | 0.67 | |
| Neutrophil (%) | 63.30 ± 10.46 | 62.44 ± 11.27 | 63.88 ± 9.89 | 0.40 | |
| Lymphocyte (%) | 30.01 ± 9.66 | 30.91 ± 9.95 | 29.41 ± 9.46 | 0.62 | |
| C3 (mg/dL) | 95.52 ± 47.86 | 96.46 ± 47.69 | 94.89 ± 48.21 | 0.90 | |
| C4 (mg/dL) | 27.94 ± 15.62 | 27.78 ± 15.78 | 28.04 ± 15.60 | 0.91 | |
| CRP (mg/L) | 2.95 ± 2.89 | 3.04 ± 3.22 | 2.88 ± 2.67 | 0.69 | |
| ESR 1st hour | 26.84 ± 13.58 | 27.67 ± 14.91 | 26.28 ± 12.65 | 0.93 | |
| ALT (U/L) | 26.61 ± 9.62 | 27.00 ± 8.32 | 26.35 ± 10.44 | 0.81 | |
| AST (U/L) | 26.50 ± 8.48 | 27.02 ± 8.49 | 26.16 ± 8.50 | 0.76 | |
| Serum creatinine (mg/dL) | 1.18 ± 1.19 | 0.99 ± 0.28 | 1.31 ± 1.51 | 0.11 | |
| Blood urea (mg/dL) | 35.11 ± 11.86 | 32.63 ± 6.96 | 36.78 ± 14.04 | 0.05 | |
| Variable | Controls | Cases | p-Value | ||
|---|---|---|---|---|---|
| Count | Proportion | Count | Proportion | ||
| HOTAIR (rs10783618) | |||||
| Allele | |||||
| T | 190 | 0.58 | 166 | 0.51 | 0.059 |
| C | 136 | 0.42 | 160 | 0.49 | |
| Genotypes | |||||
| C/C | 38 | 0.23 | 57 | 0.35 | 0.05 |
| T/C | 60 | 0.37 | 46 | 0.28 | |
| T/T | 65 | 0.4 | 60 | 0.37 | |
| LINC-ROR (rs1942347) | |||||
| Allele | |||||
| A | 194 | 0.6 | 182 | 0.56 | 0.34 |
| T | 132 | 0.4 | 144 | 0.44 | |
| Genotypes | |||||
| A/A | 69 | 0.42 | 61 | 0.37 | 0.66 |
| A/T | 56 | 0.34 | 60 | 0.37 | |
| T/T | 38 | 0.23 | 42 | 0.26 | |
| MALAT1 (rs3200401) | |||||
| Allele | |||||
| C | 210 | 0.64 | 206 | 0.63 | 0.74 |
| T | 116 | 0.36 | 120 | 0.37 | |
| Genotypes | |||||
| C/C | 59 | 0.36 | 73 | 0.45 | <0.001 |
| C/T | 92 | 0.56 | 60 | 0.37 | |
| T/T | 12 | 0.07 | 30 | 0.18 | |
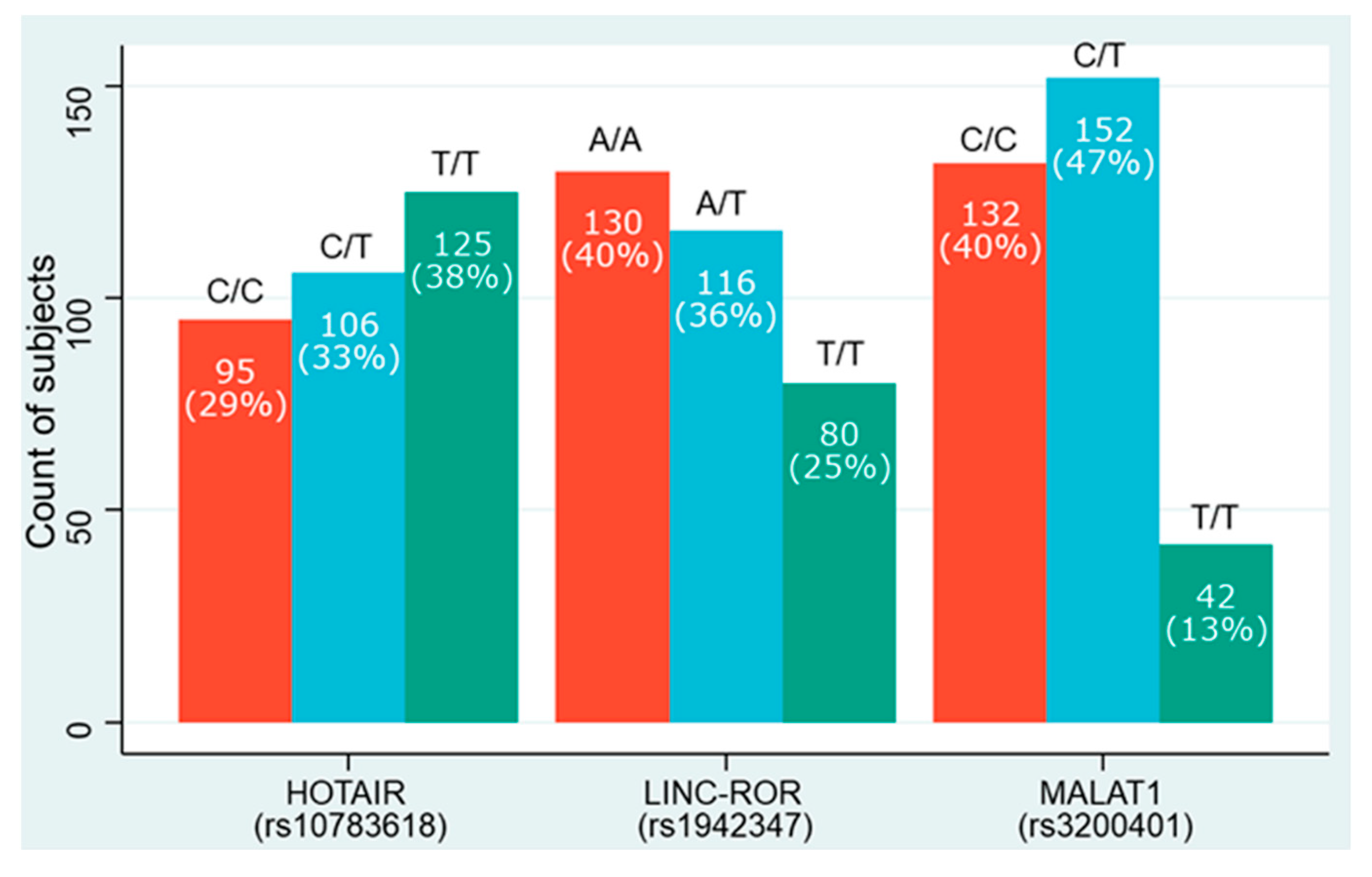
| Variables | HOTAIR | p-Value | LINC-ROR | p-Value | MALAT1 | p-Value | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| C/C | T/C | T/T | A/A | A/T | T/T | C/C | C/T | T/T | ||||
| 95 | 106 | 125 | 130 | 116 | 80 | 132 | 152 | 42 | ||||
| Early onset | 68.4% | 80.4% | 60.0% | 0.08 | 68.9% | 66.7% | 71.4% | 0.88 | 60.0% | 70.0% | 71.2% | 0.52 |
| Female gender | 89.5% | 89.1% | 91.7% | 0.89 | 93.4% | 91.7% | 83.3% | 0.21 | 96.7% | 90.0% | 87.7% | 0.38 |
| Positive FH | 36.8% | 47.8% | 25.0% | 0.05 | 47.5% | 31.7% | 23.8% | 0.034 | 36.7% | 26.7% | 42.5% | 0.16 |
| Malar rash | 63.2% | 54.3% | 80.0% | 0.016 | 62.3% | 75.0% | 61.9% | 0.24 | 66.7% | 71.7% | 63.0% | 0.57 |
| Discoid rash | 42.1% | 45.7% | 53.3% | 0.46 | 42.6% | 51.7% | 47.6% | 0.61 | 46.7% | 58.3% | 38.4% | 0.07 |
| Photosensitivity | 36.8% | 45.7% | 31.7% | 0.33 | 39.3% | 33.3% | 40.5% | 0.71 | 26.7% | 38.3% | 41.1% | 0.38 |
| Hair loss | 57.9% | 84.8% | 95.0% | <0.001 | 78.7% | 81.7% | 76.2% | 0.79 | 86.7% | 85.0% | 71.2% | 0.08 |
| Oral ulcers | 36.8% | 39.1% | 11.7% | 0.002 | 27.9% | 28.3% | 28.6% | 1.00 | 23.3% | 28.3% | 30.1% | 0.78 |
| Arthritis | 68.4% | 45.7% | 35.0% | 0.001 | 49.2% | 40.0% | 64.3% | 0.05 | 43.3% | 56.7% | 46.6% | 0.38 |
| Fever | 26.3% | 13.0% | 15.0% | 0.16 | 18.0% | 20.0% | 16.7% | 0.91 | 23.3% | 26.7% | 9.6% | 0.030 |
| Recurrent infection | 21.1% | 13.0% | 13.3% | 0.43 | 19.7% | 13.3% | 14.3% | 0.60 | 13.3% | 13.3% | 19.2% | 0.60 |
| Weight loss | 26.3% | 58.7% | 56.7% | 0.001 | 45.9% | 53.3% | 38.1% | 0.31 | 50.0% | 48.3% | 43.8% | 0.80 |
| Ecchymosis | 15.8% | 13.0% | 6.7% | 0.29 | 13.1% | 8.3% | 14.3% | 0.59 | 6.7% | 18.3% | 8.2% | 0.12 |
| Neurological | 19.3% | 13.0% | 40.0% | 0.003 | 23.0% | 28.3% | 23.8% | 0.77 | 20.0% | 41.7% | 13.7% | 0.001 |
| Hematuria | 26.3% | 26.1% | 48.3% | 0.02 | 31.1% | 40.0% | 31.0% | 0.51 | 30.0% | 43.3% | 28.8% | 0.18 |
| Lupus nephritis | 61.4% | 56.5% | 61.7% | 0.84 | 54.1% | 61.7% | 66.7% | 0.42 | 56.7% | 61.7% | 60.3% | 0.90 |
| Pulmonary | 54.4% | 54.3% | 53.3% | 0.99 | 50.8% | 58.3% | 52.4% | 0.69 | 56.7% | 60.0% | 47.9% | 0.36 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ismail, N.M.; Toraih, E.A.; Almars, A.I.; Al Ageeli, E.; Fawzy, M.S.; Maher, S.A. Genotype Triad for HOTAIR rs10783618, LINC-ROR rs1942347, and MALAT1 rs3200401 as Molecular Markers in Systemic Lupus Erythematous. Diagnostics 2022, 12, 1197. https://doi.org/10.3390/diagnostics12051197
Ismail NM, Toraih EA, Almars AI, Al Ageeli E, Fawzy MS, Maher SA. Genotype Triad for HOTAIR rs10783618, LINC-ROR rs1942347, and MALAT1 rs3200401 as Molecular Markers in Systemic Lupus Erythematous. Diagnostics. 2022; 12(5):1197. https://doi.org/10.3390/diagnostics12051197
Chicago/Turabian StyleIsmail, Nesreen M., Eman A. Toraih, Amany I. Almars, Essam Al Ageeli, Manal S. Fawzy, and Shymaa Ahmed Maher. 2022. "Genotype Triad for HOTAIR rs10783618, LINC-ROR rs1942347, and MALAT1 rs3200401 as Molecular Markers in Systemic Lupus Erythematous" Diagnostics 12, no. 5: 1197. https://doi.org/10.3390/diagnostics12051197
APA StyleIsmail, N. M., Toraih, E. A., Almars, A. I., Al Ageeli, E., Fawzy, M. S., & Maher, S. A. (2022). Genotype Triad for HOTAIR rs10783618, LINC-ROR rs1942347, and MALAT1 rs3200401 as Molecular Markers in Systemic Lupus Erythematous. Diagnostics, 12(5), 1197. https://doi.org/10.3390/diagnostics12051197








